Abstract
To identify pathways controlling prostate cancer metastasis we performed differential display analysis of the human prostate carcinoma cell line PC-3 and its highly metastatic derivative PC-3M. This revealed that a 78-kDa interferon-inducible GTPase, MxA, was expressed in PC-3 but not in PC-3M cells. The gene encoding MxA, MX1, is located in the region of chromosome 21 deleted as a consequence of fusion of TMPRSS2 and ERG, which has been associated with aggressive, invasive prostate cancer. Stable exogenous MxA expression inhibited in vitro motility and invasiveness of PC-3M cells. In vivo exogenous MxA expression decreased the number of hepatic metastases following intrasplenic injection. Exogenous MxA also reduced motility and invasiveness of highly metastatic LOX melanoma cells. A mutation in MxA that inactivated its GTPase reversed inhibition of motility and invasion in both tumor cell lines. Co-immunoprecipitation studies demonstrated that MxA associated with tubulin, but the GTPase-inactivating mutation blocked this association. Because MxA is a highly inducible gene, an MxA-targeted drug discovery screen was initiated by placing the MxA promoter upstream of a luciferase reporter. Examination of the NCI diversity set of small molecules revealed three hits that activated the promoter. In PC-3M cells, these drugs induced MxA protein and inhibited motility. These data demonstrate that MxA inhibits tumor cell motility and invasion, and that MxA expression can be induced by small molecules, potentially offering a new approach to the prevention and treatment of metastasis.
Increased understanding of the mechanisms regulating metastasis offers the potential of designing specifically targeted drugs aimed at preventing neoplastic spread. Better understanding of the genetic basis of metastasis could aid in the choice of treatment and timing of treatment modalities as well as identify molecular targets for therapy.
The clonally related pair of human prostate cancer lines, PC-3 and its more metastatic derivative, PC-3M that was derived from a liver metastasis in a nude mouse bearing a splenic explant of PC-3 (1), allowed us to explore the molecular genetic mechanisms of metastasis. To this end, we used differential display-reverse transcription-PCR (DD-RT-PCR)4 (2) to identify mRNAs with expression differences in these two lines. This study demonstrated differential expression of a DD-RT-PCR band (DD-2) that was found in the PC-3 parental cell line and not in PC-3M cells (Fig. 1B). DNA sequencing of DD-2 identified it as a portion of MxA, one of a small family of “Mx” genes (MX1 and MX2 in human and Mx1 in mouse) that encode large self-assembling dynamin-like proteins that bind and hydrolyze GTP (3). MxA transcription is inducible by types I, II, and III interferons (IFNs α/β (3), γ (4), and λ (5)), and MxA protein has been shown to be an effector of type I IFN-mediated inhibition of certain RNA viruses, including the myxoviruses. Although IFNs, both type I and II, have been used in the treatment of several forms of cancer, including melanoma, follicular lymphoma, hairy cell leukemia, chronic myelogenous leukemia, Kaposi's sarcoma, and renal cell carcinoma, the mechanisms of anticancer activity have not been fully delineated. Both direct antiproliferative effects on tumor and indirect immunomodulatory effects on the host have been reported (see Ref. 6 for review). IFNs are known to inhibit cell motility (7), and Mx proteins have significant homology to dynamin, a large GTPase involved in the scission of nascent vesicles from parent membranes. However, heretofore, MxA has been chiefly studied for its anti-viral properties (8), and it has not been associated with cell motility or metastasis. To gain a better understanding of the role of IFN and MxA in cancer biology, and to explore MxA as a new target for anti-metastatic therapy we undertook an investigation of the role of MxA in two metastatic human cancer cell lines.
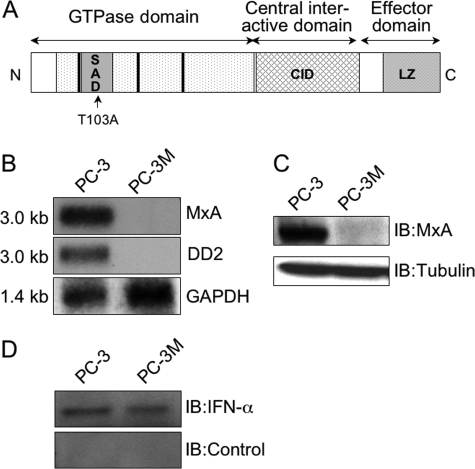
FIGURE 1.
Structure and expression of MxA. A, schematic drawing of human MxA protein (59). Amino acid residues and locations of important motifs are indicated, including the GTPase domain that contains the tripartite GTP-binding elements (thick bars) and self-assembly domain (SAD) and the C-terminal effector domain that contains the central interactive domain (CID) and a leucine zipper (LZ) region. The arrow indicates the site of the T103A mutation. B, Northern blot. 10 μg of total RNA from the prostate cancer cell lines were electrophoresed on a formaldehyde/agarose gel, blotted onto a nylon membrane, and probed as indicated: the published MxA cDNA (15); DD2, the cloned 200-bp PCR fragment isolated by differential display of mRNA from PC-3; rat glyceraldehyde dehydrogenase (GAPDH) cDNA (16) used to control for equal loading. Sizes of bands shown are indicated on the left. C, Western blot. Lysates were probed with affinity-purified goat anti-MxA antibody or mouse monoclonal anti-tubulin to control for equal loading. D, Western blot. Lysates were probed with a 1:1000 dilution of sheep anti-IFN-α globulin and sheep globulin lacking anti-IFN activity (both from the NIAID repository), as control.
EXPERIMENTAL PROCEDURES
Cell Lines—The human prostate carcinoma cell line PC-3 was obtained from ATCC (Manassas, VA). PC-3M (1), a highly metastatic subline derived from a hepatic metastasis of PC-3, was a kind gift of Dr. James Kozlowski (Dept. of Urology, Northwestern University Medical School). Both cell lines and their derivatives were grown as described previously (9). LOX is a highly metastatic human melanoma cell line (10) obtained from Dr. Dan Sackett, National Institutes of Health.
Plasmids and Transfections—PC-3M cells were initially transfected using Lipofectamine (Invitrogen) with an MxA-expressing or a β-galactosidase-expressing control vector constructed from pHβ Apr-1, in which MxA and β-galactosidase were under the control of the human α-actin promoter (11), and stable transfectants were selected in G-418. In subsequent experiments PC-3M and LOX cells were transfected with expression vectors based on pCIneo (Promega, Madison, WI) that used the cytomegalovirus immediate-early enhancer/promoter and added a FLAG tag to the vector control and wild-type (WT) MxA, and stable transfectants were selected using G-418. We used the QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) to mutate threonine 103 to alanine in the FLAG-tagged MxA-expression vector to inactivate MxA self-association and GTPase activity (12).
Antibodies and Reagents—The monoclonal anti-MxA antibody was described previously (13). Anti-FLAG antibody was obtained from Sigma. Affinity-purified polyclonal anti-MxA (14), anti-actin (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-α-tubulin (Oncogene Research Products, San Diego, CA) antibodies were used for immunocytochemistry, immunoblotting, and immunoprecipitation. Sheep anti-human IFN-α immunoglobulin (catalogue no. GO26-501-568) and control sheep immunoglobulin (catalogue no. GO27501-568) were provided by the NIAID Reference Reagent Repository, operated by Braton Biotech, Inc., Rockville, MD. Purified IFN-α was obtained from Novartis Pharma (Basel, Switzerland).
Differential Display—1 μg of poly(A)+ RNA from PC-3 and PC-3M prostate carcinoma cells was used to generate cDNAs using Superscript reverse transcriptase (Invitrogen) and anchored and arbitrary primers (Operon Biotech, Alameda, CA). The differentially expressed bands between the two templates, which ranged from 170 to 500 bp in size, were nick-translated and used to probe RNA blots that contained poly(A)+ RNA from PC-3 and PC-3M cell lines.
Northern Blot Analysis—Total RNA was prepared by standard methods (15) from cell pellets of PC-3 and PC-3M. Equal amounts (10 μg) were electrophoretically separated on a denaturing gel (1% agarose, 20 mm 3-morpholinopropanesulfonic acid, 5 mm sodium acetate, 1 mm EDTA, 18% formaldehyde, pH 7.0) and blotted onto a nylon membrane (HyBond-N, Amersham Biosciences). The blot was sequentially hybridized with 32P-labeled, nick-translated DD-2 fragment and MxA cDNA from human embryonic lung (16). We also hybridized this and other Northern blots with a 13-kb PstI fragment of rat glyceraldehyde dehydrogenase (17).
Motility and Invasion Assays—FALCON cell culture inserts with an 8-μm pore-size polyethylene terephthalate (PET) membrane (Fisher Scientific, Pittsburgh, PA) and BIOCOAT Matrigel invasion chambers (BD Biosciences, Franklin Lakes, NJ) were used for motility assays and invasion assays, respectively. For both assays, inserts were placed into the wells of a 24-well plate. Each well contained 0.5 ml of complete medium (RPMI 1640 with 10% fetal bovine serum, 1% antimycotic-antibiotic solution and 500 μg/ml G-418). Control and MxA-transfected cells were trypsinized, suspended at 1.5 × 105 cells/ml in complete medium, and 350 μl of the cell suspension was added to each insert. The plate of inserts was incubated for 24 h at 37 °C. Following incubation, cells from the upper surface of the membrane were removed by scrubbing with a cotton-tipped swab. Cells that had migrated/invaded through the insert and adhered to the bottom of the membrane were Wright stained using the CAMCO Quik Stain kit (Fisher Scientific, Pittsburgh, PA), visualized using a Leica DM IRB microscope, and counted.
Immunofluorescence Microscopy—Fluorescence immunocytochemistry was performed as described (18). For cytoskeletal preparations, the cells were permeabilized with 1% Triton X-100 in PHEM buffer (60 mm PIPES, pH 6.9, 25 mm HEPES, 2 mm MgCl2, and 10 mm EGTA, pH 6.9) for 2 min and fixed with 37% formaldehyde for 10 min at room temperature (19). Cells were incubated with primary and secondary antibodies as indicated, and nuclei were counterstained with 4,6-diamidino-2-phenylindole (DAPI). Cells were visualized using a Zeiss Axiophot microscope, and images were captured using an Optronics (East Muskegee, OK) charge-coupled device camera.
Co-immunoprecipitation and Western Blot Analysis—Immunoprecipitations were performed as described (20). Cell lysates were incubated with the indicated antibodies overnight at 4 °C. The immunocomplex was immobilized on protein A/G-Sepharose (Santa Cruz Biotechnologies, Santa Cruz, CA) and resolved on SDS-polyacrylamide gels, transferred to nitrocellulose filters and immunoblotted with the indicated antibodies.
GST Pulldown Assay—GST-MxA was constructed by standard PCR cloning techniques to fuse the GSX vector (Promega) and the MxA coding region (16). Recombinant proteins were expressed in and purified from BL21 cells. For each pulldown experiment, 2 μg of purified tubulin (Invitrogen) were incubated with GST alone or with increasing amounts of GST-MxA protein and immobilized onto glutathione-Sepharose beads (Amersham Biosciences) in NETN buffer (20 mm Tris (pH 8.0), 100 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, protease inhibitor mixture (Roche Diagnostics, Indianapolis, IN)) for 2 h at 4 °C. Unbound proteins were removed by washing six times with NETN buffer. The bound proteins were eluted by boiling in 1× SDS sample buffer, resolved on SDS-PAGE and transferred to nitrocellulose filters. Immunoblot analysis was performed with anti-α-tubulin antibody (Sigma).
Animals—Female 4- to 6-week-old beige-SCID mice (Charles River Laboratories, Wilmington, MA) were housed under pathogen-free conditions with a 12-h light/12-h dark schedule, fed autoclaved standard chow and water ad libitum. Animal care and use was in accordance with guidelines of the NIH Animal Care and Use Committee.
Animal Procedures—Surgical sites were prepared by shaving the skin and then cleansing using Betadine scrub solution (E-Z Prep, BD Biosciences) and 70% sterile alcohol. Anesthesia was induced using ketamine (0.45 mg/mouse, Ketaset, Fort Dodge Laboratories Inc., Fort Dodge, IA) and xylazine (0.45 mg/mouse, Sigma) administered intraperitoneally. Anesthesia was then maintained using methoxyflurane (Mallinckrodt Veterinary Inc., Mundelein, IL) inhalation. In vitro-passaged tumor cell lines were harvested and prepared for injection as described previously (21). Cells were brought to a final concentration of 1 × 107 cells/ml for injection in phenol red-free Hanks' balanced salt solution and kept at 4 °C. Cells were counted and their viability assessed manually after Trypan blue staining. Experiments were only continued if cell viability was >90%. Two million cells were administered using a 27-gauge needle for intrasplenic injections (hepatic metastasis assay). Splenic exposure was achieved through a high left paracostal approach to the abdomen. Each experimental group consisted of 15 beige-SCID mice. After tumor cell injection, mice were monitored at least three times weekly for evidence of tumor/metastasis-associated morbidity. The primary end point was the number of hepatic metastases detectable by visual examination 24 days after intrasplenic injection. Complete post mortem examinations were performed on all animals. Tissues obtained at necropsy were fixed in 10% formalin at room temperature. Routine histological analysis of metastases was undertaken at the conclusion of all experiments. The statistical significance of the difference in the number of experimental hepatic metastases between the mice injected with PC-3M-pCIneo cells and mice injected with PC-3M-MxA-wild-type cells was assessed by the non-parametric Welch's corrected unpaired t test using GraphPad Prism version 4.0c.
High-throughput Small Molecule Screen—To search for drugs that might induce MxA expression, the Diversity Set of 1990 pharmacophores distilled from ∼140,000 compounds by the National Cancer Institute's Developmental Therapeutics Program was screened in an assay for MxA promoter activation, using a luciferase-based assay (Promega). The MxA promoter described in Ronni et al. (22) was inserted upstream of the firefly luciferase reporter in the pGL3 plasmid (Promega). The construct was confirmed by DNA sequencing. Interferon-α was used as a positive control. The three small molecules that induced an increase in activity of >2.2-fold were also screened for their effect on MxA protein expression and on PC-3M motility, as described above.
RESULTS
Constitutive Expression of MxA in PC-3 Cells—DD-RT-PCR analysis revealed eight fragments of cDNA with possible differences in expression between PC-3 and its more metastatic derivative, PC-3M. Northern blot analysis indicated that only one of these, a 200-bp band, termed DD-2, was differentially expressed, as a strong 3.0-kb mRNA band in PC-3 but not in PC-3M (Fig. 1B). We used the DD-2 probe to screen a cDNA library generated from PC-3 mRNA, and we obtained a 2.0-kb cDNA clone that contained ∼70% of the expected 3.0-kb sequence (including 95% of the coding region) of MxA (16). As described previously (23), MxA homo-oligomerizes into ring-like and helical structures. MxA protein contains an N-terminal tripartite GTP binding motif and three regions involved in MxA self-assembly: an N-terminal self-assembly domain, a central interactive domain (CID), and an assembly domain in the C terminus that interacts with the CID and the N-terminal self-assembly domain. The assembly domain region is rich in α-helices and contains a leucine zipper motif (Fig. 1A).
There were two conservative sequence differences between MxA in PC-3 and the GenBank™ sequence. The first difference, I378V, resulted in a conservative amino acid change, while the second, at alanine 541 (GCA to GCG), was silent. There was no sequence alteration in the tripartite GTPase domain or the self-assembly domains. The difference observed may be the result of human sequence polymorphisms (24). MxA expression has only been reported following viral infection or treatment with IFNs (3-5). However, Northern blots probed with our MxA clone (data not shown) and with the original cDNA (16) displayed the same pattern of expression that was generated by our DD-2 probe (Fig. 1B): abundant expression in PC-3 but no detectible mRNA in PC-3M. Equal loading was confirmed by hybridizing the blots with a probe for glyceraldehyde-phosphate dehydrogenase.
Western blot analysis with anti-MxA antibody (13) corroborated the Northern blot expression data, demonstrating the presence of a 78-kDa MxA protein in PC-3 but not in PC-3M lysates (Fig. 1C). The same blot was probed with anti-tubulin antibody to show equal loading for both samples.
The difference between the cell lines could have been related to a high endogenous production of type-I IFN in PC-3 cells. There was, however, no difference in the amount of IFN-α present in the two cell lines, as determined by Western blot analysis (Fig. 1D).
Maintenance of Genomic Integrity at the MxA Locus—The constitutive expression of MxA in PC-3 cells, but not in PC-3M cells, could be explained by a genomic deletion or rearrangement at the MX1 locus. To explore this possibility, genomic DNA from PC-3 and PC-3M cells was digested with EcoRI, BamHI, and PstI, electrophoresed on an agarose gel and subjected to Southern blot analysis with MxA cDNA (supplemental Fig. S1). PC-3 and PC-3M showed identical patterns of hybridization, which indicated that the difference in expression of MxA in PC-3 cells and PC-3M cells was not the result of a major genomic deletion or rearrangement.
Induction of MxA Expression by Interferon—To determine whether IFN-α can induce MxA expression in PC-3 and PC-3M cells, cells were treated with recombinant IFN-α and subjected to immunocytochemical analysis using anti-MxA antibody and DAPI nuclear counterstaining (Fig. 2, A and B). Consistent with the Western blot result, this assay detected MxA protein only in untreated PC-3 and not in untreated PC-3M cells (compare upper left panels in Fig. 2, A and B). After exposure to IFN-α, the level of MxA protein increased substantially in PC-3, whereas MxA protein became detectable for the first time in PC-3M cells (compare lower left panels in Fig. 2, A and B). Western blotting (not shown) confirmed the IFN-induced increase in MxA protein expression in both cell lines. This evidence demonstrated that PC-3M cells were able to respond to IFN, consistent with the Southern blot result (supplemental Fig. S1) indicating that the MxA gene was intact in PC-3M cells. This experiment also showed that the IFN signaling pathway was still active in PC-3M cells, ruling out the possibility that lack of MxA expression in PC-3M cells was due to an inability to respond to IFN stimulation.
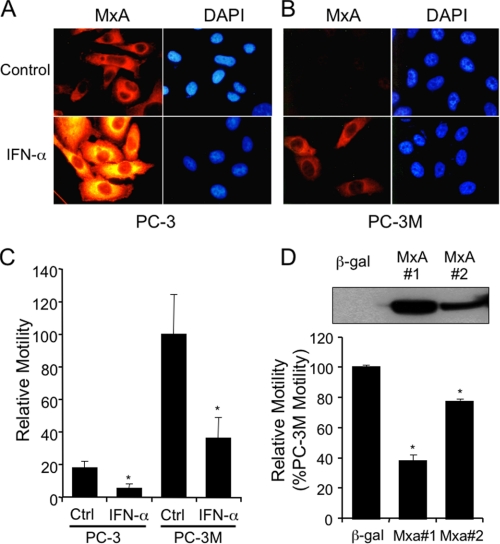
FIGURE 2.
IFN induction of MxA expression and inhibition of motility in PC-3 and PC-3M cells. A and B, fluorescence immunocytochemistry of MxA. PC-3 (panels A) and PC-3M (panels B) were grown for 24 h on coverslips in the presence or absence of 1000 international units (IU) of IFN-α/ml, fixed, permeabilized, stained with monoclonal anti-MxA and Cy-3-conjugated goat anti-mouse immunoglobulin, and counterstained with DAPI. Immunofluorescence was visualized with a Zeiss Axiophot microscope with a 40× objective, and the images were captured on an Optronics charge-coupled device camera. C, motility of PC-3 and PC-3M cells, with and without 24-h exposure to 1000 IU/ml IFN-α. Bars represent the percentage of control PC-3M mobility. Each bar represents the mean of two independent triplicate determinations ± S.E. D, MxA expression and motility. Motility of stable clones of PC-3M expressing β-galactosidase or two different levels of MxA was measured as above. To assess levels of expression in the cell lines being tested, Western blots of 50 μg of protein lysate per lane were probed with anti-MxA and shown in the inset. Each bar represents the mean ± S.E. of two independent experiments performed in triplicate. Wilcoxon rank sum test was used to compare the relative motility of control versus interferon-α-treated PC-3 and PC-3M cells (Fig. 2C) and β-gal-expressing versus two MxA-expressing PC-3M stable transfectants (Fig. 2D). *, p < 0.03.
Effect of MxA on Motility of PC-3M Cells—The constitutive expression of MxA in PC-3 cells and absence of expression in PC-3M cells suggested the possibility that MxA might suppress some aspect of metastatic behavior. Furthermore, it had been reported that type I IFN can reduce cell motility (7), one component of metastasis. To test whether PC-3 and PC-3M differed in motility, we subjected both lines to an assay that measured the ability of cells to migrate through pores in a PET membrane. Fig. 2C shows that untreated (control) PC-3M cells were considerably more motile than PC-3, and IFN-α reduced PC-3M motility to a level comparable to that of untreated PC-3. Consistent with the result seen in Fig. 2A, PC-3 cells were also responsive to IFN, which reduced their motility still further.
To test directly whether MxA played a role in regulating motility, we transfected PC-3M cells with MxA- and β-galactosidase control-expressing vectors, and stable cell lines, including PC-3M-MxA#1 and PC-3M-MxA#2, constitutively expressing full-length human MxA protein, and the control, PC-3M-β-gal, were selected. The level of MxA expression in the three transfectants was determined by Western blot (Fig. 2D, inset). MxA protein was not detected in PC-3M β-gal, whereas the other two lines expressed exogenous MxA, with a higher level of expression in MxA #1 than in MxA #2. The level of MxA in MxA clone #1 was comparable to the endogenous level of MxA in PC-3, and the level of MxA in clone #2 was lower than the level in PC-3 (supplemental Fig. S2). MxA expression inhibited motility in both MxA clone #1 and #2 (Fig. 2D), and the level of inhibition correlated with the level of MxA expression. PC-3M MxA clone #1 (PC-3M-MxA) was used in all subsequent studies.
Time-lapse Microscopy—Time-lapse video microscopy visually demonstrated the reduction of motility induced by MxA expression in PC-3M cells (supplemental Fig. S3). PC-3M-MxA cells showed markedly reduced levels of movement across the field, compared with control PC-3M cells that express the unrelated protein, β-galactosidase. Although translational motility of the PC-3M-MxA cells was markedly inhibited, the majority of the cells demonstrated membrane ruffling and several mitoses were apparent. However, in contrast to PC-3M-β-gal cells, in PC-3M-MxA cells there was an absence of translational motility of daughter cells following cytokinesis.
MxA Expression Has No Effect on Proliferation—To confirm the impression gained in the time-lapse study that expression of MxA did not impede cell division, and to investigate the role of GTPase activity in the ability of MxA to inhibit motility, another set of stable transfectants of PC-3M was made using the pCI vector that includes the CMV promoter, a neo selectable marker and a FLAG tag for immunodetection. In addition to the FLAG-tagged wild-type MxA, a FLAG-tagged MxA with a threonine to alanine mutation at residue 103 between the first and second GTP-binding consensus motif that ablates both GTPase and antiviral activity (12) was introduced into PC-3M cells. As shown in Fig. 3A, all three transfectants proliferated at the same rate, strongly suggesting that the effect of MxA on motility was unrelated to an effect on cell proliferation.
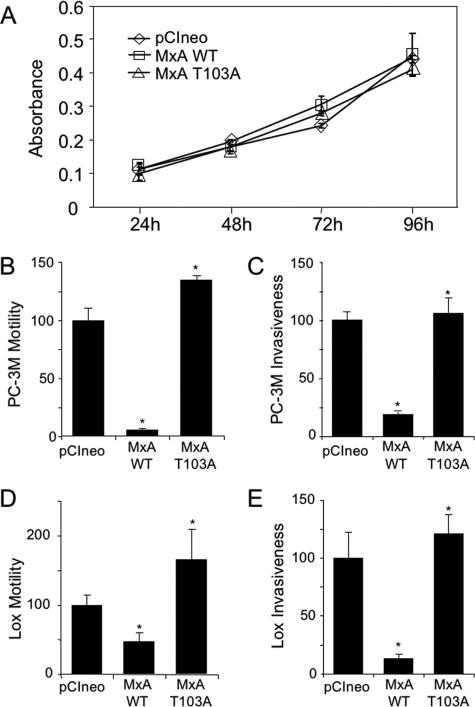
FIGURE 3.
Effect of MxA and MxA-T103A mutant on growth, motility, and invasion of tumor cells. PC-3M and LOX cells were stably transfected with wild-type MxA and MxA-T103A mutant in the pCI expression vector. A, proliferation. PC-3M cells expressing pCIneo vector, wild-type MxA, or MxA-T103A were grown in vitro for 4 days, and proliferation was assessed by 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (mean ± S.D. of triplicate observations). B and D, motility. Motility was measured, as in Fig. 2, for stable clones of PC-3M or LOX cells that expressed pCIneo vector, wild-type MxA, or MxA-T103A. The results are expressed as percentage of motility of the cells bearing the pCIneo control vector. C and E. Invasiveness. Percent of control cells (PC-3M or LOX cells with pCIneo vector alone) that invaded Matrigel and penetrated the PET membrane are shown (mean ± S.E.). Each bar represents the means ± S.E. of the mean of two independent experiments performed in triplicate in PC-3M cells (B and C), and the mean ± S.E. of three independent experiments performed in duplicate in LOX cells (D and E). Wilcoxon rank sum test was used to compare the relative motility and invasion of the stable pCIneo and MxA-T103A transfectants to the MxA-WT stable transfectants in PC-3M and LOX cells. *, p ≤ 0.01.
T103A Mutation Reverses Effects of MxA on Motility and Invasion—To confirm and extend our motility studies, we repeated them with the PC-3M MxA-FLAG transfectants. In addition, to determine if the effect of MxA was limited to PC-3M cells, stable transfectants expressing FLAG-tagged wild-type MxA and FLAG-tagged T103A-mutant MxA were created in the highly metastatic human melanoma cell line LOX (10), which, like PC-3M, does not express endogenous MxA (data not shown). The studies were also extended to test the effect of MxA on invasion, using a Transwell invasion assay in which the cells are required to cross a Matrigel layer as well as migrate through pores in the PET filter. Consistent with the results shown in Fig. 2, FLAG-tagged MxA inhibited PC-3M motility (Fig. 3B). Expression of FLAG-tagged MxA also inhibited PC-3M invasion (Fig. 3C). Similarly, the expression of exogenous MxA decreased LOX cell motility and invasiveness (Fig. 3, D and E). However, in both PC-3M and LOX cell lines, the T103A mutation in the GTPase/self-assembly region reversed the ability of MxA to suppress in vitro motility and invasiveness of these highly metastatic tumor cells.
Wild-type MxA but Not Mutant MxA Associates with Tubulin—It has been reported that MxA can transiently bind the cytoskeletal proteins actin and tubulin (3). Because elements of the cytoskeleton are instrumental in cell motility, we investigated whether MxA associated with the actin or tubulin cytoskeleton in PC-3 and LOX cells, using immunoprecipitation and immunocytochemical studies of cytoskeletal preparations. Fig. 4A demonstrates that endogenous MxA co-immunoprecipitated with tubulin, but not with actin, in PC-3 cells. In a cell-free GST pulldown experiment, GST-MxA associated with purified tubulin in a concentration-dependent manner, consistent with direct binding of MxA and tubulin (Fig. 4B).
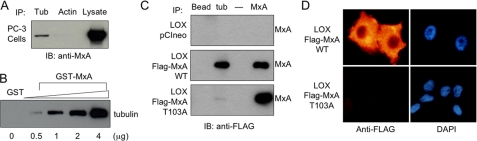
FIGURE 4.
Association of MxA with tubulin and microtubule cytoskeleton. A, co-immunoprecipitation in PC-3 cells. PC-3 cell lysates (2.5 mg of protein) were immunoprecipitated with either anti-α-tubulin or anti-actin antibodies, and the bound proteins were detected by Western blotting with anti-MxA antibody. PC-3 lysate was also included on the Western blot. B, GST pulldown. In vitro association between 2 μg of purified tubulin and GST alone or increasing amounts of GST-MxA that have been immobilized on glutathione-Sepharose beads. The bound proteins were eluted by boiling in SDS sample buffer, resolved on SDS-PAGE transferred to nitrocellulose, and probed with anti-α-tubulin antibody. C, co-immunoprecipitation in LOX cells. Cell lysates from LOX-FLAG-pCI neo, LOX-FLAG-MxA-WT, and LOX-FLAG-MxA-T103A cells (2.5 mg of protein for all immunoprecipitations except 0.5 mg of protein for immunoprecipitation with anti-MxA antibody) were immunoprecipitated with beads alone, anti-α-tubulin- or anti-MxA antibodies. The bound proteins were resolved by SDS-PAGE, transferred to nitrocellulose, and probed with anti-FLAG antibody. Lane 3 is an empty lane. D, association of MxA with the cytoskeleton. LOX cells, stably transfected with either FLAG-tagged wild-type MxA or FLAG-tagged MxA-T103A, were grown on glass coverslips, and cytoskeletal preparations were made as described under “Experimental Procedures,” and stained with anti-FLAG antibody. Nuclei were visualized by counterstaining with DAPI.
The association of wild-type MxA with tubulin was also examined in the stably transfected LOX cell line. Whole cell lysates were immunoprecipitated with anti-α-tubulin antibody, anti-MxA antibody, or protein A/G-coated Sepharose beads alone, followed by Western blotting with anti-FLAG antibody (Fig. 4C). MxA was detected in association with tubulin in LOX-FLAG-MxA-WT (panel 2, lane 2) while protein A/G alone (lane 1) did not bind MxA-containing complexes. No binding activity was detected in LOX-pCIneo control cells (panel 1), supporting the specificity of the co-immunoprecipitation.
To test whether the association of MxA with the microtubule cytoskeleton was dependent upon its GTPase/self-assembly activity, as was the ability of MxA to suppress motility and invasion, we also performed co-immunoprecipitation experiments using the LOX-FLAG-MxA-T103A stable transfectant (Fig. 4C, panel 3). In contrast to the LOX cells that expressed wild-type MxA, in LOX-FLAG-MxA-T103A cells, the binding of the T103A mutant of MxA to tubulin was virtually undetectable.
When soluble proteins were extracted from LOX melanoma cells that stably expressed wild-type MxA or T103A mutant MxA, only wild-type MxA protein remained bound to the insoluble cytoskeletal matrix (Fig. 4D). These data are consistent with the co-immunoprecipitation experiments in Fig. 4 (A and B) that showed that only wild-type MxA associated with tubulin. MxA-T103A washed out of the insoluble cytoskeleton preparation. These data are consistent with an association of tubulin and wild-type MxA but not T103A mutant MxA, suggesting that microtubules play a role in MxA-mediated reduction of motility and invasiveness of PC-3M prostate carcinoma cells and LOX melanoma cells.
GFP-MxA transiently expressed in PC-3M cells demonstrated two subcellular localizations, a diffuse cytoplasmic localization, and a punctate cytoplasmic localization that has a vesicular appearance (supplemental Fig. S4). Together with the earlier data suggesting a transient association of MxA and tubulin (3), the data suggest that MxA exists in multiple subcellular localizations and that microtubule-associated MxA is potentially associated with inhibition of motility.
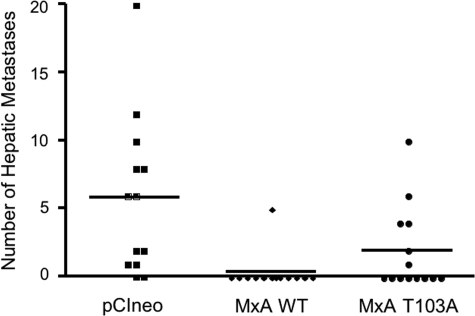
Effect of MxA on in Vivo Experimental Hepatic Metastasis—To test the effect of MxA expression in vivo, an experimental hepatic metastasis assay was used. In this assay, 2 × 106 cells from the PC-3M-pCIneo, PC-3M-MxA-wild-type, and PC-3M-MxA-T103A cell lines were injected into the spleens of beige/SCID mice. Fifteen mice per group were injected, and procedure-associated deaths occurred in two mice in the PC-3M-pCIneo and PC-3M-MxA-WT group and in one mouse in the PC-3M-MxA-T103A group. 24 days following intrasplenic injection liver metastases developed in 11/13 mice injected with PC-3M-pCIneo cells (Fig. 5). In contrast, metastases developed in 1/13 mice injected with PC-3M-MxA-WT. Liver metastases developed in 6/14 mice injected with PC-3M-MxA-T103A. Histological examination of primary tumors and metastases revealed highly epithelial neoplasia with limited stromal elements. No discrete acinar elements were noted in any samples. No significant differences were noted among tumor samples expressing MxA, MxA-T103A, or vector control. The number of hepatic metastases from PC-3M cells bearing the control pCIneo vector (mean = 5.8) was significantly greater than from PC-3M-MxA cells (mean = 0.39; p < 0.06; Fig. 5). The T103A mutation resulted in a 5-fold higher mean number of metastases than wild-type MxA (mean = 2.0). These data demonstrate that MxA inhibits the development of experimental metastases in vivo.
FIGURE 5.
Exogenous MxA expression inhibits PC-3M hepatic metastasis. Two million PC-3M cells stably transfected with pCIneo, wild-type MxA, or the MxA-T103A mutant were injected into the spleens of beige/SCID mice, and the number of liver metastases found on autopsy was determined by visual examination 24 days following intrasplenic injection. The number of hepatic metastases from mice injected with PC-3M-pCIneo (n = 13; mean number of metastases = 5.8) was significantly greater than from mice injected with PC-3M-MxA (n = 13; mean number of metastases = 0.39; p = 0.06 using Welch's corrected unpaired t test). A replicate hepatic metastasis assay yielded concordant results.
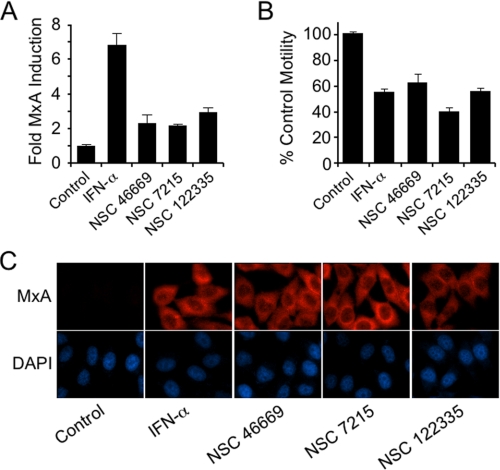
High-throughput Small Molecule Screen—The data presented thus far demonstrate the loss of expression of MxA in the highly metastatic PC-3 variant PC-3M, and that re-expression of MxA inhibits motility and metastasis. Because MxA is known to be highly inducible by a defined signaling pathway, the data suggest the potential success of targeting MxA in a small molecule screen. For this screen we cloned the human MxA promoter upstream of luciferase as described under “Experimental Procedures.” The MxA promoter-luciferase reporter construct together with a neomycin-resistance plasmid were transfected into PC-3M cells, and G418 was used to select stable transfectants. The stably transfected cells were incubated with the compounds in the NCI Diversity Set of 1900 pharmacophores at a concentration of 10 μm each for 24 h, and the cells were tested for luciferase activity using the assay kit provided by Promega, according to the manufacturer's instructions. Recombinant interferon-α was used as a positive control. Three molecules induced a >2.2-fold increase over the vehicle control (Fig. 6A). The positive control, 1000 IU/ml interferon-α, induced a 6.8-fold increase in reporter activity. The same three compounds also induced MxA protein expression in PC-3M cells in culture (Fig. 6C).
FIGURE 6.
Screen for small molecules that induce MxA and inhibit motility. A, the Diversity Set of 1990 pharmacophores from the NCI Developmental Therapeutics Program was tested in a high-throughput screen using an MxA promoter-luciferase reporter assay. PC-3M cells were incubated for 24 h with 10 μm compound. Interferon-α was used as a positive control. Results from three small molecules that induced a >2.2-fold increase in activity are shown (two independent experiments in triplicate). B, effect of the three small molecule hits on PC-3M motility (two independent experiments in duplicate). Motility of PC-3M cells, with and without 24-h exposure to 1000 international units/ml IFN-α or the small molecule hits. Bars represent the percentage of control PC-3M mobility. Motility was assayed as described under “Experimental Procedures.” C, immunocytochemistry with anti-MxA antibody confirmed induction of MxA protein by the three compounds detected in the high-throughput assay for MxA promoter activation.
These three active small molecules were also screened for their effect on PC-3M motility, as described above. In the motility assay, PC-3M cells were inhibited between 40 and 60% by these compounds (Fig. 6B). In this assay, interferon-α caused approximately a 50% inhibition of motility, indicating that the small molecules were comparable in activity to that of interferon-α.
DISCUSSION
This study began with the use of DD-RT-PCR to identify genes differentially expressed in two clonally related human prostate carcinoma cell lines differing in metastatic activity, and this revealed a dramatic difference in MxA expression. As demonstrated here, MxA mRNA and protein were abundant in PC-3 but were not detectable in its more metastatic derivative, PC-3M. To test the hypothesis that MxA plays a role in reduction of motility and metastasis of prostatic and other cancers, we expressed the full-length MxA cDNA in PC-3M prostate carcinoma cells and in LOX melanoma cells and compared its effect with that of control vectors. MxA induced a clear reduction in motility and invasiveness in both tumor types in two in vitro assays. Stable expression of exogenous MxA in PC-3M cells also caused a significant reduction in an in vivo assay of metastasis in immunocompromised beige-SCID mice: the number of hepatic metastases following intrasplenic injection.
It was unexpected to find that PC-3 expressed MxA spontaneously, because it was believed that MxA is not expressed in normal or neoplastic cells in the absence of viral infection or exposure to exogenous interferon (25, 26). However, our Western blot analysis of 27 cancer cell lines from the NCI-60, the National Cancer Institute Developmental Therapeutics Program's panel of 60 cultured human cancer cells, that had been established to screen chemical compounds for anti-cancer activity (27) detected MxA expression in 0/1 leukemia, 3/7 non-small cell lung cancer, 2/7 colon, 4/6 CNS, and 2/6 melanoma (data not shown). This demonstrates that expression of MxA is more widespread than anticipated and suggests that regulation of MxA warrants further investigation.
It has been known for almost 30 years that IFN can inhibit normal cell motility (7) but the mechanism has not been identified. MxA is known to be strongly induced by IFN, and MxA expression is a preferred marker for evidence of IFN biological activity in vivo (28). Our data show that MxA mimics the IFN effect on motility, suggesting that it might be a critical molecular mediator of the IFN effect.
Down-regulation of a number of IFN target genes has been reported in several studies of global gene expression in prostate cancer. Shou et al. (29), Nagano et al. (4), and Schulz et al. (30) showed that a significant portion of the genes whose down-regulation was associated with prostate cancer tumorigenesis or tumor progression were IFN-inducible genes, including MxA. It is also of interest that a recent study by Han and colleagues found that the genome organizer SATB1 reprograms gene expression to promote breast cancer tumor growth and metastasis, and that MX1 is a target for repression by SATB1 (31).
IFN has been used in the treatment of prostate cancer (32), melanoma (33), renal cell carcinoma (34, 35) and other human neoplasms. When expression of IFN-β was induced in PC-3M cells by transfection of an expression vector, these cells showed a reduced ability to metastasize and reduced tumorigenicity in nude mice (36). The authors demonstrated an anti-angiogenic effect of the IFN-producing tumor cells on surrounding stroma. The data in the present report demonstrate that IFN also directly inhibits PC-3M motility, indicating that IFN may affect both tumor and stroma. In aggregate, these data suggest that MxA may be a mediator of the effect of IFN on normal and tumor cell motility.
Motile cells are polarized, with a leading edge characterized by a ruffling lamellipodium and a trailing tail that retracts from substratum attachment sites. Actin polymerization is an essential force in cell propulsion, and small GTPases regulate lamellipodium function via their effects on actin. In addition, the tubulin-based cytoskeleton, specifically, via microtubule retraction, is also important to the function of the uropod that alternatively holds and releases the cell from its attachments (37). Our data demonstrate that MxA interacts with tubulin and that a point mutation of the MxA GTPase domain known to inactivate the GTPase (12) also inactivates MxA control of motility and blocks MxA association with microtubules. This confirms the earlier report (3) of transient association of MxA with subcellular components. Studies of the cytoskeleton and motility have focused more on actin and actin-regulatory small GTPases of the Rho family than on microtubules. Recent research, however, has demonstrated that, in motile cells, microtubules also regulate Rho protein activity and actin polymerization, and, thus, microtubules are important regulators of directional movement (38, 39). Furthermore, there are several recent reports implicating microtubules and the microtubule-organizing center in directing cell polarization and migration (40-42). Our data suggest that MxA may be important in this process, representing a member of a new class of microtubule-associated proteins that regulate motility. Membrane ruffling and mitosis appear uncompromised in time-lapse studies of MxA-expressing PC-3M cells. The pronounced decrease of vectorial movement of these cells compared with PC-3M cells expressing β-galactosidase suggests that MxA targets specific processes regulating motility, such as cell polarization and/or detachment from substratum adhesion sites (37, 39, 43). Reversible association with tubulin that depends on GTP binding is an attractive candidate for regulating motility in normal cells and metastasis in transformed cells. Several other metastasis-suppressing genes share with MxA a predilection for association with the cytoskeleton (44, 45). Specifically, several prostate cancer metastasis-suppressing gene products, CD44, nm23-H1, KAI1/CD82, and KiSS1 (the last two specifically demonstrated in experimental rat prostate metastasis) appear to stimulate or moderate reorganization of the actin cytoskeleton. This mechanism may also explain the observation that MxA, like certain other dynamin family members, plays a role in membrane remodeling, such as a transient perturbation of endocytosis (46, 47). Our data indicate that, unlike some dynamin family members (48), MxA does not seem to be essential for cell division, rendering it a classic metastasis-suppressor molecule.
Our goal was to identify a new pathway for the control of tumor cell motility and metastasis. This was achieved by the identification of MxA as a metastasis control gene. The level of MxA expression may be a predictor of metastatic potential. If this is verified, MxA could be recognized as a metastasis-specific component of the molecular phenotype and have an significant impact on therapeutic decisions (49). It is of interest in this regard that a bioinformatics strategy has revealed recently that ∼50% of prostate cancers from prostate-specific antigen-screened surgical cohorts are positive for a fusion of TMPRSS2 and ERG on chromosome 21 (50). This fusion, which is associated with an invasive phenotype, is frequently associated with interstitial DNA deletions that encompass the MX1 locus (51). The fusion gene can induce an invasion-associated transcriptional program. However, the finding that interstitial deletions encompassing the MxA-encoding locus are associated with increased aggressiveness of prostate cancer, together with the data presented here, suggest there may also be a contribution from the loss of MxA-mediated anti-metastatic activity to the observed correlation of the fusion protein with aggressivity.
In addition to its predictive potential, up-regulation of MxA and its metastasis-inhibiting activity is worth consideration as a new therapeutic target in tumors where the MX1 locus is intact (45). Drugs that target up-regulation of MxA might provide more specific anti-tumor activity and less toxicity than type I IFNs (32) or even a new anti-tumor interferon, limitin (52). To develop an MxA-targeted small molecule, we employed an MxA promoter-reporter system in a high-throughput format to screen for inducers of MxA expression. Our data showed that there are small molecules that can cause increased MxA expression and inhibition of motility. The MxA-promoter-luciferase reporter screen demonstrates that small molecules can up-regulate the MxA promoter and MxA expression. Studies of the effect of MxA-directed small molecules on tumor behavior in vivo await further refinement of the hits by an expanded screen and structure-activity relationship analysis.
Various expression strategies have been used to successfully identify tumor differentiation states, predict metastatic potential, and develop new therapeutic targets (53-58). The results of the present study are particularly amenable to clinical translation, because the gene identified is highly inducible. Small molecule up-regulation of MxA expression may represent a new strategy for development of anti-metastasis therapeutics.
Supplementary Material
Acknowledgments
We thank Lee Helman, NCI, NIH, for helpful discussions, Ion Gresser, Institut Curie, for interest and antibodies, Jeff Rubin and William Taylor, both from the NCI, for a Northern blot of human prostate RNAs, Marina Marini and Walter Schlapkohl, NCI, and Dan Sackett, NICHD, NIH, for critical reading of the manuscript and valuable suggestions. We thank James Kozlowski and Dan Sackett for the kind gifts of the PC-3M and LOX cell lines, respectively. We acknowledge with gratitude the reference antisera from the Reference Repository of the NIAID, NIH, maintained by Braton Biotech, Inc., Rockville, MD.
This work was supported, in whole or in part, by the National Institutes of Health NCI Intramural Program.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: DD-RT-PCR, differential display reverse transcription-PCR; IFN, interferon; WT, wild-type; DAPI, 4,6-diamidino-2-phenylindole; PET, polyethylene terephthalate; CID, central interactive domain.
References
- 1.Kozlowski, J. M., Fidler, I. J., Campbell, D., Xu, Z. L., Kaighn, M. E., and Hart, I. R. (1984) Cancer Res. 44 3522-3529 [PubMed] [Google Scholar]
- 2.Liang, P., and Pardee, A. B. (1992) Science 257 967-971 [DOI] [PubMed] [Google Scholar]
- 3.Horisberger, M. A. (1992) J. Virol. 66 4705-4709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagano, K., Masters, J. R., Akpan, A., Yang, A., Corless, S., Wood, C., Hastie, C., Zvelebil, M., Cramer, R., and Naaby-Hansen, S. (2004) Oncogene 23 1693-1703 [DOI] [PubMed] [Google Scholar]
- 5.Kotenko, S. V., Gallagher, G., Baurin, V. V., Lewis-Antes, A., Shen, M., Shah, N. K., Langer, J. A., Sheikh, F., Dickensheets, H., and Donnelly, R. P. (2003) Nat. Immunol. 4 69-77 [DOI] [PubMed] [Google Scholar]
- 6.Jonasch, E., and Haluska, F. G. (2001) Oncologist 6 34-55 [DOI] [PubMed] [Google Scholar]
- 7.Brouty-Boye, D., and Zetter, B. R. (1980) Science 208 516-518 [DOI] [PubMed] [Google Scholar]
- 8.Haller, O., and Kochs, G. (2002) Traffic 3 710-717 [DOI] [PubMed] [Google Scholar]
- 9.Lee, S. J., Ha, M. J., Lee, J., Nguyen, P., Choi, Y. H., Pirnia, F., Kang, W. K., Wang, X. F., Kim, S. J., and Trepel, J. B. (1998) J. Biol. Chem. 273 10618-10623 [DOI] [PubMed] [Google Scholar]
- 10.Fodstad, O., Aamdal, S., McMenamin, M., Nesland, J. M., and Pihl, A. (1988) Int. J. Cancer 41 442-449 [DOI] [PubMed] [Google Scholar]
- 11.Horisberger, M. A. (1995) Am. J. Respir. Crit. Care Med. 152 S67-S71 [DOI] [PubMed] [Google Scholar]
- 12.Ponten, A., Sick, C., Weeber, M., Haller, O., and Kochs, G. (1997) J. Virol. 71 2591-2599 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horisberger, M. A., and Hochkeppel, H. K. (1987) J. Interferon Res. 7 331-343 [DOI] [PubMed] [Google Scholar]
- 14.Yamada, T., Horisberger, M. A., Kawaguchi, N., Moroo, I., and Toyoda, T. (1994) Neurosci. Lett. 181 61-64 [DOI] [PubMed] [Google Scholar]
- 15.Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989) Molecular Cloning: A Laboratory Manual, 2nd Ed., Cold Spring Harbor Laboratory, Cold Spring Harbor, NY
- 16.Horisberger, M. A., McMaster, G. K., Zeller, H., Wathelet, M. G., Dellis, J., and Content, J. (1990) J. Virol. 64 1171-1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fort, P., Marty, L., Piechaczyk, M., el Sabrouty, S., Dani, C., Jeanteur, P., and Blanchard, J. M. (1985) Nucleic Acids Res. 13 1431-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi, Y. H., Lee, S. J., Nguyen, P., Jang, J. S., Lee, J., Wu, M. L., Takano, E., Maki, M., Henkart, P. A., and Trepel, J. B. (1997) J. Biol. Chem. 272 28479-28484 [DOI] [PubMed] [Google Scholar]
- 19.Hartwig, J. H. (1992) J. Cell Biol. 118 1421-1442 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bang, Y. J., Pirnia, F., Fang, W. G., Kang, W. K., Sartor, O., Whitesell, L., Ha, M. J., Tsokos, M., Sheahan, M. D., Nguyen, P., Niklinski, W. T., Myers, C. E., and Trepel, J. B. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 5330-5334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Khanna, C., Prehn, J., Yeung, C., Caylor, J., Tsokos, M., and Helman, L. (2000) Clin. Exp. Metastasis 18 261-271 [DOI] [PubMed] [Google Scholar]
- 22.Ronni, T., Matikainen, S., Lehtonen, A., Palvimo, J., Dellis, J., Van Eylen, F., Goetschy, J. F., Horisberger, M., Content, J., and Julkunen, I. (1998) J. Interferon Cytokine Res. 18 773-781 [DOI] [PubMed] [Google Scholar]
- 23.Kochs, G., Haener, M., Aebi, U., and Haller, O. (2002) J. Biol. Chem. 277 14172-14176 [DOI] [PubMed] [Google Scholar]
- 24.Tazi-Ahnini, R., di Giovine, F. S., McDonagh, A. J., Messenger, A. G., Amadou, C., Cox, A., Duff, G. W., and Cork, M. J. (2000) Hum. Genet. 106 639-645 [DOI] [PubMed] [Google Scholar]
- 25.Goetschy, J. F., Zeller, H., Content, J., and Horisberger, M. A. (1989) J. Virol. 63 2616-2622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.al-Masri, A. N., Werfel, T., Jakschies, D., and von Wussow, P. (1997) Mol. Pathol. 50 9-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Scherf, U., Ross, D. T., Waltham, M., Smith, L. H., Lee, J. K., Tanabe, L., Kohn, K. W., Reinhold, W. C., Myers, T. G., Andrews, D. T., Scudiero, D. A., Eisen, M. B., Sausville, E. A., Pommier, Y., Botstein, D., Brown, P. O., and Weinstein, J. N. (2000) Nat. Genet. 24 236-244 [DOI] [PubMed] [Google Scholar]
- 28.Roers, A., Hochkeppel, H. K., Horisberger, M. A., Hovanessian, A., and Haller, O. (1994) J. Infect. Dis. 169 807-813 [DOI] [PubMed] [Google Scholar]
- 29.Shou, J., Soriano, R., Hayward, S. W., Cunha, G. R., Williams, P. M., and Gao, W. Q. (2002) Proc. Natl. Acad. Sci. U. S. A. 99 2830-2835 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schulz, W. A., Alexa, A., Jung, V., Hader, C., Hoffmann, M. J., Yamanaka, M., Fritzsche, S., Wlazlinski, A., Muller, M., Lengauer, T., Engers, R., Florl, A. R., Wullich, B., and Rahnenfuhrer, J. (2007) Mol. Cancer 6 14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Han, H. J., Russo, J., Kohwi, Y., and Kohwi-Shigematsu, T. (2008) Nature 452 187-193 [DOI] [PubMed] [Google Scholar]
- 32.Kuratsukuri, K., Nishisaka, N., Jones, R. F., Wang, C. Y., and Haas, G. P. (2000) Urol. Oncol. 5 265-273 [DOI] [PubMed] [Google Scholar]
- 33.Cascinelli, N., Belli, F., MacKie, R. M., Santinami, M., Bufalino, R., and Morabito, A. (2001) Lancet 358 866-869 [DOI] [PubMed] [Google Scholar]
- 34.Nanus, D. M. (2000) Curr. Oncol. Rep. 2 417-422 [DOI] [PubMed] [Google Scholar]
- 35.Pastore, R. D., Pfeffer, L. M., and Nanus, D. M. (2001) Cancer Invest. 19 281-291 [DOI] [PubMed] [Google Scholar]
- 36.Dong, Z., Greene, G., Pettaway, C., Dinney, C. P., Eue, I., Lu, W., Bucana, C. D., Balbay, M. D., Bielenberg, D., and Fidler, I. J. (1999) Cancer Res. 59 872-879 [PubMed] [Google Scholar]
- 37.Ratner, S., Sherrod, W. S., and Lichlyter, D. (1997) J. Immunol. 159 1063-1067 [PubMed] [Google Scholar]
- 38.Waterman-Storer, C. M., Worthylake, R. A., Liu, B. P., Burridge, K., and Salmon, E. D. (1999) Nat. Cell Biol. 1 45-50 [DOI] [PubMed] [Google Scholar]
- 39.Wittmann, T., and Waterman-Storer, C. M. (2001) J. Cell Sci. 114 3795-3803 [DOI] [PubMed] [Google Scholar]
- 40.Lafont, F., Burkhardt, J. K., and Simons, K. (1994) Nature 372 801-803 [DOI] [PubMed] [Google Scholar]
- 41.Abal, M., Piel, M., Bouckson-Castaing, V., Mogensen, M., Sibarita, J. B., and Bornens, M. (2002) J. Cell Biol. 159 731-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Magdalena, J., Millard, T. H., and Machesky, L. M. (2003) J. Cell Sci. 116 743-756 [DOI] [PubMed] [Google Scholar]
- 43.Ballestrem, C., Wehrle-Haller, B., Hinz, B., and Imhof, B. A. (2000) Mol. Biol. Cell 11 2999-3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dolle, L., Depypere, H. T., and Bracke, M. E. (2006) Curr. Cancer Drug Targets 6 729-751 [DOI] [PubMed] [Google Scholar]
- 45.Kauffman, E. C., Robinson, V. L., Stadler, W. M., Sokoloff, M. H., and Rinker-Schaeffer, C. W. (2003) J. Urol. 169 1122-1133 [DOI] [PubMed] [Google Scholar]
- 46.Accola, M. A., Huang, B., Al Masri, A., and McNiven, M. A. (2002) J. Biol. Chem. 277 21829-21835 [DOI] [PubMed] [Google Scholar]
- 47.Jatiani, S. S., and Mittal, R. (2004) Biochem. Biophys. Res. Commun. 323 541-546 [DOI] [PubMed] [Google Scholar]
- 48.Danino, D., and Hinshaw, J. E. (2001) Curr. Opin. Cell Biol. 13 454-460 [DOI] [PubMed] [Google Scholar]
- 49.Oh, W. K., and Kantoff, P. W. (1999) J. Clin. Oncol. 17 3664-3675 [DOI] [PubMed] [Google Scholar]
- 50.Tomlins, S. A., Laxman, B., Varambally, S., Cao, X., Yu, J., Helgeson, B. E., Cao, Q., Prensner, J. R., Rubin, M. A., Shah, R. B., Mehra, R., and Chinnaiyan, A. M. (2008) Neoplasia 10 177-188 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Attard, G., Clark, J., Ambroisine, L., Fisher, G., Kovacs, G., Flohr, P., Berney, D., Foster, C. S., Fletcher, A., Gerald, W. L., Moller, H., Reuter, V., De Bono, J. S., Scardino, P., Cuzick, J., and Cooper, C. S. (2008) Oncogene 27 253-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Kawamoto, S., Oritani, K., Asakura, E., Ishikawa, J., Koyama, M., Miyano, K., Iwamoto, M., Yasuda, S., Nakakubo, H., Hirayama, F., Ishida, N., Ujiie, H., Masaie, H., and Tomiyama, Y. (2004) Exp. Hematol. 32 797-805 [DOI] [PubMed] [Google Scholar]
- 53.Reiter, R. E., Gu, Z., Watabe, T., Thomas, G., Szigeti, K., Davis, E., Wahl, M., Nisitani, S., Yamashiro, J., Le Beau, M. M., Loda, M., and Witte, O. N. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 1735-1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Yang, R. M., Naitoh, J., Murphy, M., Wang, H. J., Phillipson, J., deKernion, J. B., Loda, M., and Reiter, R. E. (1998) J. Urol. 159 941-945 [PubMed] [Google Scholar]
- 55.Amara, N., Palapattu, G. S., Schrage, M., Gu, Z., Thomas, G. V., Dorey, F., Said, J., and Reiter, R. E. (2001) Cancer Res. 61 4660-4665 [PubMed] [Google Scholar]
- 56.Argani, P., Rosty, C., Reiter, R. E., Wilentz, R. E., Murugesan, S. R., Leach, S. D., Ryu, B., Skinner, H. G., Goggins, M., Jaffee, E. M., Yeo, C. J., Cameron, J. L., Kern, S. E., and Hruban, R. H. (2001) Cancer Res. 61 4320-4324 [PubMed] [Google Scholar]
- 57.Saffran, D. C., Raitano, A. B., Hubert, R. S., Witte, O. N., Reiter, R. E., and Jakobovits, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 2658-2663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Singh, D., Febbo, P. G., Ross, K., Jackson, D. G., Manola, J., Ladd, C., Tamayo, P., Renshaw, A. A., D'Amico, A. V., Richie, J. P., Lander, E. S., Loda, M., Kantoff, P. W., Golub, T. R., and Sellers, W. R. (2002) Cancer Cell 1 203-209 [DOI] [PubMed] [Google Scholar]
- 59.Haller, O., Stertz, S., and Kochs, G. (2007) Microbes Infect. 9 1636-1643 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.