Abstract
PHLPP2 (PH domain leucine-rich repeat protein phosphatase 2) terminates Akt and protein kinase C (PKC) activity by specifically dephosphorylating these kinases at a key regulatory site, the hydrophobic motif (Ser-473 in Akt1). Here we identify a polymorphism that results in an amino acid change from a Leu to Ser at codon 1016 in the phosphatase domain of PHLPP2, which reduces phosphatase activity toward Akt both in vitro and in cells, in turn resulting in reduced apoptosis. Depletion of endogenous PHLPP2 variants in breast cancer cells revealed the Ser-1016 variant is less functional toward both Akt and PKC. In pair-matched high grade breast cancer samples we observed retention of only the Ser allele from heterozygous patients (identical results were observed in a pair-matched normal and tumor cell line). Thus, we have identified a functional polymorphism that impairs the activity of PHLPP2 and correlates with elevated Akt phosphorylation and increased PKC levels.
Breast cancer is diagnosed in ∼180,000 women and is the cause of 40,000 deaths each year in the U.S.2 A prevalent underlying mechanism driving tumorigenesis is aberrant signal transduction pathways that result in constitutive activation of cell growth, proliferation, and survival pathways (2). A well characterized signal transduction pathway in breast cancer that promotes cellular survival, growth, and proliferation is the phosphatidylinositol 3-kinase/Akt pathway (3). This pathway is activated by a number of mechanisms, including gene amplification or gain of function mutations in upstream receptor protein-tyrosine kinases (4, 5), constitutive activation of hormone receptors (6), activating mutations in phosphatidylinositol 3-kinase and Akt (7, 8), and loss of function mutations in the regulatory phosphatase PTEN3 (phosphatase and tensin homolog on chromosome ten) (9). Thus, Akt is a major regulator of breast tumorigenesis.
There are three isoforms of Akt present in humans. All three isoforms contain activating phosphorylation sites in the activation loop (Thr-308 in Akt1) and in the C-terminal hydrophobic motif (Ser-473 in Akt1) (10). Upon growth factor receptor stimulation, phosphatidylinositol 3-kinase becomes activated and phosphorylates the D3 position of, typically, phosphatidylinositol (4, 5) bisphosphate to generate phosphatidylinositol (3,4,5)-trisphosphate (11). This 3′-phosphorylated lipid recruits Akt to the plasma membrane by binding to its PH domain, resulting in conformational changes that allow access to the activation loop phosphorylation site (11). Constitutively bound phosphatidylinositol-dependent kinase-1 then phosphorylates Akt at Thr-308, accompanied by phosphorylation at Ser-473 resulting in a catalytically active kinase (12). Phosphorylation of Ser-473 depends on the mTORC2 complex (13-16). Signaling through this pathway is terminated by removal of the lipid second messenger phosphatidylinositol (3,4,5)-trisphosphate catalyzed by the phosphatase PTEN and by direct dephosphorylation of Akt by the recently-identified PHLPP family of phosphatases and protein phosphatase 2A-type phosphatases (17-20).
The PHLPP family of phosphatases comprise three variants, the alternatively spliced PHLPP1α and PHLPP1β, and PHLPP2 (21). PHLPP1 and PHLPP2 specifically dephosphorylate the hydrophobic motif of specific Akt isozymes, thus decreasing Akt activity and promoting apoptosis (18, 19). PHLPP2 binds and dephosphorylates Akt1 and Akt3, whereas PHLPP1 binds and dephosphorylates Akt2 and Akt3 (18, 22). Their role in inactivating Akt suggests that both PHLPP1 and PHLPP2 could be potential tumor suppressors. Consistent with such a role, these phosphatases also dephosphorylate the hydrophobic motif of PKC, resulting in degradation of PKC. For this kinase, phosphorylation stabilizes the enzyme, so that the effect of depletion of the PHLPP phosphatases is to increase PKC protein levels (23). PKC is a well characterized oncogene, and loss of function of the PHLPP phosphatases could increase PKC protein levels and promote tumorigenesis (24). Providing further rationale that PHLPP2 could be a potential tumor suppressor, the phosphatase is located on chromosome 16q22.3, a region that encounters frequent loss of heterozygosity (LOH) in many primary and malignant breast tumors (25).
Here we identify a non-synonymous polymorphism that results in an amino acid change from a Leu to a Ser at codon 1016 in the PP2C phosphatase domain of PHLPP2. Overexpression studies reveal the Ser-1016 variant has impaired phosphatase activity and is less effective at inducing apoptosis than the Leu-1016 variant. When comparing a pair-matched normal and breast cancer cell line or pair-matched normal and high grade tumor patient samples that are heterozygous, we observe preferential loss of the Leu allele in the tumor tissue or breast cancer cell line. This observation provides evidence that PHLPP2 could be one of the elusive tumor suppressor genes on chromosome 16q, and for heterozygous patients, loss of the more catalytically active Leu-1016 may promote breast tumorigenesis.
EXPERIMENTAL PROCEDURES
Materials—SMARTpool siRNAs against PHLPP1 and PHLPP2 were purchased from Dharmacon. The following antibodies were purchased from Cell Signaling: phospho-specific to Thr-308 and Ser-473 of Akt, Akt, PTEN, and ERK1/2. PHLPP1 and PHLPP2 isoform-specific antibodies were purchased from Bethyl Laboratories. PKCα, Annexin I, and Lamin A antibodies were purchased from Santa Cruz Biotechnology. EGF was purchased from Upstate Biotechnologies. His-tagged human Akt1 was purified from baculoviral-infected Sf21 cells as described previously (19). Breast cancer cell lines or DNA from the cell lines listed in Fig. 1B were purchased from the ATCC. Breast tumor tissue DNA was purchased from Asterand or provided by Dr. Linda Wasserman. Asterand provided histology for tumor tissue samples. 100 DNA samples extracted from tissues of healthy individuals were purchased from the Coriell Institute.
FIGURE 1.
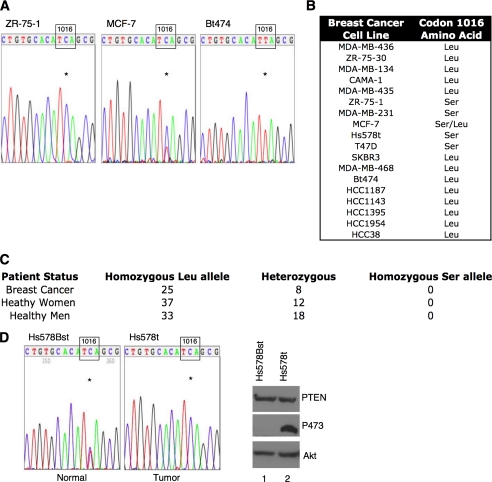
Identification of the L1016S polymorphism in the PHLPP2 phosphatase. A, chromatograms from the indicated breast cancer cell lines that possess only the Ser allele, both alleles, or only the Leu allele. Reverse transcription-PCR was performed using primers flanking the polymorphism, and PCR products were then sequenced using nested primers. The nucleotide (C or T) at position 3047 is indicated with an asterisk. B, table displaying genotype of 18 breast cancer cell lines. C, genotype of 100 healthy individuals from the Coriell Institute and 33 pair match breast cancer patients, confirming nucleotide change is indeed a polymorphism. D, comparison of the normal breast cell line Hs578Bst and the breast tumor cell line Hs578t from the same patient showing presence of T and C at position 3047 in normal cell line but only C in tumor cell line (asterisk). Western blot of lysates from Hs578Bst (lane 1) or Hs578t (lane 2) cells probed with antibodies for PTEN, Akt, or Akt phosphorylated at Ser-473.
Cell Culture, Transfections, and Immunoblotting—ZR-75-1, T47D, MDA-MB-468, and SKBR3 cell lines were maintained in RPMI 1640 (Cellgro), and all other cell lines were maintained in Dulbecco's modified Eagle's medium (Cellgro); both media were supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin. Cells were maintained at 37 °C in 5% CO2. Full-length HA-PHLPP2 variants were expressed in the pcDNA3HA vector as described previously (18). The nucleotide at position 3047 was changed fromaTtoaC using the QuikChange site-directed mutagenesis kit (Stratagene). Transient transfections and siRNA experiments were performed as previously described (18). Briefly, cells were transfected with 0.5 μg of DNA and incubated 48 h prior to lysis. Transfection efficiencies for 293T and H157 cell lines ranged from 70 to 90%. siRNA transfections were performed using 50 nm SMARTpool siRNA, and cells were incubated for 48 h and either lysed or their media was changed to low serum conditions for 12 h and then EGF (10 ng/ml) was added for the indicated times prior to lysis. For immunoblotting, transfected cells were lysed in Buffer 1 (50 mm Na2HPO4, pH 7.5, 1 mm sodium pyrophosphate, 20 mm NaF, 2 mm EDTA, 2 mm EGTA, 1% SDS, 1 mm DTT, 200 μm benzamidine, 40 μgml-1 leupeptin, and 1 mm phenylmethylsulfonyl fluoride) and sonicated for 5 s. Lysates containing equal protein (determined by Bradford assay) were analyzed on SDS-PAGE gels, and individual blots were probed using the antibodies indicated in figure legends. Densitometric analysis was performed with the NIH Image analysis software (version 1.63).
Phosphatase Assays—The activity of full-length PHLPP2 variants was assessed by expressing the HA-PHLPP2 variants and then immunoprecipitating the PHLPP2 variants from H157 cells (18). Cells were lysed in Buffer 2 (20 mm HEPES, pH 7.4, 1% Triton X-100, 1 mm DTT, 200 μm benzamidine, 40 μg ml-1 leupeptin, and 1 mm phenylmethylsulfonyl fluoride). Detergent-soluble lysates were incubated overnight at 4 °C with HA antibody and ultra-link protein A/G beads (Pierce). Beads were then washed three times with Buffer 1, three times with phosphatase buffer (containing 50 mm Tris (pH 7.4), 1 mm DTT, and 5 mm MnCl2), and incubated in phosphatase buffer with 1 μg of purified phosphorylated Akt at 30 °C for indicated times as previously described (19).
Apoptosis Assays—Apoptotic assays were performed as previously described (26). Briefly, cells were cotransfected with green fluorescent protein and HA-Leu-1016 or HA-Ser-1016 and incubated in low Serum (0.1% fetal bovine serum) media for 48 h; cells were gated based on green fluorescent protein expression as described previously and sub2n DNA was quantified (26).
DNA and RNA Isolation—Genomic DNA was isolated from human tissues using QIAamp DNA mini kit (Qiagen) following the manufacturer's protocol. Tissue samples were incubated in ATL buffer (from kit) supplemented with proteinase K for 3 h at 56 °C prior to DNA extraction. Total RNA was isolated from cell lines using RNeasy mini kit (Qiagen), following the manufacturer's protocol.
PCR and RT-PCR—One-step RT-PCR (Qiagen) was used to generate PCR products spanning the entire open reading frame of PHLPP2 in the following cell lines: H157, 293T, MDA-MB-231, Hs578t, SKBR-3, MDA-MB-468, Bt474, T47D, MCF-7, and ZR-75-1. RT-PCR products were subcloned into pGEM-T easy vector (Promega, Madison, WI) and sequenced. Following the identification of a polymorphism at Leu-1016, additional RT-PCR for all cell lines was performed by directly amplifying the region flanking the polymorphism using the following primers: 5′-gaggctcaaagggtgaagg-3′ and 5′-ggcccccagcattatgct-3′. The RT-PCR products were gel purified and sequenced directly. For sequencing genomic DNA (from Asterand, Dr. Linda Wasserman, or Coriell Institute), primers were generated to intronic and exonic sequences that flanked the polymorphism or the intronic sequences that flanked the entire exon 17. The following primers were used: 5′-gtgaatggggtaacctgctg-3′ and 5′-ctaccttgctgccattggtt-3′ (flanking polymorphism) and 5′-agtggggcagtcatagtgct-3′ and 5′-agttggctctcatcgttgct-3′ (flanking exon 17). The DNA-PCR and RT-PCR products were sequenced directly using nested primers: 5′-ctgggctgtacatacctctacc-3′ and 5′-ttcactgctcacctctgagg-3′. DNA was amplified using the following protocol: 1 ng of DNA was added to PCR sequencing buffer (5 μl of reaction mix (Stratagene), 200 nm of each primer, 1 μl of Pfu Turbo DNA polymerase (Stratagene), and purified water (5 μl of total reaction amount). The following PCR scheme was used for each reaction: 2 min at 95 °C (1 min at 94 °C, 1 min at 55 °C, 1 min at 68 °C) × 35, 10 min at 68 °C, and then 4 °C thereafter. All PCR and RT-PCR reactions were performed twice, and products were sequenced directly using nested primers.
Fractionation—Cells were transfected with non-targeting siRNA control or SMARTpool siRNA targeting PHLPP2 and incubated for 72 h at 37 °C. Cells were lysed in 200 μl of hypotonic buffer (50 mm Na2HPO4, 2 mm EDTA, 2 mm EGTA, 1 mm sodium pyrophosphate, 20 mm NaF, 1 mm DTT, 200 μm benzamidine, 40 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride), passed through a 25-gauge needle 12 times, and lysates were centrifuged at 300 × g for 1 min. The supernatant (containing the cytosolic and membrane fractions) was removed for further fractionation (below). The pellet (containing the nuclear fraction) was resuspended in 200 μl of membrane buffer (50 mm Na2HPO4, 1% Triton X-100, 2 mm EDTA, 2 mm EGTA, 1 mm sodium pyrophosphate, 20 mm NaF, 1 mm DTT, 200 μm benzamidine, 40 μg/ml leupeptin, and 1 mm phenylmethylsulfonyl fluoride), and centrifuged at 16,000 × g for 15 min at 4 °C. The resulting supernatant is defined as the nuclear fraction. The supernatant containing cytosolic and membrane fractions was centrifuged at 108,920 × g for 20 min at 4 °C. The resulting supernatant is defined as the cytosolic fraction. The pellet was resuspended in 200 μl of membrane buffer and centrifuged at 108,920 × g for 20 min at 4 °C; the resulting supernatant was the membrane fraction.
RESULTS
Identification of a Polymorphism in PHLPP2—To determine if the PHLPP2 phosphatase is mutated in breast cancer, we sequenced RNA from breast cancer cell lines. We discovered a T->C nucleotide change at position 3047 in the open reading frame, which results in an amino acid change from Leu to a Ser at codon 1016 in the PP2C phosphatase domain. We observed that 4 of 18 (22%) breast cancer cell lines possessed only the Ser allele, one breast cancer cell line possessed both the Ser and Leu alleles, and the majority of breast cancer cell lines possessed only the Leu allele (Fig. 1, A and B). To determine if this nucleotide change was a polymorphism or a somatic mutation, we genotyped 100 DNA samples from a control population from the Coriell Institute (NA17201-17300). We observed the Leu/Leu genotype in 70 individuals and the Leu/Ser genotype in 30 individuals (Fig. 1C). We never observed the Ser/Ser genotype in the control population. Additionally, we genotyped 33 breast tumor tissue samples and observed the Leu/Leu genotype in 25 patients and the Leu/Ser genotype in 8 patients; again we did not observe the Ser/Ser genotype (Fig. 1C). These results revealed that the T->C nucleotide change at position 3047 is a polymorphism present in 30% of the population.
The absence of the Ser/Ser genotype in the control population, but its presence in breast cancer cell lines, led us to ask whether this polymorphism could play a role in breast tumorigenesis. One of the most common genetic lesions in breast cancer is LOH on chromosome 16q in the chromosomal location harboring PHLPP2 (25). To determine if the presence of only the Ser allele in breast cancer cell lines results from LOH, we genotyped the normal breast cell line (Hs578Bst) that is from the same patient as the breast cancer cell line Hs578t. The genotype of the normal cell line was heterozygous (Leu/Ser); therefore, LOH explains the presence of only the Ser allele in the Hs578t breast cancer cell line (Fig. 1D). We cannot determine if LOH is the mechanism underlying the presence of only the Ser allele in the remaining breast cancer cell lines because no normal breast cell lines from the same patient are available for these cell lines. However, the Ser/Ser genotype is rare and LOH at 16q (in the region including PHLPP2) has been reported for these cell lines suggesting that the patients from whom these cell lines were derived were heterozygous (27).
We next asked whether Akt phosphorylation was altered in a tumor cell line expressing only the Ser allele compared with its normal cell counterpart with both alleles. Because the T47D, Hs578t, and ZR-75-1 breast cancer cell lines, which possess only the Ser allele, have hyperphosphorylated Akt despite expressing wild-type PTEN and having inactivated ErbB2 receptors (28-30), we hypothesized that the Ser polymorphism could impair the function of PHLPP2, thus providing a genetic mechanism driving constitutive phosphorylation of Akt in these cells. Consistent with this hypothesis, comparison of the Hs578t tumor cell line with its normal counterpart (Hs578Bst) revealed that the tumor cell line had dramatically higher levels (>45-fold) of Ser-473 phosphorylation of Akt (Fig. 1D).
Characterization of the L1016S Substitution—To determine if Ser at codon 1016 alters phosphatase activity, we substituted a Ser for a Leu in the full-length HA-PHLPP2 and expressed the Leu or Ser variants in H157 cells (possess only Leu Allele). We immunoprecipitated both variants and measured the in vitro dephosphorylation of fully phosphorylated Akt. The Ser variant of PHLPP2 dephosphorylated Ser-473 on Akt at an ∼5-fold slower rate than the Leu variant (Fig. 2A). Note that the phosphorylation of Thr-308 was relatively resistant to dephosphorylation by PHLPP2, as previously described (18). These data reveal that the amino acid change from a leucine to a serine in the PP2C domain of the phosphatase decreases the activity of PHLPP2 toward Akt.
FIGURE 2.
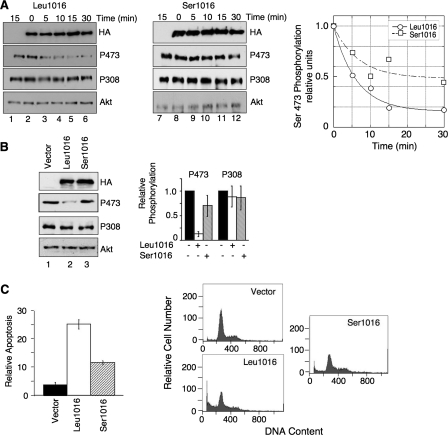
Characterization of nucleotide change in the PHLPP2 phosphatase. A, H157 non-small cell lung cancer (NSCLC) cells were transfected with vector (lanes 1 and 7), HA-Leu-1016 (lanes 2-6), or HA-Ser-1016 (lanes 8-12) under high serum conditions (10% fetal bovine serum Dulbecco's modified Eagle's medium) for 48 h; thereafter HA-PHLPP2 variants were immunoprecipitated and incubated with pure phosphorylated Akt for the indicated times. Western blots are accompanied by graphical analysis of relative Akt Ser-473 phosphorylation normalized to total Akt protein levels. Akt protein and phosphorylation were detected using Akt and phospho-specific antibodies, respectively. Western blots are representative of three independent experiments. B, H157 cells were transfected with vector (lane 1), HA-Leu-1016 (lane 2), or HA-Ser-1016 (lane 3) for 48 h under high serum conditions prior to lysis. The phosphorylation state of Akt in lysates was detected by Western blot analysis. Data from three independent experiments are summarized in the bar graph (relative phosphorylation of Akt at Ser-473 (P473) or Thr-308 (P308) was normalized to total Akt). Error bars indicate standard deviation. C, H157 cells were transfected with vector, HA-Leu-1016, or HA-Ser-1016, under low serum conditions (0.1% fetal bovine serum Dulbecco's modified Eagle's medium) for 48 h and apoptosis (sub-2N DNA content) was assessed using propidium iodide incorporation assays and flow cytometry. All assays were performed in triplicate, with error bars indicating standard deviation, and are representative of three independent experiments.
We next compared the effect of overexpressing the Ser-1016 versus the Leu-1016 variants on the basal phosphorylation of Akt in cells. Expression of the Leu-1016 construct in H157 cells resulted in a 5-fold reduction in basal phosphorylation of Akt at Ser-473 compared with control cells, with no significant effect on Thr-308 phosphorylation (Fig. 2B, lanes 1 and 2), consistent with previous results (18, 19) (levels of overexpression were comparable to those observed previously (18)). In contrast, expression of the Ser-1016 construct at a comparable level resulted in only a modest decrease in phosphorylation on Ser-473 compared with control cells (Fig. 2B, lane 3). To determine if the differential sensitivity of Akt to the two PHLPP2 variants would affect cellular processes that PHLPP2 has been demonstrated to regulate, we expressed both variants in the H157 cells and monitored apoptosis. Fig. 2C shows that expression of the Leu-1016 variant increased apoptosis 5-fold; when expressed at comparable levels (see Fig. 2B), the Ser-1016 only increased apoptosis 2-fold. These results demonstrate that the Ser-1016 variant has decreased activity toward Akt resulting in less effective induction of apoptosis.
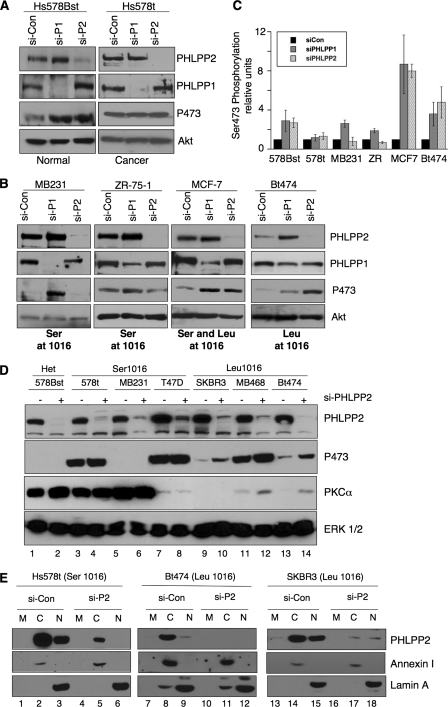
Functional Differences of Endogenous PHLPP2 Variants—To determine if endogenous PHLPP2 variants have altered activity, we compared the effect of knocking down PHLPP2 on Akt phosphorylation in the normal cell line Hs578Bst (expressing both alleles) to the pair-matched tumor cell line Hs578t (expressing only the Ser allele). siRNA validation was previously performed using various siRNAs targeting PHLPP2 to confirm observed increase in Akt phosphorylation was due to specific depletion of PHLPP2 (18). Depletion of PHLPP2 (both variants) from the Hs578Bst cell line resulted in a robust increase in phosphorylation of Akt at Ser-473 (Fig. 3A, lane 2) compared with control treated cells (Fig. 3A, lane 1). In striking contrast, depletion of PHLPP2 (Ser-1016 variant) from the tumor cell line, Hs578t, did not alter Akt phosphorylation. These data suggest that the Ser-1016 variant is less functional compared with the Leu-1016 variant and that only one functional allele is required for Akt regulation (note that Ser-473 phosphorylation was not maximal in the Hs578t cells, because addition of EGF caused an even greater increase in Ser-473 phosphorylation (data not shown; see also Fig. 4A)). Similarly, PHLPP2 depletion in the MCF-7 breast cancer cell line (possessing both alleles) increased Akt phosphorylation at Ser-473 (Fig. 3B). Surprisingly, knockdown of PHLPP1 from the Hs578t breast cancer cell line did not alter Akt phosphorylation, despite the lack of a mutation in PHLPP1 open reading frame. To confirm that the endogenous Ser-1016 variant was less functional than the Leu-1016 variant, we depleted PHLPP2 from two other breast cancer cell lines, MB-231 and ZR-75-1, which only express the Ser allele and monitored Akt phosphorylation. Depletion of the Ser-1016 variant in these breast cancer cell lines did not alter Akt phosphorylation, confirming the impaired activity of this variant compared with the Leu variant (Fig. 3B). Knockdown of PHLPP1 from these breast cancer cell lines resulted in an increase in Akt phosphorylation (Fig. 3B). Depletion of PHLPP2 from the breast cancer cell line Bt-474 (which only expresses the Leu allele) resulted in an increase in Akt phosphorylation (Fig. 3B). Similar results were observed for another cell line that expresses only the Leu allele, the SKBR-3 breast cancer cell line (18). These data reveal that the Ser-1016 variant is ineffective toward suppressing the basal phosphorylation state of Akt.
FIGURE 3.
Evaluating functionality of endogenous Ser-1016. A and B, Hs578Bst, Hs578t, MB231, ZR-75-1, MCF-7, and Bt474 cells were transfected with non-targeting siRNA control (si-Con), PHLPP1 SMARTpool siRNA (si-P1), or PHLPP2 SMARTpool siRNA (si-P2), for 48 h under high serum conditions, and lysates were analyzed by Western blot. The phosphorylation state of Akt at Ser-473, and relative protein levels of Akt, PHLPP1, and PHLPP2 were detected using the indicated antibodies. Western blots are representative of three independent experiments. C, quantification of three independent experiments showing relative phosphorylation of Akt at Ser-473; error bars indicate standard error of the mean. D, the indicated breast cancer cells were transfected with non-targeting siRNA control (si-Con) or SMARTpool siRNA targeting PHLPP2, incubated 48 h under high serum conditions, and then lysates were analyzed by Western blot. The phosphorylation state of Akt at Ser-473, and relative protein levels of Akt, PKCα, ERK1/2, and PHLPP2 were detected using the indicated antibodies. Western blots are representative of three independent experiments. E, fractionation of endogenous PHLPP2 variants into membrane (m), cytoplasmic (c), and nuclear (n) fractions analyzed by Western blot. Protein levels of PHLPP2, annexin I, and lamin A were detected using indicated antibodies. Western blots are representative of three independent experiments.
FIGURE 4.
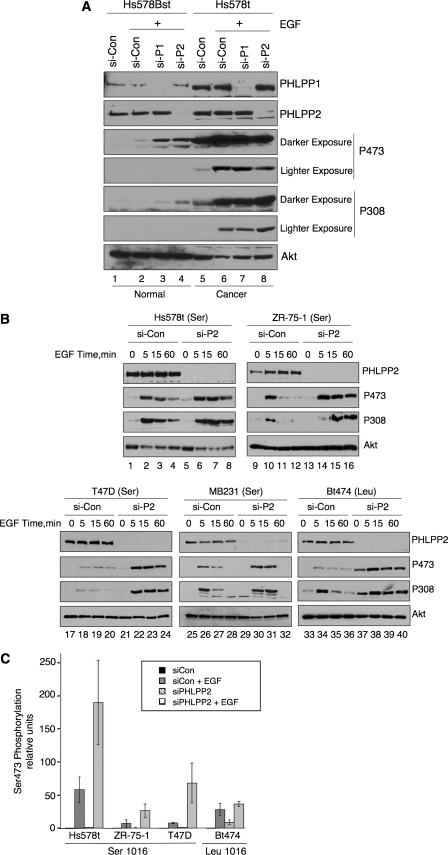
Agonist-induced regulation of Akt by PHLPP2 variants. A, Hs578Bst and Hs578t cells were transfected with non-targeting siRNA control (si-Con) or SMARTpool siRNA targeting PHLPP1 or PHLPP2 under high serum conditions and incubated for 48 h. Media was then changed to low serum conditions overnight prior to addition of EGF (10 ng/ml) for 10 min. The phosphorylation state of Akt at Ser-473 and Thr-308, and protein levels of Akt, PHLPP1, and PHLPP2 in lysates were detected by Western blot analysis. Western blots are representative of three independent experiments. B, indicated breast cancer cell lines were transfected with non-targeting siRNA control (si-Con) or SMARTpool siRNA targeting PHLPP2 under high serum conditions and incubated for 48 h. Media was then changed to low serum conditions overnight prior to addition of EGF (10 ng/ml) for the indicated times. The phosphorylation state of Akt at Ser-473 and Thr-308 and protein levels of Akt and PHLPP2 in lysates were detected by Western blot analysis. Western blots are representative of three independent experiments. C, quantification of three independent experiments showing relative phosphorylation of Akt at Ser-473 prior to EGF treatment or following 60 min of EGF treatment; error bars indicate standard error of the mean.
To determine if the Ser-1016 variant was also ineffective at regulating PKC, we depleted PHLPP2 in breast cancer cell lines expressing the Ser-1016 variant, the Leu-1016 variant, or both variants and monitored PKC protein levels. We observed an increase in PKCα protein levels following PHLPP2 depletion in breast cancer cell lines expressing the Leu-1016 variant (Fig. 3C, lanes 9-14). In contrast, depletion of PHLPP2 from cells expressing the Ser-1016 variant did not alter PKC protein levels (Fig. 3C, lanes 3-8). The up-regulation of PKC levels in cells expressing the Leu-1016 variant correlated with the increased phosphorylation of Akt (e.g. compare lanes 13 and 14 in the Ser-473 panel and the PKCα panel). Interestingly, basal Akt phosphorylation was not detectable in MB-231 cell line despite these cells expressing only the less functional Ser-1016 variant of PHLPP2; however, these cells had elevated PKCα protein expression, suggesting that the less functional PHLPP2 variant may specifically contribute to the increased expression of PKCα in this cell line.
To determine if the amino acid change from Leu to Ser at position 1016 altered the localization of endogenous PHLPP2, we fractionated breast cancer cells expressing either variant. Both the Leu-1016 and Ser-1016 variants were localized to the nucleus and cytoplasm (Fig. 3D) suggesting that this amino acid change does not play a role in regulating the global cellular localization of PHLPP2.
We previously reported that the PHLPP family of phosphatases regulate both the amplitude and duration of agonist-induced Akt activation (18). Thus, we next asked whether the Ser-1016 variant is less functional at regulating agonist-induced phosphorylation of Akt. We first compared the EGF-triggered phosphorylation of Akt in normal Hs578Bst cell line to that in its pair-matched tumor cell line, Hs578t. The Western blot in Fig. 4A reveals a dramatic increase in Akt phosphorylation in Hs578Bst cells depleted of either PHLPP1 (lane 3) or PHLPP2 (lane 4) compared with control siRNA-treated cells (lane 2) following treatment with EGF. In contrast, EGF-induced phosphorylation of Akt in Hs578t cells was not noticeably altered following knockdown of PHLPP2 (Ser-1016 variant) (compare lanes 6 and 8). Additionally, knockdown of PHLPP1 did not affect Akt phosphorylation. One possibility is that Akt is maximally phosphorylated, so phosphatase depletion has no effect on Akt phosphorylation. To address this, we measured the phosphorylation of Akt over a time course of EGF stimulation. The Western blot in Fig. 4B reveals that depletion of PHLPP2 significantly prolonged the duration of phosphorylation (compare 60-min time points, lanes 4 and 8). We then tested the effect of PHLPP2 knockdown on agonist-induced activation of Akt in the remaining breast cancer cell lines expressing only the Ser-1016 variant. The Western blots in Fig. 4B reveal that depletion of PHLPP2 in the T47D, MB231, or ZR-75-1 cells resulted in a dramatic increase in both the amplitude and duration of Akt phosphorylation. In fact, the magnitude of the knockdown effects were on the same order of magnitude as that observed in the normal breast cell line Hs578Bst (Fig. 4A) or Bt474 breast cancer cell line expressing only the Leu allele (Fig. 4B). These data reveal that the Ser-1016 variant is less functional at controlling Akt under basal or serum starvation conditions in all breast cancer cells, but it effectively suppresses agonist-evoked phosphorylation of Akt.
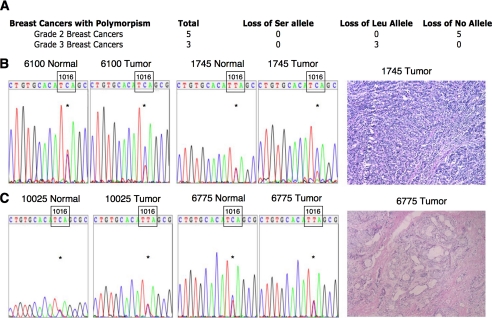
Loss of the Leu Allele from High Grade Breast Cancers—To determine if the Leu allele is lost in breast cancer tumor tissue compared with normal tissue from the same patient, similar to what we observed in the Hs578Bst-Hs578t paired cell lines, we genotyped pair-matched normal and tumor tissue samples from the same patient. Of the 33 patients we genotyped, 8 possessed the polymorphism; of these 8, 5 were grade 2 ductal breast carcinomas and 3 were grade 3 ductal breast carcinomas (Fig. 5A). Surprisingly, all three grade 3 breast cancer samples exhibited loss of the Leu allele (Fig. 5, A and B). The detection of the Leu allele is likely due to the presence of normal cells in the tumor tissue. We did not observe loss of either allele in the five grade 2 breast cancer samples (Fig. 5, A and C). These data suggest that preferential loss of the Leu allele in high grade breast cancers may contribute to the aggressive phenotype of these cancers by decreasing the basal phosphatase activity of PHLPP2, resulting in an increase in Akt phosphorylation, PKC protein levels, or both.
FIGURE 5.
Assessment of PHLPP2 variants in normal and tumor tissue from the same patient. A, table indicating genotypes of breast tumors that were heterozygous for both PHLPP2 alleles. All high grade breast cancer samples displayed LOH of the Leu-1016 variant of PHLPP2. B, chromatograms of two high grade tumors and pair-matched control tissue showing presence of T and C at position 3047 in control tissue but only C at position 3047 in tumor tissue, resulting from loss of the Leu allele in breast cancer samples (left panel). Histology displaying representative high grade breast tumor samples (right panel). C, representative chromatograms of two low grade breast tumors showing presence of both C and T at position 3047 resulting from retention of both PHLPP2 alleles (left panel). Histology of a representative low grade breast cancer (right panel).
DISCUSSION
We have identified a non-synonymous polymorphism in the PHLPP2 phosphatase that results in substitution of a Ser for a Leu at position 1016 in the PP2C domain. Biochemical analysis reveals that the less common variant, Ser-1016, has impaired phosphatase activity toward the substrate Akt in vitro. Similarly, overexpression studies reveal that the Ser-1016 variant is less effective at dephosphorylating Akt in cells, and thus less effective at inducing apoptosis compared with the Leu-1016 variant. Consistent with impaired biological function, genetic depletion of PHLPP2 in unstimulated (but not EGF-stimulated) cells expressing only the Ser-1016 allele does not significantly affect either basal Akt phosphorylation or total PKC levels, whereas depletion of PHLPP2 in cells expressing the Leu-1016 allele results in an increase in Akt phosphorylation and total PKC levels. We observe specific loss of the Leu-1016 variant from a breast cancer cell line and high-grade breast cancer tumor tissue samples when compared with controls suggesting loss of the more functional variant may contribute to tumor progression.
Identification of a Non-synonymous Polymorphism in the PHLPP2 Phosphatase—PHLPP2 is poised to be a potential tumor suppressor based on its chromosomal location and its regulation of downstream substrates, Akt and PKC, which are known oncogenes. The putative tumor suppressor genes on chromosome 16q remain elusive for ductal carcinomas. There is debate over the exact location of the smallest region of overlap that contains the putative tumor suppressor genes in ductal carcinomas, but loss of 16q is a well documented genetic event that occurs in both high grade and low grade ductal carcinomas (18). Additionally, a fragile site (FRA16B) is located within in the PHLPP2 gene, further implicating PHLPP2 as a potential tumor suppressor (NCBI Mitelman Breakpoint Map) (31). Collectively, these data prompted us to sequence breast cancer cell lines for somatic mutations and led to the discovery of the L1016S polymorphism. In our initial studies, we observed only the Ser allele in four breast cancer cell lines and confirmed that LOH is the mechanism responsible for presence of only the Ser allele in one breast cancer cell line (Hs578t); LOH may also account for the genotype of the remaining three breast cancer cell lines (27). Additionally, we observed loss of the Leu allele in high grade tumor tissue samples. These data suggest that the Ser-1016 variant plays a role in breast tumorigenesis and prompted us to investigate the functional consequences of this amino acid change.
The Ser-1016 Variant Is Less Functional than the Leu-1016 Variant—Recent studies analyzing non-synonymous polymorphic variants demonstrate that polymorphisms can play a role in the development of cancer. Characterization of the endogenous Ser-1016 variant demonstrated that it has an impaired ability to suppress basal Akt phosphorylation in all breast cancer cell lines examined. Cell lines with this variant also had elevated PKC levels, consistent with increased stability because of impaired dephosphorylation. Note that, under these basal conditions, overexpression or genetic depletion of the Leu variant of PHLPP2 selectively affects Ser-473 phosphorylation and not Thr-308 phosphorylation, consistent with direct dephosphorylation of Ser-473 by PHLPP2. In cells expressing the less active Ser-1016 variant, knockdown of PHLPP2 has little effect on Ser-473 phosphorylation, consistent with this variant no longer effectively dephosphorylating Ser-473.
Surprisingly, the Ser variant is still quite functional toward Akt under agonist-induced conditions. Under these conditions, the knockdown of the Leu variant of PHLPP2 affects both Ser-473 and Thr-308. The coupling of the phosphorylation of these two sites suggests that, under agonist-stimulation, PHLPP2 negatively regulates other signaling molecules involved in the activation of this pathway. It is quite possible that regulation of other upstream signaling molecules may not rely on phosphatase activity; therefore, either variant will regulate these molecules to a similar degree. We did observe that the Ser-1016 is less functional under agonist-induced conditions in the Hs578t breast cancer cell line, but we hypothesize that another mechanism may account for this loss of function, because PHLPP1 is also less functional in these cells, despite lack of any somatic mutation. The observation that the Ser-1016 variant still retains some biological activity suggests that additional loss of the more functional allele will only result in a modest increase in Akt phosphorylation and PKC levels and will contribute to the deregulation of these pathways in breast cancer cells. However, even a modest increase in activity and protein levels in cells with the Ser variant could contribute to the overall proliferative and metastatic potential of breast cancer cells and could explain why we observe loss of the more functional allele in breast cancer cell lines as well as tumor tissue samples. This increased activity of Akt could contribute to increased cell proliferation by increasing activity of Akt3, which has been demonstrated to play a role in breast cancer (32, 33), and lead to increased phosphorylation of p27 (18). We have previously defined this as a unique axis of signaling involving PHLPP2, Akt3, and p27 (18, 22). Depletion of PHLPP2 results in increased p27 phosphorylation, and this increase can be rescued by depletion of Akt3 in normal breast cells (18, 22). Therefore, it is not unreasonable to hypothesize that loss of a more functional PHLPP2 may increase Akt3 activity, resulting in increased p27 phosphorylation, causing cytosolic sequestration of p27 and increased proliferation. Interestingly, p27 has been demonstrated to be localized to the cytoplasm as a result of Akt phosphorylation in women with breast cancer and this correlates with poor survival rates (34, 35).
Loss of specific regions of chromosome 16q has been associated with lower grade, non-metastatic breast cancers that do not recur, but these regions do not include the PHLPP2 gene (25). Interestingly, a region that includes the PHLPP2 gene was identified to play a role in metastatic breast cancer (1). These data suggest that PHLPP2 may not be involved in the early stages of tumorigenesis but, rather, play a role in metastasis.
Acknowledgments
We thank Drs. Tony Hunter, Jack Dixon, Steve Dowdy, and Tony Wynshaw-Boris for helpful discussions. We thank Drs. Maya Kunkel, Emma Sierecki, and Tianyan Gao for helpful comments and technical assistance.
This work was supported, in whole or in part, by National Institutes of Health Grant GM067946 (to A. C. N.). This work was also supported by Department of Defense Breast Cancer Research Program Predoctoral Grant BC043239 (to J. B.), awarded and administered by the U.S. Army Medical Research Acquisition Activity, Fort Detrick, MD.
Footnotes
Recent Progress and Future Opportunities in Cancer Research, American Association of Cancer Research website.
The abbreviations used are: PTEN, phosphatase and tensin homolog on chromosome ten; PH, pleckstrin homology; PHLPP, PH domain leucinerich repeat protein phosphatase; PKC, protein kinase C; LOH, loss of heterozygosity; siRNA, small interference RNA; EGF, epidermal growth factor; HA, hemagglutinin; DTT, dithiothreitol; RT, reverse transcription.
References
- 1.Driouch, K., Dorion-Bonnet, F., Briffod, M., Champeme, M. H., Longy, M., and Lidereau, R. (1997) Genes Chromosomes Cancer 19 185-191 [DOI] [PubMed] [Google Scholar]
- 2.Blume-Jensen, P., and Hunter, T. (2001) Nature 411 355-365 [DOI] [PubMed] [Google Scholar]
- 3.Liu, W., Bagaitkar, J., and Watabe, K. (2007) Front. Biosci. 12 4011-4019 [DOI] [PubMed] [Google Scholar]
- 4.Slamon, D. J., Godolphin, W., Jones, L. A., Holt, J. A., Wong, S. G., Keith, D. E., Levin, W. J., Stuart, S. G., Udove, J., Ullrich, A., et al. (1989) Science 244 707-712 [DOI] [PubMed] [Google Scholar]
- 5.Siegel, P. M., Dankort, D. L., Hardy, W. R., and Muller, W. J. (1994) Mol. Cell Biol. 14 7068-7077 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sun, M., Paciga, J. E., Feldman, R. I., Yuan, Z., Coppola, D., Lu, Y. Y., Shelley, S. A., Nicosia, S. V., and Cheng, J. Q. (2001) Cancer Res. 61 5985-5991 [PubMed] [Google Scholar]
- 7.Samuels, Y., Diaz, L. A., Jr., Schmidt-Kittler, O., Cummins, J. M., Delong, L., Cheong, I., Rago, C., Huso, D. L., Lengauer, C., Kinzler, K. W., Vogelstein, B., and Velculescu, V. E. (2005) Cancer Cell 7 561-573 [DOI] [PubMed] [Google Scholar]
- 8.Carpten, J. D., Faber, A. L., Horn, C., Donoho, G. P., Briggs, S. L., Robbins, C. M., Hostetter, G., Boguslawski, S., Moses, T. Y., Savage, S., Uhlik, M., Lin, A., Du, J., Qian, Y. W., Zeckner, D. J., Tucker-Kellogg, G., Touchman, J., Patel, K., Mousses, S., Bittner, M., Schevitz, R., Lai, M. H., Blanchard, K. L., and Thomas, J. E. (2007) Nature 448 439-444 [DOI] [PubMed] [Google Scholar]
- 9.Li, J., Yen, C., Liaw, D., Podsypanina, K., Bose, S., Wang, S. I., Puc, J., Miliaresis, C., Rodgers, L., McCombie, R., Bigner, S. H., Giovanella, B. C., Ittmann, M., Tycko, B., Hibshoosh, H., Wigler, M. H., and Parsons, R. (1997) Science 275 1943-1947 [DOI] [PubMed] [Google Scholar]
- 10.Datta, S. R., Brunet, A., and Greenberg, M. E. (1999) Genes Dev. 13 2905-2927 [DOI] [PubMed] [Google Scholar]
- 11.Cantley, L. C. (2002) Science 296 1655-1657 [DOI] [PubMed] [Google Scholar]
- 12.Calleja, V., Alcor, D., Laguerre, M., Park, J., Vojnovic, B., Hemmings, B. A., Downward, J., Parker, P. J., and Larijani, B. (2007) PLoS Biol. 5 e95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarbassov, D. D., Guertin, D. A., Ali, S. M., and Sabatini, D. M. (2005) Science 307 1098-1101 [DOI] [PubMed] [Google Scholar]
- 14.Frias, M. A., Thoreen, C. C., Jaffe, J. D., Schroder, W., Sculley, T., Carr, S. A., and Sabatini, D. M. (2006) Curr. Biol. 16 1865-1870 [DOI] [PubMed] [Google Scholar]
- 15.Guertin, D. A., Stevens, D. M., Thoreen, C. C., Burds, A. A., Kalaany, N. Y., Moffat, J., Brown, M., Fitzgerald, K. J., and Sabatini, D. M. (2006) Dev. Cell 11 859-871 [DOI] [PubMed] [Google Scholar]
- 16.Jacinto, E., Facchinetti, V., Liu, D., Soto, N., Wei, S., Jung, S. Y., Huang, Q., Qin, J., and Su, B. (2006) Cell 127 125-137 [DOI] [PubMed] [Google Scholar]
- 17.Maehama, T., and Dixon, J. E. (1998) J. Biol. Chem. 273 13375-13378 [DOI] [PubMed] [Google Scholar]
- 18.Brognard, J., Sierecki, E., Gao, T., and Newton, A. C. (2007) Mol. Cell 25 917-931 [DOI] [PubMed] [Google Scholar]
- 19.Gao, T., Furnari, F., and Newton, A. C. (2005) Mol. Cell 18 13-24 [DOI] [PubMed] [Google Scholar]
- 20.Andjelkovic, M., Jakubowicz, T., Cron, P., Ming, X. F., Han, J. W., and Hemmings, B. A. (1996) Proc. Natl. Acad. Sci. U. S. A. 93 5699-5704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brognard, J., and Newton, A. C. (2008) Trends Endocrinol. Metab. 19 223-230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mendoza, M. C., and Blenis, J. (2007) Mol. Cell 25 798-800 [DOI] [PubMed] [Google Scholar]
- 23.Gao, T., Brognard, J., and Newton, A. C. (2007) J. Biol. Chem. 283 6300-6311 [DOI] [PubMed] [Google Scholar]
- 24.Mackay, H. J., and Twelves, C. J. (2007) Nat. Rev. Cancer 7 554-562 [DOI] [PubMed] [Google Scholar]
- 25.Rakha, E. A., Green, A. R., Powe, D. G., Roylance, R., and Ellis, I. O. (2006) Genes Chromosomes Cancer 45 527-535 [DOI] [PubMed] [Google Scholar]
- 26.Brognard, J., Clark, A. S., Ni, Y., and Dennis, P. A. (2001) Cancer Res. 61 3986-3997 [PubMed] [Google Scholar]
- 27.Callen, D. F., Crawford, J., Derwas, C., Cleton-Jansen, A. M., Cornelisse, C. J., and Baker, E. (2002) Cancer Genet. Cytogenet. 133 76-82 [DOI] [PubMed] [Google Scholar]
- 28.Nicholson, K. M., Streuli, C. H., and Anderson, N. G. (2003) Breast Cancer Res. Treat. 81 117-128 [DOI] [PubMed] [Google Scholar]
- 29.Perren, A., Weng, L. P., Boag, A. H., Ziebold, U., Thakore, K., Dahia, P. L., Komminoth, P., Lees, J. A., Mulligan, L. M., Mutter, G. L., and Eng, C. (1999) Am. J. Pathol. 155 1253-1260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Konecny, G. E., Pegram, M. D., Venkatesan, N., Finn, R., Yang, G., Rahmeh, M., Untch, M., Rusnak, D. W., Spehar, G., Mullin, R. J., Keith, B. R., Gilmer, T. M., Berger, M., Podratz, K. C., and Slamon, D. J. (2006) Cancer Res. 66 1630-1639 [DOI] [PubMed] [Google Scholar]
- 31.Freudenreich, C. H. (2007) Front. Biosci. 12 4911-4924 [DOI] [PubMed] [Google Scholar]
- 32.Faridi, J., Wang, L., Endemann, G., and Roth, R. A. (2003) Clin. Cancer Res. 9 2933-2939 [PubMed] [Google Scholar]
- 33.Nakatani, K., Thompson, D. A., Barthel, A., Sakaue, H., Liu, W., Weigel, R. J., and Roth, R. A. (1999) J. Biol. Chem. 274 21528-21532 [DOI] [PubMed] [Google Scholar]
- 34.Liang, J., Zubovitz, J., Petrocelli, T., Kotchetkov, R., Connor, M. K., Han, K., Lee, J. H., Ciarallo, S., Catzavelos, C., Beniston, R., Franssen, E., and Slingerland, J. M. (2002) Nat. Med. 8 1153-1160 [DOI] [PubMed] [Google Scholar]
- 35.Viglietto, G., Motti, M. L., Bruni, P., Melillo, R. M., D'Alessio, A., Califano, D., Vinci, F., Chiappetta, G., Tsichlis, P., Bellacosa, A., Fusco, A., and Santoro, M. (2002) Nat. Med. 8 1136-1144 [DOI] [PubMed] [Google Scholar]