Abstract
The 26 S proteasome is a large proteolytic machine, which degrades most intracellular proteins. We found that thioredoxin, Txnl1/TRP32, binds to Rpn11, a subunit of the regulatory complex of the human 26 S proteasome. Txnl1 is abundant, metabolically stable, and widely expressed and is present in the cytoplasm and nucleus. Txnl1 has thioredoxin activity with a redox potential of about-250 mV. Mutant Txnl1 with one active site cysteine replaced by serine formed disulfide bonds to eEF1A1, a substrate-recruiting factor of the 26 S proteasome. eEF1A1 is therefore a likely physiological substrate. In response to knockdown of Txnl1, ubiquitin-protein conjugates were moderately stabilized. Hence, Txnl1 is the first example of a direct connection between protein reduction and proteolysis, two major intracellular protein quality control mechanisms.
Degradation of proteins in eukaryotic cells plays a pivotal role in the regulation of several important processes, including cell division, antigen presentation, and signal transduction (1). Most intracellular proteins are degraded by the 26 S proteasome, a 2.5-MDa protease complex composed of more than 30 different subunits (2).
To become degraded, proteins are typically first conjugated to a chain of ubiquitin moieties. This reaction is catalyzed by ubiquitin ligases. The ubiquitin chains lend the proteins affinity for the 26 S proteasome (3). For efficient degradation, certain ubiquitylated proteins are shuttled to the 26 S proteasome by substrate recruiting factors, such as Rad23, Dsk2, and eEF1A (4, 5).
The 26 S proteasome is composed of two stable subcomplexes, the proteolytically active 20 S core and 19 S regulatory complexes, which bind to one or both ends of the cylindrical 20 S core particle (6). The regulatory complexes first recognize the ubiquitylated substrates (3), before the substrates are deubiquitylated (7, 8), unfolded (9, 10), and translocated into the 20 S particle for degradation.
Although the 26 S proteasome has been known for more than 20 years (11), novel subunits and cofactors have been described recently (12, 13). Here we report another novel proteasome-associated protein, Txnl1 (thioredoxin-like protein 1), that associates directly with the proteasome subunit Rpn11. Txnl1 exhibits thioredoxin activity and targets eEF1A1 in vivo. Previous reports have shown that eEF1A1 transfers misfolded nascent proteins from the ribosome to the 26 S proteasome for degradation (5, 14, 15). Accordingly, ubiquitin-protein conjugates were stabilized upon knockdown of Txnl1 expression. Txnl1 therefore directly links protein reduction and proteolysis, two major intracellular protein quality control mechanisms.
EXPERIMENTAL PROCEDURES
Buffers—The buffers were as follows: buffer A, 25 mm Tris/HCl, pH 7.6, 5 mm MgCl2, 5 mm ATP, 50 mm NaCl, 2 mm KCl, 2 mm DTT,2 10% glycerol; buffer B, 50 mm Tris/HCl, pH 8.0, 100 mm NaCl, 2 mm EDTA, 10% glycerol, 1 mm phenylmethylsulfonyl fluoride, and Complete EDTA-free protease inhibitor tablets (Roche Applied Science); buffer C, 50 mm Tris/HCl, pH 8.0, and 200 mm NaCl; buffer D, 100 mm NaH2PO4, 10 mm Tris/HCl, and 8 m urea, pH 8.0; PBS, 133 mm NaCl, 2.7 mm KCl, 6.5 mm Na2HPO4, 1.5 mm KH2PO4, pH 7.4.
Plasmids and Proteins—Full-length cDNA encoding human Txnl1 (Dr. Giannis Spyrou) was transferred to the appropriate Gateway destination vectors (Invitrogen). PCR mutagenesis was performed using QuikChange (Stratagene). The expression constructs for human Rpn11 and Csn5 were provided by Dr. Wolfgang Dubiel. The proteins were expressed in BL21*(DE3) (Invitrogen) and purified by standard methods.
Cell Culture—MelJuSo cells, stably transfected to express CD3δ-yellow fluorescent protein (Dr. Nico P. Dantuma), HeLa cells, and HEK293 cells were maintained at 37 °C in Dulbecco's modified Eagle's minimal essential medium supplemented with 10% newborn calf serum (Invitrogen) in a humidified atmosphere with 5% CO2.
Electrophoresis and Blotting—Proteins were separated on 12.5% acrylamide gels with SDS or in nondenaturing 5% acrylamide gels (16). Proteins were transferred to BA83 (Schleicher & Schuell) nitrocellulose membranes and probed with antibodies as indicated. For quantitative blotting, the blots were developed using alkaline phosphatase-coupled secondary antibodies, and the color intensities were determined using UnScanIt version 6.1 (Silk Scientific).
Antibodies—His6-tagged Txnl1 protein was used for generation of monoclonal antibodies essentially as described (17). Monoclonal antibodies to proteasome subunits were from BIOMOL. A monoclonal antibody of the same isotype but reacting with human α2-macroglobulin (18) was used as a negative control. Antibodies and their sources were as follows: anti-human Trx1 (Dr. Arne Holmgren), anti-His antibodies (Qiagen), anti-GST and anti-β-actin (Sigma), anti-eEF1A1 (Abcam), and secondary antibodies and antibody specific for ubiquitin (Dako).
Assays—Cell protein was determined (19) with bovine serum albumin as a standard. Concentrations of purified recombinant proteins were determined from A280. The thioredoxin activity of Txnl1 was determined as described (20).
Reverse Transcription-PCR—For analyses of the Txnl1 expression, HeLa cells were treated with or without 0.1 mm H2O2, 10 mm DTT, or 10 μm MG132 for 4 h, before mRNA was isolated using TRIzol (Invitrogen) and Turbo DNA-free (Ambion). The RNA was reverse transcribed using Transcriptor first strand cDNA synthesis (Roche Applied Science), purified using GFX PCR DNA (GE Healthcare), and used for reverse transcription-PCR. Actin was used as a negative control. The primers specific for Txnl1 were as follows: forward, CCGTGGTCAAGTTCACCATGAG; reverse, GGTTCACTTCTTTCTGCCTCTTC.
Determination of Txnl1 Redox Potential—Txnl1 was purified from HeLa cells on a MonoQ column (GE Healthcare) and a column of Sephacryl S300 (GE Healthcare). Fractions were assayed by dot blotting. Aliquots of 0.1 μg of Txnl1 were incubated in 100 μl of redox buffer in 100 mm potassium phosphate, pH 7.0, 1 mm EDTA, 10 mm redox buffer. Redox potentials of buffers were calculated according to the Nernst equation, E = E0 + RT/nF × ln([GSSG]/[GSH]2) for glutathione buffers and E = E0 + RT/nF × ln([LA]/[DHLA]) for buffers with lipoic acid (LA)/dihydrolipoic acid (DHLA). E0 was taken to be -0.240 V for GSH/GSSG (21) and -0.290 V for DHLA/LA (22). Fully reduced and oxidized Txnl1 were produced by substituting the redox buffers by 25 mm DTT or 50 mm N,N,N′,N′-tetramethylazodicarboxamide (diamide), respectively. After overnight incubation at 4 °C in a helium atmosphere, the redox states of Txnl1 were determined by a gel shift assay after modification with 4-acetamido-4-maleimidylstilbene-2,2-disulfonic acid (AMS) (23). Briefly, Txnl1 was precipitated by the addition of 250 μl of 10% (w/v) trichloroacetic acid, and 40 μl of 0.15% deoxycholic acid. After 10 min, precipitates were collected by centrifugation at 13,000 × g for 5 min and washed in 3 × 1 ml of ice-cold acetone. Final pellets were dissolved into 20 μl of 80 mm Tris/HCl, pH 6.8, 2% SDS, 25 mm AMS and incubated for 1 h at room temperature. Samples were separated by nonreducing SDS-PAGE and analyzed by immunoblotting.
Determination of the in Vivo Redox State—HeLa cells (1.4 × 106 cells/5-cm dish) were treated with or without 10 mm DTT or 10 mm diamide for 5 min at 37 °C. The medium was removed, and the cells were washed rapidly with 20 mm N-ethylmaleimide (NEM) in ice-cold PBS and incubated for 20 min with 20 mm NEM in PBS on ice. NEM was removed, and the cells were scraped into 150 μl of 80 mm Tris/HCl, pH 6.8, 0.2 mm phenylmethylsulfonyl fluoride. After brief sonication, SDS was added to 1%. Protein was reduced for 15 min at room temperature with 10 mm Tris(2-carboxyethyl) phosphine, treated with 12 mm AMS at room temperature for 1 h and analyzed by nonreducing SDS-PAGE and immunoblotting, using antibodies specific for Txnl1.
Transfection—Interfering RNAs toward Txnl1 were purchased from Qiagen, Txnl1-1 (SI00302064) and Txnl1-2 (SI03068394). Control small interfering RNAs (siRNAs) were siCONTROL siRNA#1 RNA from Dharmacon. Attached and exponentially growing MelJuSo cells were washed in PBS and transfected with 10 nm siRNA using Lipofectamine RNAiMAX (Invitrogen). The cultures were used 5 days after transfection.
Immunoprecipitation of 26 S Proteasomes for Mass Spectrometry—26 S proteasomes were isolated from HeLa cells by immunoprecipitation and analyzed by two-dimensional gel electrophoresis and mass spectrometry as described (13).
Txnl1-Proteasome Co-precipitation—HeLa cells were homogenized by sonication in 4 volumes of buffer A. After centrifugation (11,300 × g, 30 min), aliquots of 250 ml of supernatant were incubated with 20 μl of Ni2+-NTA-agarose beads (Qiagen) loaded with 5 μg of purified His6-Txnl1 or, as a control, His6-dihydroxyfolate reductase (DHFR). For GST precipitation experiments, purified GST-tagged Txnl1, truncations or GST alone, was loaded on glutathione S-Sepharose (GE Healthcare) beads and incubated with 3 μg of purified 26 S proteasomes. After incubation at 4 °C for 4 h with gentle agitation, the beads were washed in 6 × 1 ml of buffer A. Bound proteins were analyzed by SDS-PAGE and immunoblotting.
Differential Centrifugation—Differential centrifugation of HeLa lysates was performed as described previously (13).
Txnl1 Stability—The stability of Txnl1 in HeLa cells was assessed by following its degradation in cycloheximide-treated cultures, essentially as described (13).
Txnl1 Substrate Capture—For capturing Txnl1 substrates in vitro, His6-tagged Txnl1C34S, His6-tagged Txnl1C37S, or bovine serum albumin (Sigma), were conjugated to CNBr-activated Sepharose (GE Healthcare) to a concentration of about 4 mg/ml gel. HeLa cells (∼5 g) were harvested and lysed by sonication in 4 volumes of buffer B. The extracts were cleared by centrifugation (13,000 × g, 30 min). The supernatant was tumbled at room temperature for 2 h with 40 μl of the Sepharose beads and washed three times in buffer C, followed by three washes in buffer D, before a final wash in buffer C. Bound proteins were eluted for 30 min at room temperature with 10 mm DTT in buffer C and resolved by SDS-PAGE before the proteins were identified by mass spectrometry.
For detecting Txnl1 substrates in vivo, HEK293 cells were stably transfected to express RGSHis6-tagged Txnl1C37S. About 3 g of these cells and mock-transfected control cells were treated with NEM as described above and lysed by sonication in 4 volumes of buffer B. The extracts were cleared by centrifugation (13,000 × g, 30 min). The supernatant was tumbled with 40 μl of Sepharose beads (GE Healthcare) containing immobilized anti-RGSHis6 antibodies (Qiagen) at 4 °C for 4 h. The beads were washed three times in buffer B. Bound protein was eluted by the addition of buffer D and centrifugation. The supernatant was tumbled for 4 h at room temperature with 40 μl of Ni2+-NTA-agarose beads (Qiagen) and washed extensively with buffer D and then in buffer B. Protein was eluted in SDS-PAGE sample buffer with or without 2-mercaptoethanol. The samples were separated by SDS-PAGE and analyzed by immunoblotting.
Protein Identification by Mass Spectrometry—The protein was identified using trypsin digestion, as described previously (24). In short, the relevant bands were excised from the gel. The proteins were reduced, alkylated using iodoacet-amide, and digested by trypsin. The resulting fragments were extracted, purified using C18 ZipTip (Millipore), and measured by matrix-assisted laser desorption ionization time-of-flight mass spectrometry on an AutoFlex II instrument equipped with TOF-TOF facility (Bruker). Mass spectra of the tryptic fragments were used for searching against data bases. To confirm identifications, sequence information was obtained for several fragments, again by data base searching.
Pulse-Chase Experiments—Pulse-chase experiments were performed as described previously (25, 26).
Fluorescence Microscopy—The localization of green fluorescent protein-tagged Txnl1 was determined in HeLa cells transiently transfected with pcDNA-DEST53-Txnl1, as described (26).
RESULTS
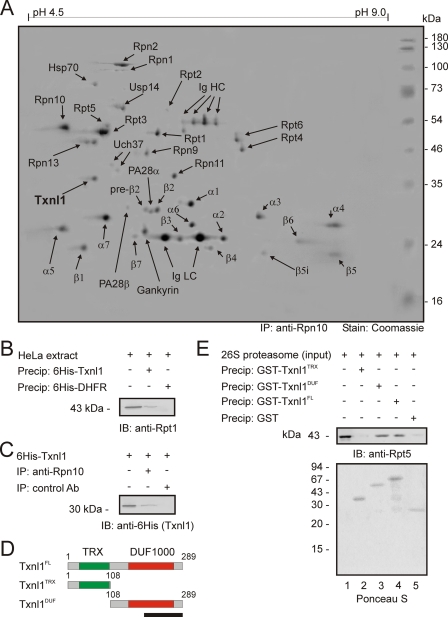
Txnl1 Associates with 26 S Proteasomes—26 S proteasomes from HeLa cells were analyzed by two-dimensional PAGE and mass spectrometry. Most known proteasome subunits were detected (Fig. 1A), except Rpn3, Rpn5, Rpn7, and Rpn12, which were not resolved, perhaps because of poor solubility of these proteins (13). We also confirmed the proteasome association of several known cofactors, including PA28 (27), Hsp70 (28), gankyrin (29), and Usp14/Ubp6 (28, 30). In addition to these proteins, Txnl1, a thioredoxin-like protein implicated in endocytosis (31) and reactions to glucose deprivation (32), was consistently found in the proteasome preparations (Fig. 1A) but not in precipitations with a control antibody (Fig. S1).
FIGURE 1.
Txnl1 associates with 26 S proteasomes. A, proteasomes were precipitated from HeLa cells with an antibody to subunit Rpn10 and analyzed by two-dimensional PAGE. Proteins were stained by Coomassie Brilliant Blue and identified by mass spectrometry. The heavy and light chain immunoglobulins of the antibody used for precipitation are visible (Ig HC and Ig LC). Txnl1 (boldface type), which was identified with 49% sequence coverage, is a novel proteasome component. B, 26 S proteasomes from HeLa cell extracts (lane 1) were precipitated using His6-tagged Txnl1 (lane 2) or His6-tagged DHFR as a control (lane 3). The precipitated material was separated by SDS-PAGE and immunoblotting. Proteasomes were detected with an antibody to subunit Rpt1. C, purified His6-tagged Txnl1 (lane 1) was added in excess to HeLa cell extracts before precipitation (IP) with an antibody to the subunit Rpn10 (lane 2) or with a nonspecific control antibody of the same isotype (lane 3). Precipitated material was separated by SDS-PAGE and immunoblotted (IB) using an antibody to the His6 tag. D, domain organization of full-length Txnl1 and the N- and C-terminal truncations utilized in the binding studies. The N terminus of Txnl1 contains a TRX domain (green), whereas the C-terminal part contains a DUF1000 domain (red). The black bar indicates the length of 100 amino acids. E, purified 26 S proteasomes (lane 1) were precipitated with the indicated GST fusion proteins. As a control, GST alone was used. Precipitated material was separated by SDS-PAGE and immunoblotted using an antibody to the subunit Rpt5. Equal loading was checked by Ponceau S staining.
The position of Txnl1 in two-dimensional gels corresponded with its predicted molecular mass of 32.2 kDa and isoelectric point of 4.8. Previous studies have shown that 26 S proteasomes are able to remodel scrambled RNase A (33), which could indicate that proteasomes were associated with a thioredoxin function. We sought to confirm the association of proteasomes and Txnl1 using a different approach. His6-tagged Txnl1 and, as a control, His6-DHFR were bound to Ni2+-NTA-agarose beads. When incubated with HeLa lysates, His6-Txnl1 precipitated 26 S proteasomes, whereas His6-DHFR did not (Fig. 1B). Conversely, if 26 S proteasomes were precipitated with antibodies to Rpn10, added His6-Txnl1 was co-precipitated (Fig. 1C).
Some cofactors, including Rad23, associate with proteasomes in an ATP-dependent manner (28). We found that the Txnl1-proteasome interaction was independent of ATP (Fig. S1). However, binding was salt-sensitive, since it was totally abrogated in buffers containing 500 mm NaCl (Fig. S1). The interaction between Txnl1 and the 26 S proteasome was not sensitive to the redox potential of the buffer (Fig. S2).
Txnl1 has an N-terminal thioredoxin (TRX) domain and a C-terminal domain of unknown function 1000 (DUF1000) (Fig. 1D). Precipitation experiments showed (Fig. 1E) that the DUF1000 domain of Txnl1 was necessary and sufficient to mediate the interaction with the 26 S proteasome.
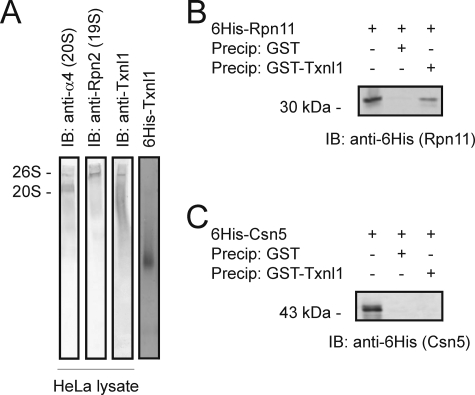
Txnl1 Is a Component of the 19 S Regulatory Complex—The 26 S proteasome is composed of two stable subparticles, the 20 S cylinder and the 19 S regulatory complex. Nondenaturing electrophoresis and immunoblotting of HeLa cell lysates confirmed (34) that HeLa cells contain a significant fraction of free 20 S proteasomes (Fig. 2A). Txnl1 co-migrated with 26 S proteasomes but not with 20 S proteasomes (Fig. 2A), indicating that Txnl1 associates with a component of the 19 S regulatory complex. The smear of Txnl1, seen below the band corresponding to 26 S proteasomes, is probably caused by continuous release of Txnl1 from 26 S proteasomes during electrophoresis. This is consistent with the lability of the interaction between Txnl1 and 26 S proteasomes. His6-tagged and wild type Txnl1 have pI values of 5.3 and 4.8, respectively, and should therefore migrate at almost the same rate in nondenaturing electrophoresis (pH 8.3). Free His6-tagged Txnl1 migrated much faster than the 26 S proteasome (Fig. 2A), and no free Txnl1 was observed in the lysate. Therefore, most or all of the Txnl1 must initially have been associated with proteasomes.
FIGURE 2.
Txnl1 is associated with the 19 S regulatory complex. A, HeLa cell extracts were separated by nondenaturing PAGE and analyzed by immunoblotting (IB) using antibodies against a 20 S subunit (α4), a 19 S subunit (Rpn2), and Txnl1. As a control, a purified sample of His6-Txnl1 was also subjected to nondenaturing PAGE and Coomassie-stained. His6-tagged Rpn11 (B) or His6-tagged Csn5 (C)(lane 1) was precipitated with glutathione-Sepharose beads loaded with GST (lane 2) and GST-Txnl1 (lane 3). Precipitated material was separated by SDS-PAGE and analyzed by immunoblotting with an antibody to the His6 tag.
Txnl1 Is a Nearly Stoichiometric Component of the 26 S Proteasome—In accordance with the results from the nondenaturing electrophoresis (above), at least 85% of the Txnl1 in HeLa cell extracts was co-precipitated with 26 S proteasomes, as determined by quantitative immunoblotting (Fig. S3). We cannot ascertain if the remaining 15% of the total cellular Txnl1 was released from proteasomes during the experiment or if it is found in the free form also in the intact cells.
The amounts of the 26 S proteasome subunit Rpt5 and Txnl1 in HeLa cells were determined by quantitative immunoblotting of dilution series of total cell protein and His6-tagged Rpt5 and Txnl1 as standards. The cells were dissolved in 2% SDS to allow determination of the protein concentration before the buffer was reconstituted to normal SDS sample buffer. Care was taken to use data only from regions where the curves, describing the correlation between color development and dilution of the cell extracts, were parallel with the standard curves (data not shown). The results revealed that HeLa cells contain around 220 μg of Txnl1 and 230 μg of Rpt5/g of cell protein. Txnl1 and Rpt5 are therefore abundant proteins present in roughly equimolar amounts.
Txnl1 Binds Directly to Rpn11—To determine direct interactions between Txnl1 and proteasome subunits, GST and GST-tagged Txnl1 were incubated with several His6-tagged subunits of the 19 S regulatory complex expressed in Escherichia coli and then precipitated. Only subunit Rpn11 bound to Txnl1 (Fig. 2B). Structurally, Rpn11 somewhat resembles the Csn5 subunit of the COP9 signalosome. However, we did not observe any interaction between Txnl1 and Csn5 (Fig. 2C).
Txnl1 Is a Stable Protein—The stability of Txnl1 was analyzed by immunoblotting of HeLa cells, treated with cycloheximide to stop protein synthesis. The level of Txnl1 did not significantly decrease after 8 h (Fig. S4), indicating that Txnl1 is a relatively stable protein.
Txnl1 Expression—Like most other proteasome components, Txnl1 was expressed at roughly similar levels in all tested tissues (Fig. 3A).
FIGURE 3.
Txnl1 is a widely expressed cytoplasmic and nuclear protein. A, mouse tissues were analyzed by SDS-PAGE and blotting (IB) with anti-Txnl1 antibodies. Txnl1 was found in all tested tissues at about equal abundance. B, a HeLa cell homogenate was separated by centrifugation into pellet (P) and supernatant (S). The pellet was then resuspended in the same volume as the supernatant before analysis of the fractions by SDS-PAGE and blotting using antibodies specific for Txnl1. C, samples of spent HeLa media (extracellular (E)) and HeLa cells (intracellular (I)) were separated by SDS-PAGE and analyzed by blotting with antibodies specific for thioredoxin (anti-Trx1), the proteasome (anti-Rpn2), and Txnl1 (anti-Txnl1). D, confocal micrographs of HeLa cells transiently transfected to express green fluorescent protein-tagged Txnl1 (green). The cells were stained with phalloidin (red) and 4′, 6-diamidino-2-phenylindole (DAPI)(blue) to detect actin and the nucleus, respectively.
Components of the ubiquitin-proteasome system are up-regulated at the mRNA level in response to proteasome inhibition (35, 36).
Neither oxidative nor reductive stress significantly altered the Txnl1 mRNA level as determined by reverse transcription-PCR. However, proteasome inhibition significantly induced Txnl1 mRNA (Fig. S5).
Txnl1 Is a Soluble Cytoplasmic and Nuclear Protein—26 S proteasomes and other components of the ubiquitin system are mostly soluble proteins localized in the cytoplasm and nucleus (37). Similarly, upon centrifugation of HeLa cell extracts, Txnl1 was found only in the supernatant (Fig. 3B).
Another thioredoxin, Trx1, is secreted via an endoplasmic reticulum-Golgi-independent pathway (38). However, although we could confirm the secretion of Trx1 into the growth medium, Txnl1, like the proteasome, was exclusively present in the interior of HeLa cells (Fig. 3C).
Our antibodies were not suitable for immunolocalization studies, but green fluorescent protein-tagged Txnl1 was present in HeLa cells throughout the cytoplasm and nucleus (Fig. 3D). The homologous Trx1 translocates to the nucleus in response to protein kinase C stimulation (39). However, no change in the subcellular localization of Txnl1 was observed in the presence of 12-O-tetradecanoylphorbol-13-acetate and bisindolylmaleimide (data not shown).
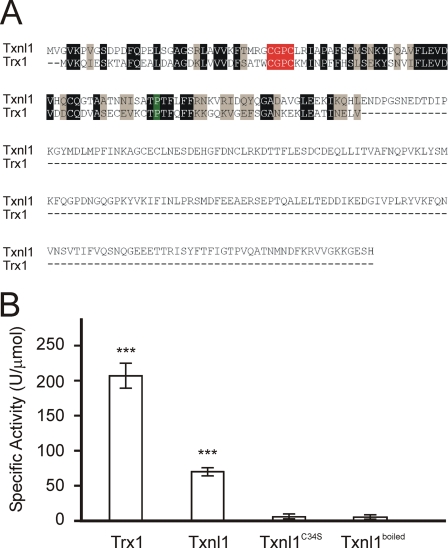
Txnl1 Displays Thioredoxin Activity—Txnl1 contains a thioredoxin domain, which is 41% identical to that of human Trx1. Both the active site CGPC motif and the proline loop are identical (Fig. 4A). With insulin as substrate (20), the specific activity of Txnl1 was about 30% of that of E. coli thioredoxin (Fig. 4B). The activity was completely lost when the first active site cysteine was altered to a serine (C34S) (Fig. 4B).
FIGURE 4.
Txnl1 is an active thioredoxin. A, Txnl1 and Trx1 were aligned using T-Coffee version 1.41. Identical and homologous residues have been shaded. The active site CGPC motif is shown in red, and the conserved proline is shown in green. B, specific activities of Txnl1, the Txnl1C34S mutant, and a sample of Txnl1 that had been boiled were determined. The E. coli thioredoxin (Trx) was included for comparison. ***, p < 0.01 (t test).
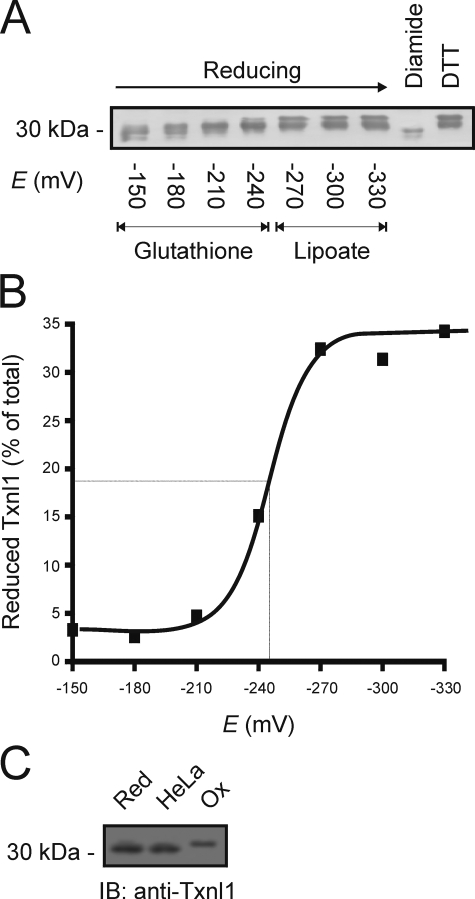
Redox Properties of Txnl1—The redox potential of Txnl1 and the redox state of Txnl1 in the cell might provide us with clues about the function of Txnl1 in vivo. Purified Txnl1 from HeLa cells was incubated with redox buffers and then allowed to react with the thiol-specific reagent AMS, which adds 450 Da for each free cysteine in the protein (23). AMS-modified (reduced) and unmodified (oxidized) Txnl1 were separated by SDS-PAGE and detected by immunoblotting. Curiously, even with DTT, we were never able to completely reduce the purified Txnl1 (Fig. 5A), indicating that during purification, a fraction of Txnl1 underwent irreversible oxidation. This fraction was ignored in the following calculation. To get a rough estimate of the Txnl1 redox potential, the ratios of reduced to oxidized Txnl1 were plotted against the E0 of the buffers (Fig. 5B). The curve follows the Nernst equation and shows that Txnl1 has a redox potential of about -250 mV, comparable with the redox potential of -270 mV for thioredoxin (40).
FIGURE 5.
Redox properties of Txnl1. A, purified Txnl1 was incubated with glutathione and dihydrolipoate redox buffers, as indicated, or with the oxidizing agent diamide or the reducing agent DTT. Reduced cysteine residues were then modified with AMS, and the proteins were resolved by SDS-PAGE and blotting (IB) using antibodies specific for Txnl1. B, the fractions of reduced Txnl1, determined from A, were plotted versus the redox potentials of the buffers. At about -250 mV, half of Txnl1 was reduced. C, HeLa cells were preincubated with NEM and lysed. The extracts were reduced with DTT and treated with AMS before being resolved by SDS-PAGE and immunoblot analysis. Lysates from cells pretreated with DTT or diamide were used as markers for the completely reduced and oxidized protein, respectively.
Next, we wanted to establish the redox state of Txnl1 in vivo. To prevent active site cysteines from engaging in thiol-disulfide exchange reactions during or after cell lysis, HeLa cells were incubated with the cell-permeable alkylating agent NEM. Subsequently, lysates were treated with DTT under denaturing conditions to reduce disulfide bonds (23). The resulting cysteines were then detected by AMS treatment and SDS-PAGE, followed by immunoblotting, as before. Control samples were generated by incubating cells under reducing conditions, using 10 mm DTT, or under oxidizing conditions with 10 mm diamide, prior to treatment with NEM. Txnl1 was found to be fully reduced in untreated HeLa cells (Fig. 5C).
eEF1A1 Is a Substrate for Txnl1 in Vivo—As part of the catalytic mechanism, reduced thioredoxin and oxidized target proteins form mixed disulfide intermediates. Subsequently, thioredoxin itself forms an intramolecular disulfide bridge, releasing the reduced target. The intermediate mixed disulfide state is stabilized when the thioredoxin lacks the second cysteine in the active site CXXC motif. Such mutant thioredoxin can therefore trap prospective substrates (41, 42), which will be released upon reduction of the disulfide bond (Fig. 6A).
FIGURE 6.
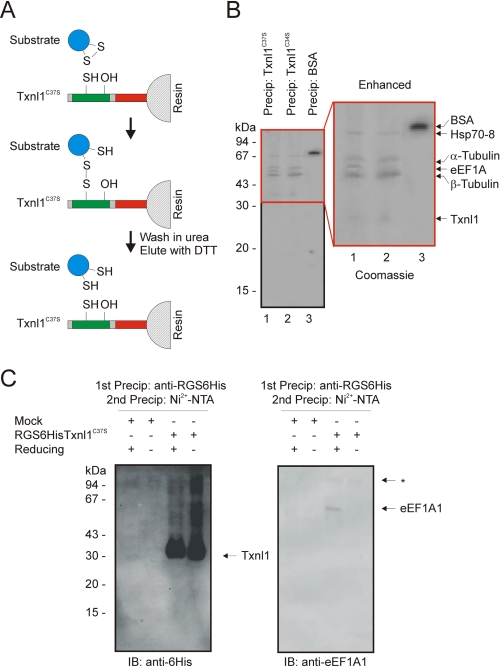
Isolation of eEF1A1 as a Txnl1 substrate. A, potential substrates for Txnl1 were isolated. Single point mutants in the active site CXXC motif are prone to form a disulfide bond with the substrate, which can subsequently be eluted by reduction of the disulfide bond with DTT. The beads were washed in urea to ensure that only covalently bound proteins were isolated. B, the isolated proteins were resolved by SDS-PAGE and stained with Coomassie. Proteins were identified by mass spectrometry as indicated. C, HEK293 cells mock-transfected or transfected to express RGSHis6-tagged Txnl1C37S were used in denaturing precipitation experiments to isolate in vivo Txnl1 substrates. Left, in the absence of reducing agent, Txnl1 formed a slow migrating smear, probably due to formation of mixed disulfides with various cellular substrates. Right, in the presence of reducing agent eEF1A1 was released. *, an unknown contaminant. IB, immunoblot.
Txnl1 active site point mutant proteins were covalently coupled to Sepharose beads and incubated with HeLa cell lysates. After incubation, the beads were washed extensively in a buffer containing 8 m urea to ensure that only covalently bound proteins were isolated. Then the bound substrates were released with DTT and resolved by SDS-PAGE. Mass spectrometry of the excised bands identified the proteins, as indicated (Fig. 6B). With the Txnl1C34S and Txnl1C37S mutant thioredoxins, a pattern of four or five bands was visible on the gel. In a control experiment with bovine serum albumin coupled to Sepharose beads, only bovine serum albumin was released from the resin (Fig. 6B). Although thioredoxins preferably form mixed disulfides with the first cysteine of the active site CXXC motif, some may utilize either cysteine for the initial reaction at the active site (43). All of the isolated proteins may therefore be substrates of Txnl1. Nonetheless, of the isolated proteins, eEF1A1 was mainly associated with the Txnl1C37S mutant (Fig. 6B). To analyze if eEF1A1 is also associated with a Txnl1C37S mutant in vivo, HEK293 cells were stably transfected to express RGSHis6-tagged Txnl1C37S. The cells were treated with NEM to block free cysteines before cell lysis. Then the RGSHis6-Txnl1C37S was isolated by first a native immunoprecipitation using anti-RGSHis6 antibodies. The precipitate was dissolved in 8 m urea, and the RGSHis6-tagged Txnl1 was again precipitated, this time with Ni2+-NTA-agarose beads. Immunoblotting showed that the Txnl1C37S formed mixed disulfides with various cell proteins in vivo (Fig. 6C, left), which disappeared when β-mercaptoethanol was added to the sample prior to electrophoresis. Release of eEF1A1 from Txnl1C37S in the presence of reducing agent could also be shown by immunoblotting (Fig. 6C, right), confirming that eEF1A1 is a substrate for Txnl1 also in vivo.
Effect of Knockdown of Txnl1 Expression—eEF1A1 plays a role in protein degradation (14) by targeting misfolded nascent proteins to the 26 S proteasome in collaboration with Rad23 (5, 15). Proteolysis might therefore be impaired in cells with a decreased content of Txnl1. To check this prediction, Txnl1 expression in MelJuSo cells was knocked down with siRNA (Fig. 7). The amounts of Rpn2, a subunit of the regulatory complex of 26 S proteasomes, and of Trx1 (Fig. 7) were the same as in the control cultures. Thus, cells do not compensate for the lack of Txnl1 by inducing proteasomes or Trx1.
FIGURE 7.
Knockdown of Txnl1 expression. HeLa cells were transfected with two different siRNAs specific for Txnl1 and with a random nonspecific siRNA. After 5 days, the cells were dissolved in 1% SDS and analyzed by SDS-PAGE and blotting (IB). The amount of Txnl1 was much reduced, whereas the level of 26 S proteasomes, detected with an antibody to Rpn2, was not affected. The level of Trx1 was also not altered. β-Actin served as a loading control. The level of ubiquitin-protein conjugates was higher in cells transfected with Txnl1 siRNA.
In order to test if protein degradation was affected, we performed pulse-chase analyses on cells transfected with Txnl1-specific siRNA or with control siRNA. However, no significant changes were observed in the degradation of bulk protein (Fig. S6). The degradation kinetics of model proteasome substrates of the secretory pathway, CD3δ-yellow fluorescent protein and α1-antitrypsin (25), were also not significantly affected by Txnl1 knockdown (Figs. S7 and S8), indicating that Txnl1 is important only for the degradation of a subset of cell proteins.
However, in response to Txnl1 knockdown, we did observe a moderate increase in the amount of ubiquitin-protein conjugates (Fig. 7), revealing that the ubiquitin-proteasome system is indeed compromised in cells lacking Txnl1.
DISCUSSION
Txnl1 (thioredoxin-like protein 1) (44), also known as TRP32 (thioredoxin-related protein of 32 kDa) (45), is a relatively uncharacterized but widely expressed thioredoxin (44, 45). We confirmed that Txnl1 is soluble and present in the nucleus and cytoplasm (45), a localization that it shares with Trx1. However, unlike Trx1 (38, 39), Txnl1 is not secreted from cells, and its subcellular localization is not regulated by protein kinase C activity.
Previous studies have revealed that Txnl1 is involved in endocytosis (31) and in protection against glucose deprivation-induced cytotoxicity (32). Such effects are probably indirect for a proteasome component.
In C. elegans, Txnl1 expression is up-regulated by the unfolded protein response (46). We did not observe any change in the Txnl1 mRNA level when unfolded protein response was induced with DTT. However, as shown by both Lundgren et al. (47) and us, Txnl1 is induced by inhibition of proteasomes, like other components of the ubiquitin-proteasome system (35, 36). Previous studies have shown that 26 S proteasomes are able to remodel scrambled RNase A (33), which could indicate that proteasomes were associated with a thioredoxin function.
Txnl1 has thioredoxin activity (45). We found that Txnl1 displays a specific activity of about 30% of that of E. coli thioredoxin and has a reduction potential of about -250 mV. In HeLa cells, Txnl1 is present almost exclusively in a reduced form, like thioredoxin.
Txnl1 is an abundant protein, present in amounts roughly equimolar with subunits of the 26 S proteasome. At least 85% of cellular Txnl1 is proteasome-associated, indicating that Txnl1 must be closely tied to proteolysis. Since Txnl1 associated with preexisting 26 S proteasomes, it may be better defined as a proteasome-interacting protein, rather than an actual subunit.
We show that Txnl1 specifically utilizes the DUF1000 domain to associate with the 26 S proteasome. We therefore suggest that the DUF1000 domain be instead named the proteasome-interacting thioredoxin (PITH) domain. The only other human protein that contains a PITH domain is C1ORF128, which is uncharacterized and does not contain any other known protein domains but has recently been found to associate with 26 S proteasomes (48). It therefore appears that the PITH domain is a general proteasome-interacting module.
Due to its association with the proteasome, one would expect Txnl1 knockdown to result in impaired intracellular protein degradation. Many proteins in the endoplasmic reticulum are stabilized by disulfide bonds. However, in response to Txnl1 siRNA, we did not observe any significant change in the degradation of bulk protein or model substrates of the endoplasmic reticulum-associated degradation pathway.
Recently, the Txnl1 orthologue in fission yeast, Txl1, was characterized. The null mutant did not display any specific responses upon glucose deprivation but was moderately sensitive to alkyl hydroperoxide (49). However, under normal growth conditions, the mutant did not exhibit any obvious phenotypes (49), corroborating the modest phenotype we found upon knockdown of Txnl1 in mammalian tissue culture cells. Curiously, the budding yeast genome does not encode a Txnl1 orthologue, so perhaps the other cytoplasmic thioredoxins, Trx1 and Trx2, maintain Txnl1 function in this organism. Accordingly, by proteomics, both Trx1 and Trx2 were recently found to associate with 26 S proteasomes in budding yeast (50).
Potential substrates for a proteasome-associated thioredoxin encompass the proteasome itself, proteasome substrates, and proteasome cofactors. Oxidation and glutathionylation itself may affect proteasome activity (51, 52). Previous studies have shown that proteasomes are prone to oxidation (53). Especially, subunit Rpt3 (51) and the subunits Rpt3 (54) and Rpn12 (55) have been shown to be targets of thioredoxin.
The experiments with active site mutated Txnl1 showed that α- and β-tubulin, Hsp70, and eEF1A1 formed mixed disulfides with cysteine in the active site. With basically the same technique, Wiseman et al. (56) were unable to capture such covalently bound substrates, perhaps because of an unfavorable ratio of cell extract to mutant Txnl1 or other experimental details.
Certainly α- and β-tubulin are subject to oxidative modification and are targeted by thioredoxins (57). Hsp70-type chaperones are also prone to oxidative inactivation and may be subject to redox regulation (58). However, for these substrates, it is presently not clear why a proteasome-associated thioredoxin such as Txnl1 should target these proteins. Most thioredoxins possess very broad substrate specificities. It is therefore not unexpected that Txnl1 can also have several substrates.
eEF1A1 seems to be a substrate for Txnl1 both in vitro and in vivo and can easily be placed in a physiological context. Besides of its well described function in recruiting codon-specific aminoacyl-tRNAs to the ribosome, eEF1A1 also possesses chaperone-like activity (59, 60) and functions in targeting of newly synthesized, damaged proteins to the proteasome (5, 14, 15), perhaps through its binding to subunits Rpn2 and Rpt4 (61). Previously, eEF1A1 was shown to be prone to oxidative modification and to be a substrate for the Dictyostelium discoideum thioredoxin Ddtrx1 (62). Perhaps during oxidative stress, when nascent protein folding is challenged, Txnl1 functions to ensure that co-translationally damaged proteins are efficiently degraded by protecting eEF1A from oxidative inactivation.
In conclusion, the results presented here agree with and complement those obtained by Wiseman et al. (56). We show that Txnl1 is a novel redox active and nearly stoichiometric component of the 26 S proteasome, which interacts with the 26 S proteasome via its C-terminal PITH domain. Thus, Txnl1 equips 26 S proteasomes with a hitherto unrecognized enzymatic function, namely protein disulfide reduction. However, despite our efforts, the exact molecular mechanism for Txnl1 function remains elusive. Hopefully, future studies can utilize the data presented here for more in depth analyses of Txnl1 function.
Supplementary Material
Acknowledgments
We thank Dr. N. P. Dantuma, Dr. W. Dubiel, Dr. A. Holmgren, and Dr. G. Spyrou for sharing valuable reagents; A. M. B. Lauridsen, K. Dissing, A. Kastrup, and Dr. P. Kristensen for help with experiments; and Dr. J. Riemer, Dr. L. Ellgaard, and Dr. J. R. Winther for helpful discussions.
This work was supported by grants from the Danish Research Academy, Lundbeck Foundation, Novo Nordisk Foundation, and Carlsberg Foundation (to R. H.-P.).
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S8.
Footnotes
The abbreviations used are: DTT, dithiothreitol; PBS, phosphate-buffered saline; LA, lipoic acid; DHLA, dihydrolipoic acid; AMS, 4-acetamido-4-male-imidylstilbene-2,2-disulfonic acid; NEM, N-ethylmaleimide; siRNA, small interfering RNA; NTA, nitrilotriacetic acid; DHFR, dihydroxyfolate reductase; GST, glutathione S-transferase; TRX, thioredoxin; DUF1000, domain of unknown function 1000; PITH, proteasome-interacting thioredoxin.
References
- 1.Glickman, M. H., and Ciechanover, A. (2002) Physiol. Rev. 82 373-428 [DOI] [PubMed] [Google Scholar]
- 2.Hendil, K. B., and Hartmann-Petersen, R. (2004) Curr. Protein Pept. Sci. 5 135-151 [DOI] [PubMed] [Google Scholar]
- 3.Thrower, J. S., Hoffman, L., Rechsteiner, M., and Pickart, C. M. (2000) EMBO J. 19 94-102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hartmann-Petersen, R., Seeger, M., and Gordon, C. (2003) Trends Biochem. Sci. 28 26-31 [DOI] [PubMed] [Google Scholar]
- 5.Chuang, S. M., Chen, L., Lambertson, D., Anand, M., Kinzy, T. G., and Madura, K. (2005) Mol. Cell Biol. 25 403-413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Voges, D., Zwickl, P., and Baumeister, W. (1999) Annu. Rev. Biochem. 68 1015-1068 [DOI] [PubMed] [Google Scholar]
- 7.Yao, T., and Cohen, R. E. (2002) Nature 419 403-407 [DOI] [PubMed] [Google Scholar]
- 8.Ambroggio, X. I., Rees, D. C., and Deshaies, R. J. (2004) PLoS Biol. 2 0113-0119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Braun, B. C., Glickman, M., Kraft, R., Dahlmann, B., Kloetzel, P. M., Finley, D., and Schmidt, M. (1999) Nat. Cell Biol. 1 221-226 [DOI] [PubMed] [Google Scholar]
- 10.Strickland, E., Hakala, K., Thomas, P. J., and DeMartino, G. N. (2000) J. Biol. Chem. 275 5565-5572 [DOI] [PubMed] [Google Scholar]
- 11.Hough, R., Pratt, G., and Rechsteiner, M. (1987) J. Biol. Chem. 262 8303-8313 [PubMed] [Google Scholar]
- 12.Sone, T., Saeki, Y., Toh-e, A., and Yokosawa, H. (2004) J. Biol. Chem. 279 28807-28816 [DOI] [PubMed] [Google Scholar]
- 13.Jørgensen, J. P., Lauridsen, A. M., Kristensen, P., Dissing, K., Johnsen, A. H., Hendil, K. B., and Hartmann-Petersen, R. (2006) J. Mol. Biol. 360 1043-1052 [DOI] [PubMed] [Google Scholar]
- 14.Gonen, H., Smith, C. E., Siegel, N. R., Kahana, C., Merrick, W. C., Chakraburtty, K., Schwartz, A. L., and Ciechanover, A. (1994) Proc. Natl. Acad. Sci. U. S. A. 91 7648-7652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen, L., and Madura, K. (2005) Cancer Res. 65 5599-5606 [DOI] [PubMed] [Google Scholar]
- 16.Hoffman, L., Pratt, G., and Rechsteiner, M. (1992) J. Biol. Chem. 267 22362-22368 [PubMed] [Google Scholar]
- 17.Hendil, K. B. (2005) Methods Enzymol. 398 439-453 [DOI] [PubMed] [Google Scholar]
- 18.Slot, L. A., and Hendil, K. B. (1987) Scand. J. Clin. Lab. Invest. 147 393-397 [DOI] [PubMed] [Google Scholar]
- 19.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265-275 [PubMed] [Google Scholar]
- 20.Holmgren, A. (1979) J. Biol. Chem. 254 9627-9632 [PubMed] [Google Scholar]
- 21.Rost, J., and Rapoport, S. (1964) Nature 201 185. [DOI] [PubMed] [Google Scholar]
- 22.Ke, B. (1957) Biochim. Biophys. Acta 25 650-651 [DOI] [PubMed] [Google Scholar]
- 23.Haugstetter, J., Blicher, T., and Ellgaard, L. (2005) J. Biol. Chem. 280 8371-8380 [DOI] [PubMed] [Google Scholar]
- 24.Lyngholm, J. M., Nielsen, H. V., Holm, M., Schiøtz, P. O., and Johnsen, A. H. (2001) Allergy 56 21-28 [DOI] [PubMed] [Google Scholar]
- 25.Menendez-Benito, V., Verhoef, L. G., Masucci, M. G., and Dantuma, N. P. (2005) Hum. Mol. Genet. 14 2787-2799 [DOI] [PubMed] [Google Scholar]
- 26.Madsen, L., Andersen, K. M., Prag, S., Moos, T., Semple, C. A., Seeger, M., and Hartmann-Petersen, R. (2008) Int. J. Biochem. Cell Biol. 40 2927-2942 [DOI] [PubMed] [Google Scholar]
- 27.Ma, C.-P., Slaughter, C. A., and DeMartino, G. N. (1992) J. Biol. Chem. 267 10515-10523 [PubMed] [Google Scholar]
- 28.Verma, R., Chen, S., Feldman, R., Schieltz, D., Yates, J., Dohmen, J., and Deshaies, R. J. (2000) Mol. Biol. Cell 11 3425-3439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dawson, S., Apcher, S., Mee, M., Higashitsuji, H., Baker, R., Uhle, S., Dubiel, W., Fujita, J., and Mayer, R. J. (2002) J. Biol. Chem. 277 10893-10902 [DOI] [PubMed] [Google Scholar]
- 30.Leggett, D. S., Hanna, J., Borodovsky, A., Crosas, B., Schmidt, M., Baker, R. T., Walz, T., Ploegh, H., and Finley, D. (2002) Mol. Cell 10 495-507 [DOI] [PubMed] [Google Scholar]
- 31.Felberbaum-Corti, M., Morel, E., Cavalli, V., Vilbois, F., and Gruenberg, J. (2007) PLoS ONE 2 e1144, 1-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jiménez, A., Pelto-Huikko, M., Gustafsson, J. A., and Miranda-Vizuete, A. (2006) FEBS Lett. 580 960-967 [DOI] [PubMed] [Google Scholar]
- 33.Liu, C. W., Millen, L., Roman, T. B., Xiong, H., Gilbert, H. F., Noiva, R., DeMartino, G. N., and Thomas, P. J. (2002) J. Biol. Chem. 277 26815-26820 [DOI] [PubMed] [Google Scholar]
- 34.Tanahashi, N., Murakami, Y., Minami, Y., Shimbara, N., Hendil, K. B., and Tanaka, K. (2000) J. Biol. Chem. 275 14336-14345 [DOI] [PubMed] [Google Scholar]
- 35.Xie, Y., and Varshavsky, A. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 3056-3061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Meiners, S., Heyken, D., Weller, A., Ludwig, A., Stangl, K., Kloetzel, P. M., and Krüger, E. (2003) J. Biol. Chem. 278 21517-21525 [DOI] [PubMed] [Google Scholar]
- 37.Brooks, P., Fuertes, G., Murray, R. Z., Bose, S., Knecht, E., Rechsteiner, M. C., Hendil, K. B., Tanaka, K., Dyson, J., and Rivett, J. (2000) Biochem. J. 346 155-161 [PMC free article] [PubMed] [Google Scholar]
- 38.Rubartelli, A., Bajetto, A., Allavena, G., Wollman, E., and Sitia, R. (1992) J. Biol. Chem. 267 24161-24164 [PubMed] [Google Scholar]
- 39.Hirota, K., Matsui, M., Iwata, S., Nishiyama, A., Mori, K., and Yodoi, J. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3633-3638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Krause, G., Lundström, J., Barea, J. L., de la Cuesta, P., and Holmgren, A. (1991) J. Biol. Chem. 266 9494-9500 [PubMed] [Google Scholar]
- 41.Motohashi, K., Kondoh, A., Stumpp, M. T., and Hisabori, T. (2001) Proc. Natl. Acad. Sci. U. S. A. 98 11224-11229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Balmer, Y., Koller, A., del Val, G., Manieri, W., Schürmann, P., and Buchanan, B. B. (2003) Proc. Natl. Acad. Sci. U. S. A. 100 370-375 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kouwen, T. R., Andréll, J., Schrijver, R., Dubois, J. Y., Maher, M. J., Iwata, S., Carpenter, E. P., and van Dijl, J. M. (2008) J. Mol. Biol. 379 520-534 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Miranda-Vizuete, A., Gustafsson, J. A., and Spyrou, G. (1998) Biochem. Biophys. Res. Commun. 243 284-288 [DOI] [PubMed] [Google Scholar]
- 45.Lee, K. K., Murakawa, M., Takahashi, S., Tsubuki, S., Kawashima, S., Sakamaki, K., and Yonehara, S. (1998) J. Biol. Chem. 273 19160-19166 [DOI] [PubMed] [Google Scholar]
- 46.Urano, F., Calfon, M., Yoneda, T., Yun, C., Kiraly, M., Clark, S. G., and Ron, D. (2002) J. Cell Biol. 158 639-646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lundgren, J., Masson, P., Mirzaei, Z., and Young, P. (2005) Mol. Cell Biol. 25 4662-4675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wang, X., and Huang, L. (2008) Mol. Cell Proteomics 7 46-57 [DOI] [PubMed] [Google Scholar]
- 49.Jiménez, A., Mateos, L., Pedrajas, J. R., Miranda-Vizuete, A., and Revuelta, J. L. (2007) Yeast 24 481-490 [DOI] [PubMed] [Google Scholar]
- 50.Guerrero, C., Milenkovíc, T., Przulj, N., Kaiser, P., and Huang, L. (2008) Proc. Natl. Acad. Sci. U. S. A. 105 13333-13338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ishii, T., Sakurai, T., Usami, H., and Uchida, K. (2005) Biochemistry 44 13893-13901 [DOI] [PubMed] [Google Scholar]
- 52.Demasi, M., Silva, G. M., and Netto, L. E. (2003) J. Biol. Chem. 278 679-685 [DOI] [PubMed] [Google Scholar]
- 53.Grune, T., Jung, T., Merker, K., and Davies, K. J. (2004) Int. J. Biochem. Cell Biol. 36 2519-2530 [DOI] [PubMed] [Google Scholar]
- 54.Wong, J. H., Balmer, Y., Cai, N., Tanaka, C. K., Vensel, W. H., Hurkman, W. J., and Buchanan, B. B. (2003) FEBS Lett. 547 151-156 [DOI] [PubMed] [Google Scholar]
- 55.Yamazaki, D., Motohashi, K., Kasama, T., Hara, Y., and Hisabori, T. (2004) Plant Cell Physiol. 45 18-27 [DOI] [PubMed] [Google Scholar]
- 56.Wiseman, R. L., Chin, K.-T., Haynes, C. M., Stanhill, A., Xu, C.-F., Roguev, A., Krogan, N. J., Neubert, T. A., and Ron, D. (2009) J. Biol. Chem. 284 15233-15245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Landino, L. M., Iwig, J. S., Kennett, K. L., and Moynihan, K. L. (2004) Free Radic. Biol. Med. 36 497-506 [DOI] [PubMed] [Google Scholar]
- 58.Vignols, F., Mouaheb, N., Thomas, D., and Meyer, Y. (2003) J. Biol. Chem. 278 4516-4523 [DOI] [PubMed] [Google Scholar]
- 59.Malki, A., Caldas, T., Parmeggiani, A., Kohiyama, M., and Richarme, G. (2002) Biochem. Biophys. Res. Commun. 296 749-754 [DOI] [PubMed] [Google Scholar]
- 60.Lukash, T. O., Turkivska, H. V., Negrutskii, B. S., and El'skaya, A. V. (2004) Int. J. Biochem. Cell Biol. 36 1341-1347 [DOI] [PubMed] [Google Scholar]
- 61.Davy, A., Bello, P., Thierry-Mieg, N., Vaglio, P., Hitti, J., Doucette-Stamm, L., Thierry-Mieg, D., Reboul, J., Boulton, S., Walhout, A. J., Coux, O., and Vidal, M. (2001) EMBO Rep. 2 821-828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brodegger, T., Stockmann, A., Oberstrass, J., Nellen, W., and Follmann, H. (2004) Biol. Chem. 385 1185-1192 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.