Abstract
PsbS plays a major role in activating the photoprotection mechanism known as “non-photochemical quenching,” which dissipates chlorophyll excited states exceeding the capacity for photosynthetic electron transport. PsbS activity is known to be triggered by low lumenal pH. However, the molecular mechanism by which this subunit regulates light harvesting efficiency is still unknown. Here we show that PsbS controls the association/dissociation of a five-subunit membrane complex, composed of two monomeric Lhcb proteins (CP29 and CP24) and the trimeric LHCII-M. Dissociation of this supercomplex is indispensable for the onset of non-photochemical fluorescence quenching in high light, strongly suggesting that protein subunits catalyzing the reaction of heat dissipation are buried into the complex and thus not available for interaction with PsbS. Consistently, we showed that knock-out mutants on two subunits participating to the B4C complex were strongly affected in heat dissipation. Direct observation by electron microscopy and image analysis showed that B4C dissociation leads to the redistribution of PSII within grana membranes. We interpreted these results to mean that the dissociation of B4C makes quenching sites, possibly CP29 and CP24, available for the switch to an energy-quenching conformation. These changes are reversible and do not require protein synthesis/degradation, thus allowing for changes in PSII antenna size and adaptation to rapidly changing environmental conditions.
Photosynthetic reaction centers exploit solar energy to drive electrons from water to NADP+. This transport is coupled to H+ transfer from chloroplast stroma to thylakoids lumen, building a proton gradient for ATP synthesis (1). The capacity for light absorption is increased by pigment-binding proteins that compose the antenna system. In higher plants, the antenna is composed of the nuclear-encoded Chl4 a/b-binding light-harvesting complexes (Lhc). The major constituent of the photosystem II (PSII) outer antenna is LHCII, a heterotrimer composed by different combinations of Lhcb1, Lhcb2, and Lhcb3 gene products (2). Three additional monomeric antenna complexes (CP29, CP26, and CP24) encoded by the lhcb4, lhcb5, and lhcb6 genes, respectively, are localized in between the core complex and LHCII (3). Similarly, PSI has four Lhca antenna proteins, yielding a total of 10 distinct Lhc isoforms (2). Differences between the mentioned isoforms have been largely conserved in all higher plants during at the last 350 million years of evolution, strongly indicating that each pigment-protein complex has a specific function (4), although the specific role of each gene product in light harvesting and/or photoprotection is still under debate (5). Their topological organization into the supercomplex has been analyzed by electron microscopy and biochemical methods (3, 6-8) showing that Lhcb subunits are organized into two layers around the PSII core. The inner layer is composed of CP29, CP26, and the S-type LHCII trimer, forming, together with the PSII core, the so-called C2S2 particle (9, 10). The outer layer is made of LHCII trimers and CP24, to build up the larger C2S2M2Lx complexes (9) in which the number of LHCII-L trimers depends on the light intensity during growth (11, 12).
This structural organization responds to the requirements of light harvesting regulation; in high light, when absorbed energy does not limit growth, the PSII antenna loses the components of the external antenna layer, namely CP24, LHCII-M, and LHCII-L (12), whereas the internal antenna components, CP26, CP29, and LHCII-S, are always retained in a 1:1 stoichiometry with the PSII core complex. This is consistent with the composition of a mutant exhibiting chronic plastoquinone reduction, mimicking overexcitation (10). Such an acclimation to contrasting light conditions, however, requires days to weeks (12, 13), whereas plants are often exposed to rapid changes in light intensity, temperature, and water availability. In these conditions, incomplete photochemical quenching leads to an increased Chl excited state (1Chl*) lifetime and increased probability of Chl a triplet formation (3Chl*) by intersystem crossing. Chl triplets react with oxygen (3O2) and form harmful reactive oxygen species, responsible for photoinhibition and oxidative stress (14). These harmful events are counteracted by photoprotection mechanisms consisting either in the scavenging of generated reactive oxygen species (15) or in prevention of their production through dissipation of the 1Chl* in excess (16, 17). This latter process is known as non-photochemical quenching (NPQ) and is observed as light-dependent quenching of Chl fluorescence. NPQ has been shown to be composed by at least two components with different activation time scales. The first, feedback de-excitation quenching (qE), is rapidly activated upon increasing light intensity, whereas a second component (qI) is slower. Full NPQ activation requires zeaxanthin synthesis (18, 19) and the PsbS protein (20) that senses low lumenal pH through two lumen-exposed protonatable residues (21, 22). Mutants lacking Chl b, and thus lacking Lhc proteins, or exhibiting alterations in the topological organization of PSII antenna also undergo a strong reduction in NPQ, demonstrating the involvement of antenna proteins in its activation (23-25).
In this work we analyzed the changes in the organization of the PSII antenna during exposure to strong light and NPQ development. We found that a supramolecular complex, named B4C, composed of CP29, CP24, and LHCII-M, connects the inner and outer antenna moieties, dissociates during light exposure, and reassociates during subsequent dark recovery. Dissociation of the B4C complex appears necessary for the establishment of non-photochemical fluorescence quenching. Upon illumination, PSII distribution within grana membranes was also affected, and we observed a shorter distance between PSII reaction centers, implying enrichment in C2S2 complexes and depletion in the outer LHCII components. These results suggest that the NPQ process includes a rapid and reversible change in the organization of grana membranes with disconnection of a subset of Lhcb proteins from the PSII reaction center.
EXPERIMENTAL PROCEDURES
Plant Growth, Light Treatment, and Thylakoid Isolation—WT plants of Arabidopsis thaliana ecotype Columbia and mutants npq1, npq2, npq4, koLhcb3, koCP24, and koCP26 were obtained from the Arabidopsis Biological Resource Center; the complemented npq4 mutant with PsbS WT and E122Q, E226Q, and double E122Q/E226Q mutants were obtained as described previously (21); the npq2/npq4 double mutant was obtained by crossing npq2 and npq4 mutants; the koCP29 triple mutant was obtained by crossing the single mutants Lhcb4.1, Lhcb4.2, and Lhcb4.3 obtained from the Arabidopsis Biological Resource Center. WT plants and mutants were grown for 4-6 weeks at 100 μmol of photons m-2 s-1, 21 °C, 90% humidity, and 8 h of daylight. Leaves detached from plants grown for 3 weeks were adapted to 1 h in the dark and eventually treated for 30 min at 1500 μmol of photons m-2 s-1 or for different times as indicated under “Results.” Unstacked thylakoids were isolated from dark-adapted or light-treated leaves as described previously (26). Membranes corresponding to 150 mg of chlorophyll were washed with 5 mm EDTA and then solubilized with 0.6% α-DM. Solubilized samples were then fractionated by ultracentrifugation in a 0.1-1 m sucrose gradient containing 0.03% α-DM and 20 mm HEPES, pH 7.5 (SW60ti rotor, 5 h 30 min at 60,000 rpm, 4 °C).
Grana Membrane Isolation—Grana membranes were isolated from dark- and light-adapted samples using α-DM solubilization of stacked thylakoids as described elsewhere (10).
Quantification of B4C Dissociation—The image of each sucrose gradient fractionation was analyzed by using Gel-Pro Analyzer© software to determine the integrated optical density (IOD) of the different bands. B4C was quantified as the ratio between the IOD of the corresponding band and the sum of all bands corresponding to antenna proteins (band 2, 3, and 4).
Pigment Analyses—Pigments were extracted from leaf discs or grana membranes (see below and Table 1) with 80% acetone (v/v) and then separated and quantified by HPLC (27).
TABLE 1.
Pigment-binding properties of light-treated leaves and isolated grana membranes
The pigment composition of leaves during light treatment was analyzed by HPLC. Values (bold numbers) are reported as normalized to 100 total Chl molecules. S.D. is also indicated.
| Chl a/b | Neoxanthin | Violaxanthin | Anteraxanthin | Lutein | Zeaxanthin | β-Carotene | |
|---|---|---|---|---|---|---|---|
| Grana | |||||||
| WT dark | 2.19 | 4.9 | 3.1 | 0.0 | 12.1 | 0.0 | 6.5 |
| S.D. | 0.04 | 0.4 | 0.2 | 0.3 | 0.4 | ||
| WT light (30 min) | 2.16 | 5.3 | 1.6 | 0.5 | 12.0 | 0.9 | 6.0 |
| S.D. | 0.03 | 0.5 | 0.2 | 0.1 | 0.3 | 0.1 | 0.6 |
| WT light (90 min) | 2.33 | 4.8 | 1.4 | 0.4 | 11.2 | 1.1 | 5.9 |
| S.D. | 0.03 | 0.3 | 0.1 | 0.1 | 0.2 | 0.1 | 0.2 |
| WT recovery (90 min) | 2.42 | 4.8 | 1.9 | 0.8 | 10.9 | 0.7 | 6.2 |
| S.D. | 0.07 | 0.4 | 0.1 | 0.1 | 0.5 | 0.1 | 0.1 |
| N4 dark | 2.37 | 5.2 | 2.4 | 0.0 | 12.0 | 0.0 | 6.6 |
| S.D. | 0.02 | 0.7 | 0.1 | 0.1 | 0.1 | ||
| N4 light (30 min) | 2.31 | 5.7 | 1.5 | 0.3 | 12.2 | 0.6 | 6.1 |
| S.D. | 0.04 | 1.1 | 0.2 | 0.1 | 0.1 | 0.1 | 0.3 |
| N4 light (90 min) | 2.22 | 6.2 | 1.5 | 0.3 | 12.5 | 0.8 | 5.5 |
| S.D. | 0.04 | 1.3 | 0.2 | 0.1 | 0.4 | 0.1 | 0.3 |
| Leaves | |||||||
| WT dark | 3.12 | 4.5 | 4.3 | 0.0 | 12.7 | 0.0 | 7.1 |
| S.D. | 0.26 | 0.4 | 0.3 | 1.1 | 0.1 | ||
| WT light (30 min) | 3.31 | 4.9 | 1.5 | 0.9 | 11.5 | 2.3 | 7.6 |
| S.D. | 0.02 | 0.4 | 0.1 | 0.1 | 0.1 | 0.1 | 0.3 |
| WT light (90 min) | 3.20 | 4.3 | 1.2 | 0.6 | 12.3 | 2.4 | 6.5 |
| S.D. | 0.07 | 0.3 | 0.1 | 0.1 | 1.0 | 0.2 | 1.5 |
| WT recovery (90 min) | 3.20 | 4.3 | 2.9 | 1.4 | 12.5 | 1.1 | 6.7 |
| S.D. | 0.03 | 0.1 | 0.4 | 0.3 | 1.1 | 0.1 | 0.3 |
In Vivo Fluorescence Analysis—Non-photochemical quenching of Chl fluorescence was measured on whole leaves at room temperature with a PAM 101 fluorimeter (Walz, Effeltrich, Germany). Minimum fluorescence (F0) was measured with a 0.15 μmol m-2 s-1 beam, and maximum fluorescence (Fm) was determined with a 0.6-s light pulse (4500 μmol m-2 s-1). White continuous light (100-2000 μmol m-2 s-1) was supplied by a KL1500 halogen lamp (Schott, Mainz, Germany). NPQ was calculated according to the following equation (28): NPQ = (Fm - F′m)/F′m, where Fm is the maximum Chl fluorescence from dark-adapted leaves and F′m the maximum Chl fluorescence under actinic light exposition. For photoinhibition analyses, Fv/Fm was determined after 5 or 15 h of dark incubation following light treatment with equivalent results.
Electron Microscopy and Image Analysis—Electron microscopy (EM) on isolated grana membranes was conducted using an FEI Tecnai T12 electron microscope operating at 100 kV accelerating voltage. Samples were applied to glow-discharged carbon-coated grids and stained with 2% uranyl acetate. Images were recorded using a charge-coupled device camera. Best stained grana patches were analyzed and PSII core positions were identified using Boxer software, manually edited in case of uncertain attributions. Distribution of PSII cores within the image distribution was analyzed by a homemade procedure written in MATLAB© (available upon request), which determines the distance between each core and the closest (or n closest) neighbor. In total around 1000 points, resulting from at least three independent biological replications, were considered for the analysis for each sample.
RESULTS
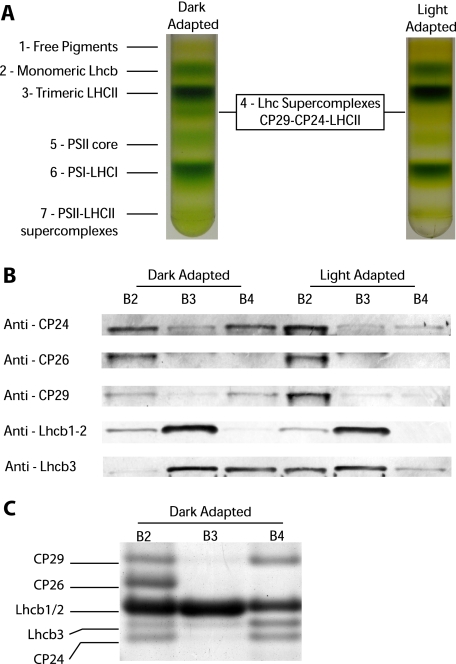
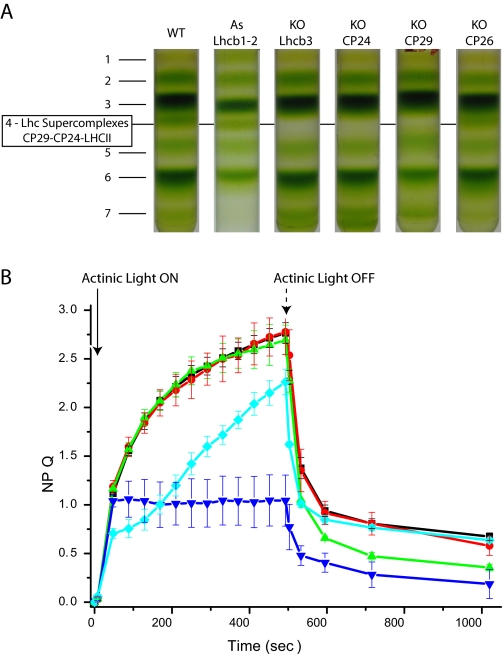
A Pentameric Lhcb Complex (B4C) Is Dissociated upon Light Treatment—PSII antenna organization has been shown to be fundamental for the full establishment of NPQ, which, in fact, is significantly impaired in plants depleted of antenna proteins or where the antenna organization is affected (23-25). To test whether a reorganization of grana membranes is involved in NPQ, we analyzed the interactions between the PSII-LHCII supercomplex subunits using sucrose gradient ultracentrifugation upon mild solubilization of thylakoid membranes. To this aim, Arabidopsis plants were either dark-adapted or illuminated with saturating light for 30 min. Upon illumination, leaves were cooled in ice water slurry, and thylakoids were isolated, solubilized with α-DM, and fractionated by ultracentrifugation. Different pigment-protein complexes migrated as green bands depending on their sizes, as shown in Fig. 1A. Light treatment affected the band pattern, and sucrose band 4 (B4) disappeared. This band is made of a Lhc supercomplex composed of monomeric CP24, CP29, and a LHCII trimer (6). Here-after we will refer to this pentameric Lhc complex as B4C. The distribution of individual Lhcb proteins among different green bands was analyzed in detail by using specific antibodies. As shown in Fig. 1B, a large fraction of the CP24 and CP29 subunits is present in the B4C from the dark-adapted sample, whereas the rest, in monomeric form, is found in fraction 2 (B2). After light treatment, CP24 and CP29 were barely detectable in the gradient fraction corresponding to B4C, whereas they increased in the B2 fraction. These results show that the reduction of B4C in the gradients is indeed due to the dissociation of this oligomeric antenna complex. Among the other polypeptides analyzed, it was very interesting to observe that Lhcb3 is an enriched component of B4C in the dark. Upon light treatment, Lhcb3 was instead found in the monomeric fraction, confirming that this subunit participates in light-dependent dissociation of the B4C supercomplex. Lhcb1-2, the major components of LHCII trimers, are found mostly in B3, as expected, and in the monomeric B2 fraction. A fraction of Lhcb1-2 is also detected in B4C; their relative abundance appears to be rather low. However, this is only because of their high enrichment in B2 and B3 fractions. In fact, by loading equal Chl amounts of B2, B3, and B4 in a Coomassie-stained gel (Fig. 1C), it is clear that Lhcb1-2 are indeed present in significant amounts in B4C, in agreement with to a previous report (6). To further confirm that Lhcb1-2 are genuine B4C components, we observed that their content in B4C was reduced upon light treatment, just as all of the other previously mentioned components of this complex (Fig. 1B). CP26, instead, is not involved in B4C formation, as shown by the fact that it is found in B2, in a monomeric state, irrespective of the treatment.
FIGURE 1.
Light-dependent dissociation of B4C protein complex in A. thaliana. A, sucrose gradient fractionations of mildly solubilized thylakoids membranes purified from dark- and light-adapted (30 min at 1500 μE) leaves. In the dark thylakoid pigment-binding complexes separate into seven distinct bands, the fourth one (B4C) being depleted in light-treated sample. B, distribution of monomeric antenna proteins in sucrose gradients between bands 2, 3, and 4 from dark- and light-adapted samples. Western blotting analysis was carried out using specific antibodies against CP24, CP26, CP29, Lhcb1-2, and Lhcb3, respectively. Samples from different bands were loaded in amounts proportional to their abundance in the sucrose gradient. In Lhcb1-2 blotting, each band was loaded with five times less protein to avoid antibody signal saturation. C, Coomassie-stained SDS-PAGE loading of equal Chl amounts (2 μg) from sucrose gradient bands 2, 3, and 4 from dark-adapted samples. Bands corresponding to CP29, CP26, Lhcb1/2, Lhcb3, and CP24, as identified by Western blotting, are indicated. Only the gel region where antenna polypeptides are migrating is shown.
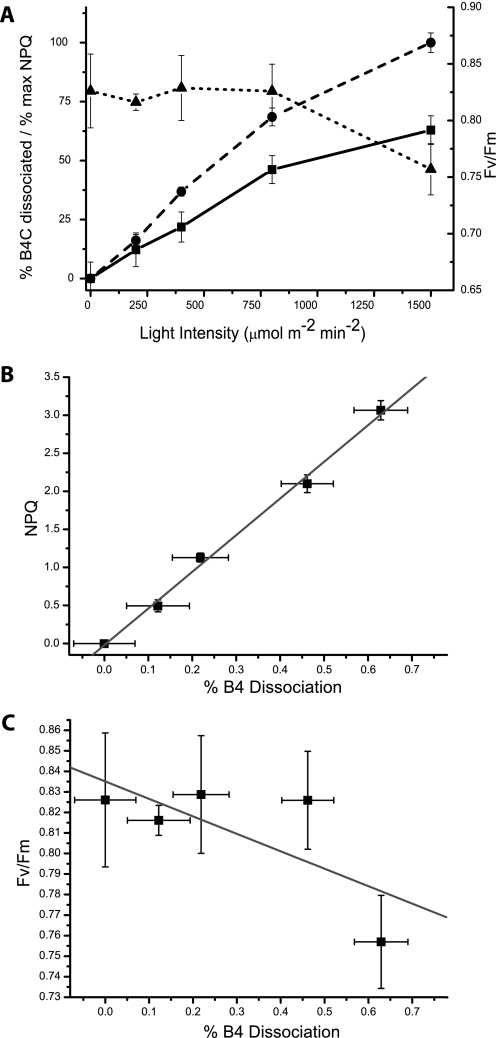
Thereafter we analyzed the dependence of B4C dissociation on light intensities; dissociation increased with the level of illumination, reaching saturation above 1500 μE (Fig. 2A). State transitions are known to involve dissociation of LHCII subunits from PSII reaction centers (29) and cannot be excluded as the cause of B4C complex dissociation. However, state transitions are saturated at low light (200 μE) and are inhibited by high light, in agreement with their role in balancing PSI versus PSII photon trapping under limiting light conditions (30, 31) and thus are unlikely to be correlated with B4C dissociation.
FIGURE 2.
A, comparison of B4C dissociation (solid line, squares), NPQ (dashed line, circles), and PSII yield (Fv/Fm, dotted line, triangles). All parameters were evaluated after 30 min of light treatment at the indicated intensity. B4C is quantified as the ratio of the band with respect to the sum of all bands containing antenna upon separation of pigment-binding complexes in sucrose gradient or nondenaturing gel. NPQ values, in order to have a scale comparable with B4C dissociation, are expressed as a fraction of the maximal value (3.1). Fv/Fm values where measured after 15 min of dark incubation following light treatment to allow for PSII reduction and NPQ relaxation but not recovery from photoinhibition. B, correlation between B4C dissociation and NPQ measured with different light intensities is analyzed in more detail. Line shows the linear fitting of reported points (R-value is 0.997). C, correlation analysis of Fv/Fm and B4C dissociation determined using different illumination intensities. Line shows the best linear fitting of reported points (R-value is 0.71).
Also, we did not observe any increase in phosphorylated CP29 in light-treated samples, consistent with previous results (30). This is an indication that this phenomenon is also unrelated to B4C dissociation.
At least two processes can be active under high light conditions leading to B4C dissociation: one is photoinhibition, meaning light-induced damages in PSII, and the other is non-photochemical quenching. To test whether one of these processes (or both) is involved in B4C dissociation, we measured the recovery of variable fluorescence, a marker of photoinhibition, upon illumination with different intensities (1500 μE; Fig. 2A). It clearly appears that Fv/Fm is significantly affected only with the highest light intensity, whereas B4C dissociation, instead, occurs with weaker levels of illumination, suggesting that the two phenomena are not correlated. Instead, the level of B4C dissociation at different light intensities closely matches the amplitude of NPQ obtained under the same light conditions (Fig. 2A); in fact, both NPQ and B4C show activation with increasing illumination. This correlation is evidenced in Fig. 2B, where it can be appreciated how B4C dissociation and NPQ have a similar dependence on light intensity, different from Fv/Fm (Fig. 2C). We thus tentatively concluded that B4C dissociation correlates with NPQ rather than with photoinhibition.
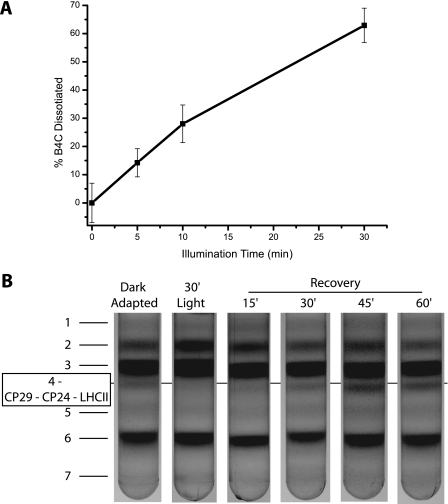
We analyzed the time dependence on light of B4C dissociation; as shown in Fig. 3A, dissociation increases with time. This process is reversible, and following dark incubation, B4C slowly reassociates. Upon a lag phase of 15 min, B4C can be detected again at 30 min, and the dark control level is reached upon 45 min of dark adaptation (Fig. 3B). We also verified that these results were not due to the thylakoid fractionation technique used, by analyzing the kinetics of B4C dissociation by nondenaturing Deriphat-PAGE. We obtained results fully consistent with sucrose gradient analysis (data not shown).
FIGURE 3.
Time dependence of B4C dissociation and reassociation in A. thaliana. A, B4C dissociation was determined from sucrose gradients after different durations of 1500 μE illumination. B4C is quantified as described under “Experimental Procedures.” B, reassociation of B4C complex in samples left in the dark for 15, 30, 45, and 60 min after a 30-min-long illumination at 1500 μE.
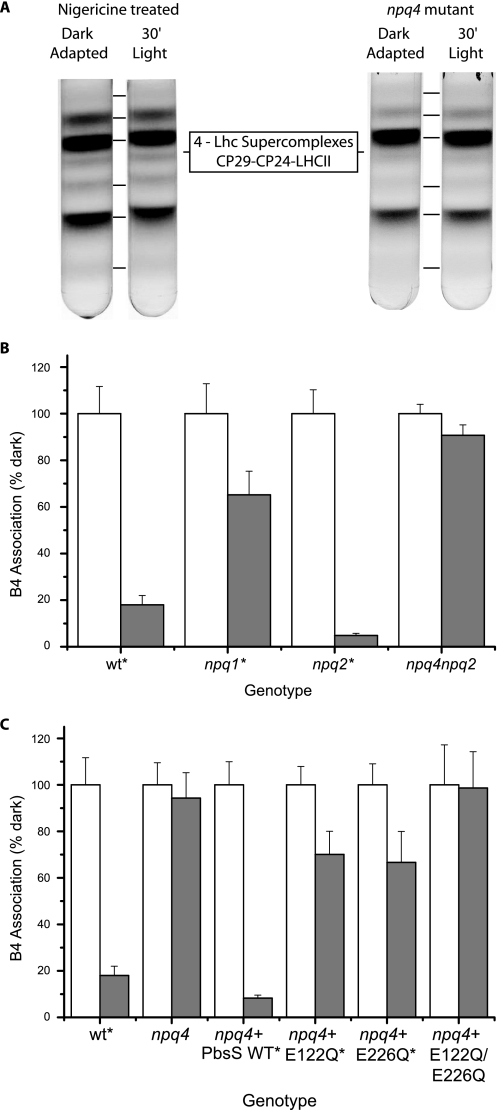
Pharmacologic and Genetic Analysis of B4 Complex Dissociation—To investigate in more detail the correlation between NPQ and B4C dissociation, light treatment was performed on leaves treated with nigericin, a NPQ inhibitor through its uncoupling effect on the trans-membrane pH gradient (Fig. S1A). As shown in Fig. 4A, in nigericin-treated leaves, the B4C complex is stable, implying that its light-dependent dissociation requires the presence of a trans-membrane pH gradient.
FIGURE 4.
Dependence of B4C dissociation on ΔpH, PsbS, and zeaxanthin. A, light dependent B4C dissociation in nigericin-treated leaves (left) and PsbS-depleted mutant (npq4, right); B, quantification of B4C in leaves that are either dark-adapted (white) or treated with 1500 μE for 30 min (gray). WT is compared with mutants affected by zeaxanthin biosynthesis (npq1) or constitutively accumulating zeaxanthin (npq2) and in an npq2/npq4 dark-adapted double mutant in (white). C, quantification of B4C in leaves that are either dark-adapted (white) or treated with 1500 μE for 30 min (gray). WT is compared with npq4, also complemented with PsbS WT or mutated E122Q and/or E226Q. Quantifications in B and C were performed as described in the legend for Fig. 2. All genotypes where dark- and light-treated samples are significantly different (p > 0.05, n = 3) are indicated by an asterisk. NPQ measurements for all genotypes are reported in supplemental Fig. S1.
To dissect the components of the relation between B4C and NPQ, we analyzed a series of mutants that are affected in NPQ at different levels, namely npq4 impaired in qE, the fastest NPQ component due to the absence of PsbS (20); npq1 (no zeaxanthin and reduced NPQ (32)); and npq2 with constitutive zeaxanthin and accelerated NPQ (32); as well as the double npq2/npq4 mutant. The NPQ kinetics of these genotypes have been reported in earlier work and are shown here in supplemental Fig. S1 (20, 32, 33). The results from Fig. 4, A and B, show that the npq4 mutation completely abolishes the light-dependent B4C dissociation, even in the presence of constitutive zeaxanthin, implying a key role for the PsbS protein in the process. Zeaxanthin is also a factor in B4C dissociation; in fact, in its absence, the npq1 mutant undergoes only 30% dissociation of B4C upon illumination, whereas, when present constitutively, as in the npq2 mutant, B4C is dissociated faster than in the WT (not shown).
PsbS activity in NPQ has been reported to be dependent on the protonation of two lumen-exposed glutamate residues (21, 22). The analysis of B4C dissociation in mutants at these Glu residues thus allows verification of the hypothesis that PsbS activity in B4C dissociation and in NPQ is mediated by the same mechanism, i.e. the protonation of Glu-122 and Glu-226 residues. To this aim, we analyzed npq4 mutants complemented with PsbS-encoding sequences carrying either WT, E122Q, E226Q or the double E122Q/E226Q mutant sequence (21, 22). The data in Fig. 4C show that the double mutant is unable to dissociate B4C, whereas each of the single glutamate mutants undergoes ∼40% B4C dissociation. The complementation with the WT PsbS sequence instead fully restored the capacity for light-dependent B4C dissociation. Thus, the extent of B4C dissociation was completely correlated with the NPQ activity of each genotype (all kinetics are shown in supplemental Fig. S1). It is worth mentioning that PsbS protein was detected in sucrose gradients in several of the pigmented fractions, as reported previously (34, 35), and that this distribution did not change upon light treatment.
Specific Lhcb Mutants Show Constitutive B4C Dissociation—The crucial role of PsbS in NPQ has been known for several years (20). After intense debate, it is now widely recognized that its role is to sense the low lumenal pH, thus inducing a quenching conformation of specific antenna complexes where the quenching reaction(s) occur (36-38). Because antenna polypeptides are involved both in structurally building the B4C and in catalyzing the quenching reactions, we investigated the effect of knocking out individual components of the PSII antenna system on the levels and kinetics of NPQ and on the structural stability of B4C. In Fig. 5A, sucrose gradient fractionation patterns of thylakoids purified from dark-adapted plants depleted in CP24, CP26 (25), Lhcb1 and -2 (39), CP29, and Lhcb3 (this work) are shown. We observed that knock-out mutants for some B4C components, namely CP29, CP24, and Lhcb3, do actually miss B4C. Instead, depletion of Lhcb1 and -2 does not affect B4C abundance in the sucrose gradient, implying that Lhcb3 is not equivalent to the other LHCII components as far as its contribution to the building of B4C. On this basis, we propose that B4C complex formation requires the direct interaction of Lhcb3, CP29, and CP24. On the contrary, Lhcb1 and Lhcb2, although participating in the trimeric LHCII M component, are not indispensable for B4C formation. Consistently, B4 is present in koCP26 plants.
FIGURE 5.
Dependence of B4C and NPQ on the presence of individual Lhc proteins. A, the capacity of B4C formation was analyzed in plants depleted of individual Lhcb proteins by separating different pigment-binding complexes from dark-adapted thylakoid membranes. Genotypes analyzed were WT, koLhcb3, koCP24, koCP29 (which is a triple knock-out in all CP29 Arabidopsis isoforms, Lhcb4.1, Lhcb4.2, and Lhcb4.3) and koCP26. B, NPQ dependence on antenna composition, analyzed by comparing WT (black) with plants depleted of CP26 (green), CP24 (blue), CP29 (knock-out in Lhcb4.1, Lhcb4.2, and Lhcb4.3 (cyan)), and Lhcb3 (red).
Analysis of the above genotypes by pulse fluorometry during light treatment showed that the mutants on B4C components were not equivalent as far as their NPQ behavior; ko-Lhcb3 did not show any effect, whereas CP29 and CP24 were deeply affected, showing a significant decrease in NPQ amplitude and slower induction kinetics. Although the effect of CP24 depletion on the NPQ process has recently been well described (24, 25), the NPQ kinetics of koCP29 reveals that also this subunit has a relevant role in NPQ induction, different from previous suggestions coming from the study of antisense plants (42). Lhcb1 and -2-depleted plants showed very similar kinetics but a slightly lower amplitude of NPQ with respect to WT (not shown), in agreement with previous reports (39). CP26 is the only Lhcb complex not involved directly in B4C assembly, in agreement with its location in a different domain of the PSII supercomplex (3). The NPQ kinetic of koCP26 plants is unaffected in the onset during light treatment but has a faster recovery of fluorescence in the dark, implying an effect on the qI component (Fig. 5B). We concluded that the presence of a stable B4C complex is not a prerequisite for NPQ and that the NPQ properties of the mutants depend on the residual composition in Lhcb complexes of the different genotypes. However, when B4C complex is present, its dissociation is needed for NPQ expression.
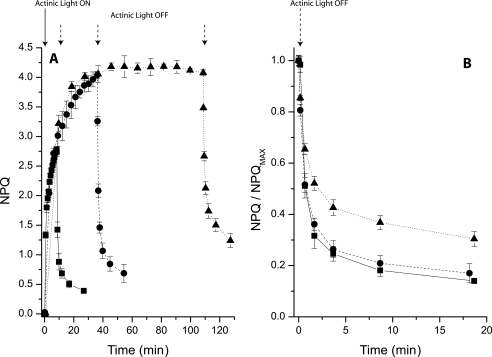
NPQ upon Prolonged Illumination—NPQ and B4C dissociation have in common a dependence on light intensity and the presence of PsbS. These phenomena, however, are activated in different time scales. The literature shows that the onset of NPQ requires only a few minutes, whereas B4C dissociation needs a longer period of illumination. In order to compare the two processes over the same time span, we measured NPQ kinetics using a longer exposure to actinic light than what is usually reported in the literature. In Fig. 6 we show the NPQ kinetics in WT Arabidopsis plants upon illumination for 8, 30, and 90 min. These kinetics show that the largest part of NPQ is activated within a few minutes of illumination (t½ is 3 min) but continues to rise for up to 30 min. After 90 min of illumination NPQ amplitude is stable at its saturated level. However, the kinetics of dark relaxation was slower upon 90 min of illumination than what observed after 8 or 30 min of light. This is evidenced in Fig. 6B, where the NPQ decay curves are normalized to the maximal NPQ value. Although relaxation kinetics are similar upon 8 or 30 min of illumination, they are slower upon 90 min of treatment. We can exclude the suggestion that this effect is due to the normalization procedure, because the level of quenching is very similar following 30 and 90 min of illumination.
FIGURE 6.
Long term NPQ induction and recovery. A, NPQ kinetics in WT leaves was recorded using different durations of actinic light. For all samples NPQ was recorded at 12 points during illumination. Curves with 8, 36, and 108 min of actinic light are shown, respectively, as a solid line with squares, a dashed line with circles, and a dotted line with triangles. In all samples, six points were recorded at the same time intervals during dark recovery. B, enlargement of recovery kinetics shown in A after normalization to maximal NPQ.
The possibility that photoinhibition could be a reason for the slowdown of fluoresce recovery was assayed by measuring dark recovery of PSII quantum yield after light treatment. As this was essentially the same in plants that were treated with light for 30 or 90 min, we concluded that differential photoinhibition is not the cause of the different kinetics of fluorescence recovery after NPQ (supplemental Fig. S2). Alternatively, sustained quenching could be due to zeaxanthin accumulation (33). However, no difference was observed in the Zea content of leaves treated with 30 or 90 min of actinic light, and the recovery of violaxanthin content was very similar (supplemental Fig. S2).
We concluded that, in addition to the two well known phases of NPQ consisting of a rapidly developing step dominated by qE and a slower phase depending on zeaxanthin synthesis (19), we could distinguish a later phase with sustained quenching. During this later phase the properties of the photosynthetic apparatus undergo changes that cannot be explained in terms of photoinhibition or zeaxanthin quenching, and yet they significantly affect the recovery of fluorescence.
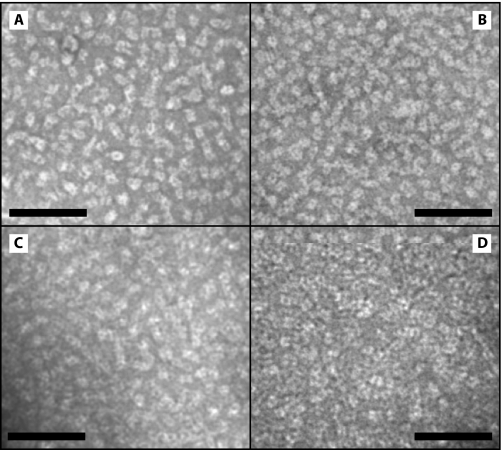
The Organization of Grana Membranes Is Modified upon PsbS-dependent Dissociation of the B4C Complex—In order to test the hypothesis that the slowly developing changes of the slowest NPQ phase described above involve a reorganization of the thylakoid membrane, we studied the organization of the PSII components in grana membranes through direct observation by electron microscopy. In grana partitions, the PSII complex forms C2S2 particles (3), which constitute the inner antenna system and are maintained even in high light-adapted plants (12) and include a subset of the B4C complex, namely CP29. The rest of the B4C components are part of the outer antenna, which is assembled under low light conditions, leading to the formation of larger C2S2M2LX particles (3). This suggests that the integrity of B4C is required for the formation of large supercomplexes and its dissociation could lead to a reorganization of the antenna system with a consequent reduction of antenna size. To test this hypothesis, we analyzed the organization of grana membranes in WT and selected mutants upon treatment with light. To this aim we isolated grana membranes from plants illuminated for different periods, using a mild solubilization with α-DM, which preserves the interaction between PSII components and the overall mutual organization of PSII units (10). Photosystem II forms tightly packed domains in grana partition membranes with very little lipid interspersed between protein complexes (43). When observed by transmission electron microscopy upon negative staining, PSII membranes were characterized by stain-excluding particles with a tetrameric structure that could be readily identified as PSII cores (Fig. 7). Lhc protrude less from the membrane plane than PSII reaction centers and are thus covered by an electron-dense stain. The observation of grana membranes isolated from leaves treated with different light intensities did not show any eye-catching difference (Fig. 7).
FIGURE 7.
EM of grana membranes isolated upon light treatment. EM of grana membranes isolated from WT leaves that were dark-adapted (A) or treated with 30 min of light (B), 90 min of light (C), or 90 min of light and 90 min of dark (D). Space bar represents 100 nm.
Searching for more subtle differences, we identified in each image the position of the PSII cores and calculated the distance of the closest neighbor. The distribution of the distances is reported in supplemental Fig. S3. In most cases, the closest PSII core is located at 16-17 nm. Two shoulders in the distribution are also visible, one at 14 nm and a second at 20-21 nm. This distribution, with one main peak and two symmetric shoulders, is close to that obtained previously using different microscopic methods, AFM and freeze fracture (44, 45). This allowed us to conclude that, despite our using a different approach, the results obtained for the dark-adapted sample were quite consistent with published data, strongly supporting the reliability of grana isolation and image analysis procedures.
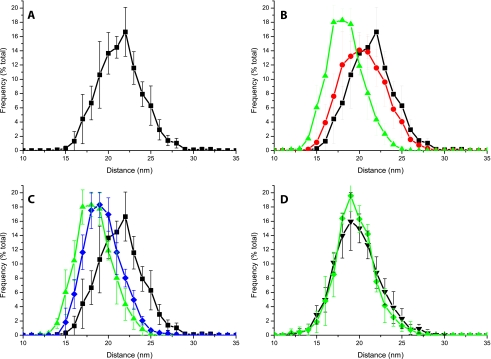
To reduce the dependence of the results on the possible imprecise assignment of PSII cores, we repeated the analysis by calculating for each core the average distance from the four closest PSII cores rather than from the closest neighbor only. As shown in Fig. 8, data deviations in this case were smaller, because the error in PSII core location is partially compensated for by the larger sampling. This method was thus employed to analyze grana membranes isolated from light-treated leaves (data considering only the closest neighbor are shown in the supplemental material). As shown in Fig. 8B, upon 30 min of illumination the PSII distribution shows a decrease in the population with an average distance of 22 nm and a compensating increase of the 18-19 nm population. The difference is significantly greater in the sample illuminated for 90 min; in that case, there is a far lesser abundance of PSII cores, with an average distance of 22 nm and the highest frequency found at 18 nm.
FIGURE 8.
Analysis of PSII distribution in grana isolated from dark/light-treated leaves. A, distribution of PSII complexes in grana membranes from dark-adapted leaves expressed as the average distance from the four nearest PSII core complexes. B, comparison of PSII distribution in dark-adapted samples (black squares) or those treated with 30 min (red circles) and 90 min (green triangles) of light before grana isolation. C, comparison of light-adapted samples (green triangles) with samples treated successively with 90 min of dark following light treatment (blue diamonds). D, effect of light treatment on PSII distribution in npq4 grana membranes. Dark-adapted sample is shown as black triangles and 90-min light-treated sample as green diamonds. S.D. in the case of the x axes is the maximum 1.5 nm.
We also verified the reversibility of the phenomenon by analyzing samples that, after 90 min of illumination, were further incubated in the dark for 90 min before grana isolation. In this case (Fig. 8C), we observed that the average distance between PSII increased, showing a decrease in the content of PSII cores of <17 nm and an increase in abundance for all points of >19 nm. Thus, although the recovery was partial and likely required a longer time to complete, these data strongly suggest the phenomenon is reversible. As a control, it is interesting to observe the phenotype of PsbS-less mutant npq4. Data from mutant grana isolated before and after 90 min of light treatment are shown in Fig. 8D. Remarkably, the distribution is substantially unaffected by light treatment. These results thus allowed us to conclude that light induces a redistribution of PSII cores within grana membranes but only in the presence of PsbS protein.
DISCUSSION
B4C Complex Has a Key Role in Building the PSII Supercomplex—CP29 and CP26 are located in direct contact with the PSII core complex (9, 46, 47) and interact with the LHCII-S trimer to form the so-called C2S2 particles (9). This PSII complex organization is found in plants grown under any conditions, even very intense light, and in mutants with constitutively reduced plastoquinone where PSII is constantly overexcited (10). C2S2 particles thus represent the minimal PSII supercomplex.
Growth in low or moderate light leads to the accumulation of additional antenna complexes, CP24, LHCII-M, and LHCII-L trimers (12, 48), and the formation of an outer antenna system in C2S2M2 particles or higher mass supercomplexes (9). PSII-LHCII supramolecular complexes thus appear to include two layers of Lhc proteins: the inner layer, composed by CP26, CP29, and the LHCII-S trimer, and the outer layer, including CP24 and at least two LHCII trimers (M and L).
B4C is a Lhc supercomplex composed of CP29 (inner layer), CP24, and LHCII-M (outer) (46). B4C has been described (6) as a product of incomplete solubilization of PSII membranes with mild detergents. It was later understood to represent a structural brick of the PSII-LHCII supercomplexes as shown by electron microscopy and cross-linking studies of the interactions of these subunits (3, 9, 49). Its presence is fundamental for the structure of PSII supercomplexes with larger antenna sizes. A further confirmation of this view comes from the observation that green algae, which do not have CP24 and Lhcb3 subunits, are not able to form B4C complex. In fact, green algae show the presence of C2S2 particles only (47). It can be asked whether all CP29 and CP24 subunits are organized into a B4 supercomplex with a LHCII trimer. Fig. 1 shows that a non-negligible amount CP29 and CP24 is found as monomers. However, in a previous report from Zea mays, 100% of the CP29 and CP24 was found in the B4C (6). These considerations suggest that monomerization of CP24 and CP29 is most likely because of partial destabilization of the B4C supercomplex due to detergent treatment during thylakoids solubilization.
Dissociation of the B4C Complex Is Correlated to Non-photochemical Quenching—In this work we have reported on the light-dependent dissociation of the supramolecular pentameric complex composed of CP29, CP24, and the LHCII-M trimer (6), which connects the inner and outer moieties of the light-harvesting system. We noticed that the fractionation pattern of PSII subunits in sucrose gradients or nondenaturing PAGE was modified upon strong illumination; an oligomeric green band, B4C, present in dark-adapted leaves, dissociated from its monomeric and trimeric components (Fig. 1).
Pharmacologic and genetic analysis showed that B4C dissociation correlates fully with NPQ. In fact, we observed that this event was sensitive to the uncoupler nigericin and was up-modulated by zeaxanthin, as shown by the npq1 mutant, which cannot synthesize Zea and is inefficient in both B4C dissociation and NPQ. The most striking evidence of this correlation, however, comes from the phenotype of PsbS-depleted plants, which are unable to activate NPQ and do not show the light-dependent dissociation of B4C (Fig. 4). Furthermore, the mutation of each of two lumen-exposed glutamate residues in PsbS, essential for NPQ activation (20, 21), leads to an NPQ decrease of ∼50% (21). Here, we show that the same mutation impairs the dissociation of B4C to approximately the same extent (Fig. 4), making it very unlikely that these two phenomena are independent but occur simultaneously upon illumination.
Functional Role of B4C Dissociation in Non-photochemical Quenching—The above results suggest a tight relationship between B4C dissociation and NPQ. It is, however, unclear whether dissociation of B4C is sufficient for feedback de-excitation or if additional operations are involved in the process that might well be fulfilled by PsbS. One way to address this question is to compare the WT with mutants in which B4C is constitutively dissociated, such as koCP24 (Fig. 5). In this mutant, dark-adapted plants have a higher fluorescence level than the WT, implying that the disconnected LHCII is not efficiently quenched (24, 25). Furthermore, koCP24 plants are able, despite the complete absence of B4C, to activate NPQ, although at a reduced level with respect to WT. By analyzing different mutants depleted in antenna proteins, koLhcb3 and koCP29 in Fig. 5, we noticed that the absence of B4C in a number of these mutants did not completely abolish NPQ, although the kinetics were differently affected depending on the residual complement of Lhcb proteins in each mutant.
B4C dissociation by itself is thus not sufficient for NPQ. However, in all cases where B4C was present in the dark, we never observed NPQ without B4C dissociation. This suggests that B4C stabilizes an unquenched state and that its dissociation is required for quenching activation. On these bases, we hypothesized that the quenching activity is performed by one or more B4C components. The integrity of B4C prevents quenching, which can be triggered only when this supercomplex is dissociated. It is not clear at present whether the role of PsbS is limited to the dissociation of B4C or if it extends to establishing an interaction with the resulting dissociated Lhcb subunits. We favor the latter hypothesis because in koCP24 the disconnected LHCII-M + L are clearly detectable; however, they are not quenched (24). Indeed, the koCP24 genotype mimics the constitutive dissociation of B4C, and EM analysis of its grana membranes clearly shows that in the absence of B4C, PSII core is nonhomogeneously distributed in grana membrane disks with PSII-rich areas and LHCII-enriched stain-depleted domains (supplemental Fig. S4).
It might be asked which of the different subunits are the “quenching sites.” Considering the similarity between antenna proteins it is unlikely that quenching occurs in only one subunit, and they may well all contribute. Nevertheless, recent reports are helpful in identifying some preferential quenching sites based on the in vivo effect of knocking out individual gene products on NPQ amplitude/kinetics and on the capacity of catalyzing the transient formation of carotenoid radical cation, a chemical species proposed to be responsible for excess energy quenching (50) by isolated pigment-protein complexes. Thus, depletion of Lhcb1 and -2 (39) or Lhcb3 (Fig. 5) has little effect on NPQ in vivo, and these antenna subunits cannot produce any carotenoid radical cation in vitro (51).
Monomeric Lhcb4, -5, and -6 do indeed perform charge transfer quenching in vitro (37, 51), whereas in vivo depletion strongly affects NPQ components: qE in the case of CP24 and CP29 (25) (Fig. 5) and qI in the case of CP26. The involvement of two components of B4C complex in qE, together with the NPQ depletion in all mutants/conditions where B4C complex cannot be dissociated, is strong evidence for the direct interaction of a PsbS-minor Lhc complex in triggering NPQ.
An NPQ Model Based on the Rearrangement of PSII Supercomplex Antenna—A possible discrepancy between B4C dissociation and NPQ can be found in their activation time scales. In fact, a large fraction of NPQ is activated within a few minutes after the dark to light transition (t½ ∼ 3 min; Fig. 6), whereas we observed that B4C dissociation is developed with a t½ of 12-15 min (Fig. 3). This phenomenon thus appears to have the same time scale of the slow phase of NPQ activation. However, the method we used for biochemical detection of B4C dissociation, i.e. sucrose gradient ultracentrifugation, is all but rapid and sensitive. It is likely that dissociated B4C need to be accumulated in order to be detected by biochemical fractionation and/or that unstable B4C dissociation is involved in the fastest phase of NPQ. In fact, after light treatment, thylakoid isolation and solubilization requires about 60 min. Thus, despite the fact that all operations were performed in ice water, we cannot exclude the possibility that early changes were reverted to before thylakoid solubilization and gradient loading. On the contrary, the physiological effects that elicit NPQ might be activated in a few seconds by a partial and unstable B4C dissociation, which, if the high light conditions are maintained, is increased by a complete B4C dissociation and stabilized by the structural redistribution of complexes within the grana membrane.
Our results provide experimental support of previous suggestions that (i) PsbS functions by interacting with other antenna proteins; (ii) PsbS controls the grana membrane organization; and (iii) structural modification in photosystems are implied in NPQ (35, 38, 52, 53). Such an extensive PSII reorganization might appear surprising. Nevertheless, recently reported data are consistent with such a model; FRAP measurements show that the greater part of complexes (75%) within grana membranes is substantially immobile. However, the remaining complexes, likely identified as antenna complexes, have instead a high diffusion coefficient. According to this hypothesis, an antenna complex might travel from the center to the limit of the grana membrane in about 2 s (54). These results thus suggest that a rearrangement of antenna complexes, also implying protein displacement, would be compatible with NPQ kinetics. Furthermore, the latter study suggests that this movement is not only diffusion-driven but is likely to be “oriented”; in this respect, a role could be played by PsbS activity, which might help in creating the so-defined diffusion channels (54). Concerning the role of PsbS, it interesting that we observed a small but significant difference between WT and npq4 in PSII core distribution in the dark. This is consistent with the observation that npq4 mutants grow better in low light conditions than do WT (33), suggesting that PsbS activity is not fully restricted to excess light conditions.
B4C Dissociation Induces a Rapid Decrease in the Antenna Size of PSII Supercomplex—Structural analysis of grana membranes clearly showed that light treatment induces a decrease in the distance between PSII reaction centers. This effect was reversible upon dark recovery, in parallel with the dissociation and reassociation of B4C. Grana membranes are densely packed with PSII components and contain little free lipids (43, 55). Thus, the distance between PSII core complex is determined mainly by the Lhcb protein complement of photosystems (10, 24, 44, 47). We concluded that light treatment, with the involvement of PsbS activity, leads to a reversible dissociation of the outer layer of the PSII antenna system from the reaction center complex, thus reducing the size of the antenna system, which feeds excitation energy to PSII reaction centers.
We observed that such a PSII redistribution requires more than 30 min (Fig. 8). Thus, it correlates with the stabilization of NPQ occurring in the 30-90-min interval (Fig. 6), suggesting that NPQ stabilization occurs by rearranging PSII complexes and their antenna.
The advantage of such a regulatory mechanism is clear if we consider that overexcitation of PSII is the major source of photoinhibition (14). Long term acclimation to high light is well known to lead to a reduction of peripheral antenna size (11, 12). However, the acclimatory process requires protein degradation and takes several days, and it is not useful or energetically affordable when light intensity varies within minutes. NPQ, which is instead the major regulatory mechanism acting within these time scales (17), could thus work by functionally dissociating part of the antenna from the core complex while avoiding degradation and resynthesis.
The Fate of Lhcb upon Disconnection from PSII Reaction Centers—It might be asked where antenna complexes travel to upon disconnection from the PSII core particles. Possible destinations are the grana periphery and the stroma lamellae. The former hypothesis is likely for at least a fraction of the antenna, as the PSII distribution was shown to be reversible. In the case of koCP24 it was also observed that regions with lower PSII core density were generally found in the grana periphery (supplemental Fig. S4). On a longer time scale (hours or even days), it is also possible that antenna complexes may migrate even further. Early work in fact showed that in light-treated leaves, LHCII isoforms disappear from grana membranes and are found in stroma membranes (56). Because proteolysis is particularly active in stroma membranes, antenna migrating here might be degraded, leading to a stable downsizing of PSII antenna size. This hypothesis suggests the idea that the domain segregation process described here under prolonged illumination contributes to the acclimatory modulation of the PSII antenna size.
Evolution of NPQ Mechanisms in Viridiplantae—It is interesting to point out that CP24 and Lhcb3, two of the three components of the B4C complex, are late additions to the PSII antenna system, being present only in land plants (4) and never detected in algae. Arabidopsis mutants that are missing any of these polypeptides are also missing B4C (Fig. 5), demonstrating that they are fundamental for the formation of the supercomplexes. Algae and plants also differ as to their PsbS content. In fact, although the gene is present in several algal genomes, the corresponding polypeptide is not accumulated in any algal species analyzed thus far (40, 41). Furthermore, in algae, only C2S2 supercomplexes have been found, while larger supercomplexes, such as those of land plants, have never been observed (47). This suggests the hypothesis that the regulatory mechanism we described in this work might have evolved upon land colonization in order to defend plants from the highly variable conditions found in the land environment (17). On the contrary, algal water environment is more stable in temperature, light, and CO2 supply, thus making a fast quenching mechanism less important in algae with than in plants (29).
Conclusion—We have described herein new experimental evidence on the function of PsbS, consisting of the reorganization of the protein domains in grana membranes as suggested recently (53). This reorganization is obtained by controlling the assembly of a pentameric complex, called B4C, which includes components of both the inner (CP29) and outer (CP24, LHCII-M) moieties of PSII antenna system. In low light the complex is assembled and PSII antenna size is maximal; in high light the complex dissociates, and we proposed that C2S2 particles dissociates from CP24/LHCII-M/LHCII-L complexes that migrates to the periphery or the grana. Upon dissociation of PSII supercomplexes the formation of quenching sites in PSII antenna, which we propose to be CP24 and CP29, is allowed by the dissociation of B4C and accessibility of these minor Lhc to the interaction with PsbS. We also suggest that this regulation of PSII supercomplex assembly is part of a control cycle connecting short term energy dissipation to long term regulation of PSII antenna size upon acclimation.
Supplementary Material
Acknowledgments
We thank G. Tognon for technical assistance with EM and Prof. Niyogi for the kind gift of npq4 mutants.
The on-line version of this article (available at http://www.jbc.org) contains supplemental Figs. S1-S4.
Footnotes
The abbreviations used are: Chl, chlorophyll; PSI and PSII, photosystem I and II, respectively; LHCII, light-harvesting complex II; NPQ, non-photochemical quenching; WT, wild type; IOD, integrated optical density; EM, electron microscopy; μE, microeinstein; HPLC, high pressure liquid chromatography; α-DM, n-dodecyl-α-d-maltoside.
References
- 1.Nelson, N., and Ben Shem, A. (2004) Nat. Rev. Mol. Cell Biol. 5 971-982 [DOI] [PubMed] [Google Scholar]
- 2.Caffarri, S., Croce, R., Cattivelli, L., and Bassi, R. (2004) Biochemistry 43 9467-9476 [DOI] [PubMed] [Google Scholar]
- 3.Boekema, E. J., van Roon, H., Calkoen, F., Bassi, R., and Dekker, J. P. (1999) Biochemistry 38 2233-2239 [DOI] [PubMed] [Google Scholar]
- 4.Alboresi, A., Caffarri, S., Nogue, F., Bassi, R., and Morosinotto, T. (2008) PLoS ONE 3 e2033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Horton, P., and Ruban, A. (2005) J. Exp. Bot. 56 365-373 [DOI] [PubMed] [Google Scholar]
- 6.Bassi, R., and Dainese, P. (1992) Eur. J. Biochem. 204 317-326 [DOI] [PubMed] [Google Scholar]
- 7.Boekema, E. J., Hankamer, B., Bald, D., Kruip, J., Nield, J., Boonstra, A. F., Barber, J., and Rögner, M. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 175-179 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nield, J., Orlova, E. V., Morris, E. P., Gowen, B., van Heel, M., and Barber, J. (2000) Nat. Struct. Biol. 7 44-47 [DOI] [PubMed] [Google Scholar]
- 9.Boekema, E. J., van Roon, H., van Breemen, J. F., and Dekker, J. P. (1999) Eur. J. Biochem. 266 444-452 [DOI] [PubMed] [Google Scholar]
- 10.Morosinotto, T., Bassi, R., Frigerio, S., Finazzi, G., Morris, E., and Barber, J. (2006) FEBS J. 273 4616-4630 [DOI] [PubMed] [Google Scholar]
- 11.Bailey, S., Walters, R. G., Jansson, S., and Horton, P. (2001) Planta 213 794-801 [DOI] [PubMed] [Google Scholar]
- 12.Ballottari, M., Dall'Osto, L., Morosinotto, T., and Bassi, R. (2007) J. Biol. Chem. 282 8947-8958 [DOI] [PubMed] [Google Scholar]
- 13.Lindahl, M., Yang, D. H., and Andersson, B. (1995) Eur. J. Biochem. 231 503-509 [DOI] [PubMed] [Google Scholar]
- 14.Barber, J., and Andersson, B. (1992) Trends Biochem. Sci. 17 61-66 [DOI] [PubMed] [Google Scholar]
- 15.Asada, K. (1999) Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 601-639 [DOI] [PubMed] [Google Scholar]
- 16.Niyogi, K. K. (2000) Curr. Opin. Plant Biol. 3 455-460 [DOI] [PubMed] [Google Scholar]
- 17.Kulheim, C., Agren, J., and Jansson, S. (2002) Science 297 91-93 [DOI] [PubMed] [Google Scholar]
- 18.Niyogi, K. K., Björkman, O., and Grossman, A. R. (1997) Plant Cell 9 1369-1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Demmig-Adams, B. (1990) Biochim. Biophys. Acta 1020 1-24 [Google Scholar]
- 20.Li, X. P., Bjorkman, O., Shih, C., Grossman, A. R., Rosenquist, M., Jansson, S., and Niyogi, K. K. (2000) Nature 403 391-395 [DOI] [PubMed] [Google Scholar]
- 21.Li, X. P., Phippard, A., Pasari, J., and Niyogi, K. K. (2002) Funct. Plant Biol. 29 1131-1139 [DOI] [PubMed] [Google Scholar]
- 22.Li, X. P., Gilmore, A. M., Caffarri, S., Bassi, R., Golan, T., Kramer, D., and Niyogi, K. K. (2004) J. Biol. Chem. 279 22866-22874 [DOI] [PubMed] [Google Scholar]
- 23.Briantais, J.-M. (1994) Photosynth. Res. 40 287-294 [DOI] [PubMed] [Google Scholar]
- 24.Kovacs, L., Damkjaer, J., Kereiche, S., Ilioaia, C., Ruban, A. V., Boekema, E. J., Jansson, S., and Horton, P. (2006) Plant Cell 18 3106-3120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Bianchi, S., Dall'Osto, L., Tognon, G., Morosinotto, T., and Bassi, R. (2008) Plant Cell 20 1012-1028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bassi, R., Rigoni, F., Barbato, R., and Giacometti, G. M. (1988) Biochim. Biophys. Acta 936 29-38 [Google Scholar]
- 27.Gilmore, A. M., and Yamamoto, H. Y. (1991) Plant Physiol. 96 635-643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Kooten, O., and Snel, J. F. H. (1990) Photosynth. Res. 25 147-150 [DOI] [PubMed] [Google Scholar]
- 29.Wollman, F. A. (2001) EMBO J. 20 3623-3630 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tikkanen, M., Piippo, M., Suorsa, M., Sirpio, S., Mulo, P., Vainonen, J., Vener, A. V., Allahverdiyeva, Y., and Aro, E. M. (2006) Plant Mol. Biol. 62 779-793 [DOI] [PubMed] [Google Scholar]
- 31.Bellafiore, S., Barneche, F., Peltier, G., and Rochaix, J. D. (2005) Nature 433 892-895 [DOI] [PubMed] [Google Scholar]
- 32.Niyogi, K. K., Grossman, A. R., and Björkman, O. (1998) Plant Cell 10 1121-1134 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dall'Osto, L., Caffarri, S., and Bassi, R. (2005) Plant Cell 17 1217-1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dominici, P., Caffarri, S., Armenante, F., Ceoldo, S., Crimi, M., and Bassi, R. (2002) J. Biol. Chem. 277 22750-22758 [DOI] [PubMed] [Google Scholar]
- 35.Teardo, E., De Laureto, P. P., Bergantino, E., Dalla, V. F., Rigoni, F., Szabo, I., and Giacometti, G. M. (2007) Biochim. Biophys. Acta 1767 703-711 [DOI] [PubMed] [Google Scholar]
- 36.Ruban, A. V., Berera, R., Ilioaia, C., van Stokkum, I. H., Kennis, J. T., Pascal, A. A., Van Amerongen, H., Robert, B., Horton, P., and van Grondelle, R. (2007) Nature 450 575-578 [DOI] [PubMed] [Google Scholar]
- 37.Ahn, T. K., Avenson, T. J., Ballottari, M., Cheng, Y. C., Niyogi, K. K., Bassi, R., and Fleming, G. R. (2008) Science 320 794-797 [DOI] [PubMed] [Google Scholar]
- 38.Bonente, G., Howes, B. D., Caffarri, S., Smulevich, G., and Bassi, R. (2008) J. Biol. Chem. 283 8434-8445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Andersson, J., Wentworth, M., Walters, R. G., Howard, C. A., Ruban, A. V., Horton, P., and Jansson, S. (2003) Plant J. 35 350-361 [DOI] [PubMed] [Google Scholar]
- 40.Finazzi, G., Johnson, G. N., Dall'Osto, L., Zito, F., Bonente, G., Bassi, R., and Wollman, F. A. (2006) Biochemistry 45 1490-1498 [DOI] [PubMed] [Google Scholar]
- 41.Bonente, G., Passarini, F., Cazzaniga, S., Mancone, C., Buia, M. C., Tripodi, M., Bassi, R., and Caffarri, S. (2008) Photochem. Photobiol. 84 1359-1370 [DOI] [PubMed] [Google Scholar]
- 42.Andersson, J., Walters, R. G., Horton, P., and Jansson, S. (2001) Plant Cell 13 1193-1204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tremolieres, A., Dainese, P., and Bassi, R. (1994) Eur. J. Biochem. 221 721-730 [DOI] [PubMed] [Google Scholar]
- 44.Kirchhoff, H. (2008) Trends Plant Sci. 13 201-207 [DOI] [PubMed] [Google Scholar]
- 45.Kirchhoff, H., Lenhert, S., Buchel, C., Chi, L., and Nield, J. (2008) Biochemistry 47 431-440 [DOI] [PubMed] [Google Scholar]
- 46.Yakushevska, A. E., Keegstra, W., Boekema, E. J., Dekker, J. P., Andersson, J., Jansson, S., Ruban, A. V., and Horton, P. (2003) Biochemistry 42 608-613 [DOI] [PubMed] [Google Scholar]
- 47.Dekker, J. P., and Boekema, E. J. (2005) Biochim. Biophys. Acta 1706 12-39 [DOI] [PubMed] [Google Scholar]
- 48.Frigerio, S., Campoli, C., Zorzan, S., Fantoni, L. I., Crosatti, C., Drepper, F., Haehnel, W., Cattivelli, L., Morosinotto, T., and Bassi, R. (2007) J. Biol. Chem. 282 29457-29469 [DOI] [PubMed] [Google Scholar]
- 49.Harrer, R., Bassi, R., Testi, M. G., and Schäfer, C. (1998) Eur. J. Biochem. 255 196-205 [DOI] [PubMed] [Google Scholar]
- 50.Holt, N. E., Zigmantas, D., Valkunas, L., Li, X. P., Niyogi, K. K., and Fleming, G. R. (2005) Science 307 433-436 [DOI] [PubMed] [Google Scholar]
- 51.Avenson, T. J., Ahn, T. K., Zigmantas, D., Niyogi, K. K., Li, Z., Ballottari, M., Bassi, R., and Fleming, G. R. (2008) J. Biol. Chem. 283 3550-3558 [DOI] [PubMed] [Google Scholar]
- 52.Horton, P., Johnson, M. P., Perez-Bueno, M. L., Kiss, A. Z., and Ruban, A. V. (2008) FEBS J. 275 1069-1079 [DOI] [PubMed] [Google Scholar]
- 53.Kiss, A. Z., Ruban, A. V., and Horton, P. (2008) J. Biol. Chem. 283 3972-3978 [DOI] [PubMed] [Google Scholar]
- 54.Kirchhoff, H., Haferkamp, S., Allen, J. F., Epstein, D. B., and Mullineaux, C. W. (2008) Plant Physiol. 146 1571-1578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tremmel, I. G., Kirchhoff, H., Weis, E., and Farquhar, G. D. (2003) Biochim. Biophys. Acta 1607 97-109 [DOI] [PubMed] [Google Scholar]
- 56.Kyle, D. J., Staehelin, L. A., and Arntzen, C. J. (1983) Arch. Biochem. Biophys. 222 527-541 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.