Abstract
Osteoblasts are the primary cells responsible for bone formation. They also support osteoclast formation from bone marrow precursors in response to osteotropic factors by inducing receptor activator of NF-κB ligand (RANKL) expression and down-regulating osteoprotegerin (OPG) production. In addition to the RANKL-RANK-OPG signaling axis, other factors produced by osteoblasts/stromal cells are involved in osteoclastogenesis. Here, we describe the identification and characterization of leucine-rich repeat-containing 17 (LRRc17), a member of the LRR superfamily that acts as a negative regulator of RANKL-induced osteoclast differentiation. Osteoblasts showed high levels of LRRc17 expression, which was down-regulated in response to the pro-osteoclastogenic factor 1,25-dihydroxyvitamin D3. Recombinant LRRc17 protein inhibited RANKL-induced osteoclast differentiation from bone marrow precursors, whereas it did not affect the differentiation or activation of macrophages and dendritic cells. These results suggest that among the cell types derived from common myeloid precursors, LRRc17 specifically regulates osteoclasts. Further analysis revealed that LRRc17 attenuated RANKL-induced expression of NFATc1 by blocking phospholipase C-γ signaling, which, in turn, inhibited RANKL-mediated osteoclast differentiation. Taken together, our results demonstrated a novel inhibitory activity of LRRc17 in RANKL-induced osteoclastogenesis.
Bone remodeling continuously renews the skeleton and maintains its structure through a spatially coordinated balance between bone resorption and bone formation. This process involves the synthesis of organic matrix by osteoblasts and bone resorption by osteoclasts. The development and functions of both cell types are tightly regulated by various osteotropic factors and hormones. Mature matrix-secreting osteoblasts are derived from mesenchymal stem cells through a series of progenitor stages before being progressively transformed into osteocytes. On the other hand, multinucleated mature osteoclasts differentiate from macrophage/monocyte lineage precursor cells following a sequential process that includes proliferation, differentiation, fusion, and activation (1-4).
Receptor activator of NF-κB ligand (RANKL)4 induces osteoclast formation from hematopoietically derived, myeloid lineage monocyte precursor cells (1, 2, 4, 5). The binding of RANKL to its receptor, receptor activator of NF-κB (RANK), activates NF-κB, c-Jun N-terminal kinase (JNK), p38, extracellular signal-related kinase (ERK), and Akt, which mediate the differentiation, activation, and survival of osteoclasts (4, 6, 7). RANKL activates and/or induces the expression of transcription factors known to be important for osteoclastogenesis in vitro and in vivo, including c-Fos, microphthalmia transcription factor (MITF), and NFATc1 (1, 3, 4, 8). In particular, NFATc1 is thought to be a master downstream regulator of RANKL-induced osteoclastogenesis because ectopic expression of NFATc1 causes precursors to efficiently differentiate into osteoclasts in the absence of RANKL, whereas NFATc1-deficient embryonic stem cells fail to differentiate into osteoclasts in response to RANKL (8-10).
Osteoblasts support osteoclast formation from monocyte precursors in response to such osteotropic factors as 1,25-dihydroxyvitamin D3 (1,25(OH)2D3) and parathyroid hormone (PTH) (2). Most of these catabolic factors induce osteoclast formation by up-regulating the expression of M-CSF and RANKL, whereas concomitantly reducing the levels of the soluble RANKL decoy receptor osteoprotegerin (OPG) in osteoblasts (2, 4). In addition to the essential roles of these molecules in osteoclastogenesis, a number of other proteins act as modulators of osteoclast differentiation. Mice lacking ITAM-harboring DAP12 and FcRγ, which activate calcium signaling in osteoclast precursors, exhibit an osteopetrotic phenotype due to the defective osteoclast formation (8, 11, 12). These findings suggest that DAP12- and FcRγ-mediated ITAM signaling is critical for RANKL-induced osteoclastogenesis. It has been reported that costimulatory receptors, such as triggering receptor expressed in myeloid cells 2 (TREM2), signal-regulatory protein β1 (SIRPβ1), paired Ig-like receptor A (PIR-A), and osteoclast-associated receptor (OSCAR), interact with DAP12 or FcRγ (11); these receptors enhance calcium signaling and subsequent RANKL-induced osteoclastogenesis (4, 8, 11, 13). Despite progress in understanding such costimulatory receptors, OPG is the only well characterized inhibitory regulator of RANKL-induced osteoclast differentiation. Thus, we screened osteoblasts, important mediators of pathophysiological bone remodeling, for additional inhibitors of osteoclastogenesis and identified a leucine-rich repeat-containing protein of previously unknown function (LRRc17). This protein is highly expressed in osteoblasts, and its expression is markedly suppressed in response to the pro-osteoclastogenic factor 1,25(OH)2D3. Moreover, we show that recombinant LRRc17 inhibits RANKL-induced osteoclast differentiation, demonstrating that LRRc17 is a novel negative regulator of osteoclastogenesis.
EXPERIMENTAL PROCEDURES
PCR-mediated cDNA Subtraction—Primary osteoblasts were obtained from calvarias of newborn C57BL/6 mice using conventional methods and collagenase as described previously (14). In brief, calvarial cells were harvested from periosteum following incubation with 0.2% collagenase/dispase in serum-free α-minimum essential medium (α-MEM). The calvaria-derived cells were cultured for 3 days in α-MEM containing 10% fetal bovine serum (FBS). The 3-day-old immature osteoblasts were cultured with 5 × 10-8 m 1,25(OH)2D3 for 2 days and used to prepare a cDNA library. The enriched cDNA library was generated using poly(A)+ RNA from the 1,25(OH)2D3-treated cells and a fibroblastic cell line (NIH3T3) as described previously (13). Briefly, 2 μg of poly(A)+ RNA from the 1,25(OH)2D3-treated and NIH3T3 cells was used to produce tester and driver cDNA, respectively. A subtractive PCR was performed using a PCR-Select cDNA subtraction kit (Clontech) according to the manufacturer's protocol (13, 15).
Identification of LRRc17—We compared the mRNA expression profile of primary osteoblasts treated with 1,25(OH)2D3 with that of NIH3T3 cells using a PCR-based cDNA subtraction technique. The subtracted cDNA library was constructed as described previously (13, 15). We randomly selected and tested 171 clones from the cDNA library. Among the 171 clones tested, 85 clones showed higher expression levels in 1,25(OH)2D3-treated cells than in NIH3T3 cells, suggesting that ∼50% of the cDNA library was derived from genes that were preferentially expressed in 1,25(OH)2D3-treated cells, a result that agrees with our previous experiences (13, 15). Sequence analysis of the 85 clones revealed that they represented 42 independent genes. To examine the mRNA expression patterns of the 42 selected genes, we performed Northern analyses. mRNA was prepared from NIH3T3 cells, primary calvarial osteoblasts, and primary calvarial osteoblasts stimulated with 5 × 10-8 m 1,25(OH)2D3 for 2 days. Among the 42 independent genes, 29 showed higher mRNA expression levels in osteoblasts than in NIH3T3 cells and no change in response to 1,25(OH)2D3. Eleven genes showed higher mRNA expression levels in osteoblasts than in fibroblasts, differences that were potentiated by 1,25(OH)2D3. Interestingly, the mRNA levels of two genes that were more highly expressed in osteoblasts than in NIH3T3 cells were significantly suppressed by 1,25(OH)2D3 treatment. In this study, we further characterized one of these genes (initially described as clone OB86). Sequence analysis showed that OB86 contained a cDNA fragment representing nucleotides 1961-2150 in the 3′-untranslated region of the LRRc17 gene, the function of which was previously unknown.
Osteoclast Formation—Murine osteoclasts were prepared from bone marrow cells as described previously (16). Bone marrow cells were cultured in α-MEM containing 10% FBS with M-CSF (50 ng/ml) for 3 days, and the attached bone marrow-derived macrophages (BMMs) were used as osteoclast precursors. To generate osteoclasts, BMMs were cultured with M-CSF (50 ng/ml) and RANKL (150 ng/ml) for 3 days. To generate osteoclasts from cocultures containing osteoblasts and bone marrow cells, primary osteoblasts were prepared from calvarias of newborn mice as described previously (13). Bone marrow cells and primary osteoblasts were cocultured for 6 days in the presence of 1,25(OH)2D3 (1 × 10-8 m) or PTH (1 × 10-8 m)/prostaglandin E2 (5 × 10-7 m). Cultured cells were fixed and stained for TRAP as described previously (17). TRAP-positive multinucleated cells (TRAP+ MNCs) that contained more than three nuclei were counted.
Soluble LRRc17—A soluble form of LRRc17 was produced by creating an LRRc17-Fc fusion protein in which LRRc17 was fused to the constant region of human IgG1. LRRc17-Fc was produced in Escherichia coli and purified using protein G affinity column chromatography. In brief, E. coli BL21 (DE3) cells transformed with LRRc17-Fc expression vector were incubated at 37 °C in LB broth. When the absorbance at 600 nm reached 0.8, 1 mm isopropyl-1-thio-β-galactopyranoside was added to the culture to induce protein production, and the cells were incubated for an additional 4 h. To isolate the insoluble protein fraction containing LRRc17-Fc, cells were resuspended in lysis buffer (50 mm Tris-Cl (pH 8.0), 100 mm NaCl, 5 mm EDTA, 0.1 mm phenylmethylsulfonyl fluoride, 1 mm dithiothreitol, and 0.5% Triton X-100), ultrasonicated at 150 watts for 15 min, and centrifuged at 4500 × g for 15 min. This process was repeated three times. Then, the separated intracellular insoluble protein fraction was solubilized in 8 m urea, 100 mm Tris-Cl (pH 8.0), 50 mm glycine, 5 mm GSH, and 0.5 mm GSSG at 25 °C. After solubilization, the proteins were refolded using stepwise dialysis with refolding buffer containing 100 mm Tris-Cl (pH 8.0), 400 mm l-arginine, 1 mm EDTA, and 0.2 mm phenylmethylsulfonyl fluoride. To purify soluble LRRc17-Fc, affinity chromatography was performed using protein G-agarose. Heat inactivation of LRRc17-Fc was performed by incubating the samples at 95 °C for 15 min. We used human IgG and heat-inactivated LRRc17-Fc as control samples.
Retroviral Infection—To generate retrovirus stock, retroviral vectors were transfected into the Plat E packaging cell line using Lipofectamine 2000 (Invitrogen). Viral supernatant was collected from cultured media 24-48 h after transfection. BMMs were incubated with viral supernatant for 8 h in the presence of Polybrene (10 μg/ml). After removing the viral supernatant, BMMs were further cultured with M-CSF (50 ng/ml) and RANKL (150 ng/ml) for 3 days.
Measurement of Alkaline Phosphatase (ALP) Activity—Stromal cells derived from bone marrow were cultured in α-MEM containing 10% FBS. On the third day of culture, the initial plating medium was replaced with α-MEM containing 10% FBS, ascorbic acid (50 μg/ml), β-glycerophosphate (10 mm), and BMP-2 (100 ng/ml) in the absence or presence of LRRc17-Fc (1 μg/ml). After 7 days of culture, cells were washed with phosphate-buffered saline and lysed with 400 μl of lysis buffer (0.5 m Tris (pH 9.0), 150 mm NaCl, and 1% Triton X-100) for 16 h at 4 °C. Fifty microliters of each lysate was incubated with 100 μl of p-nitrophenyl phosphate substrate (Sigma-Aldrich) for 1 h at room temperature. The absorbance was measured at 570 nm using a microplate reader.
Nodule Formation Assay—Stromal cells derived from bone marrow were cultured for 21 days as described above. Cells were fixed with 10% formalin for 30 min at room temperature. After washing with distilled water, samples were stained with 1% Alizarin Red S (Sigma-Aldrich) for 30 min at room temperature and then were washed three times with distilled water.
Phagocytosis Assay—BMMs were cultured for 3 days with M-CSF (50 ng/ml) or M-CSF (50 ng/ml) and RANKL (150 ng/ml) in the absence or presence of LRRc17-Fc (1 μg/ml). Fluorescein-conjugated zymosan A (Saccharomyces cerevisiae) BioParticles (Molecular Probes) were added to the cultured cells in 96-well culture plates (20 μg/0.2 ml in each well). After 1 h of incubation, cells were washed with phosphate-buffered saline to remove the particles that were not incorporated by the cells. Cells were fixed and observed with UV illumination under the microscope.
Dendritic Cell Culture and FACS Analysis—Dendritic cells were prepared from BMMs as described previously with minor modifications (18). The harvested cells were resuspended in RPMI 1640 medium supplemented with 5% FBS, and BMMs were seeded in 24-well plates with GM-CSF (10 ng/ml). Cells were cultured for 4 days; cells were transferred to fresh medium containing the same concentration of GM-CSF on day 2. On day 4, 1 ml of fresh medium containing 1 μg/ml LPS (Sigma-Aldrich) was added to the cultures to stimulate the maturation of dendritic cells. The next day, the cells were harvested and analyzed with anti-CD86, anti-I-Ab, and anti-CD11c antibodies.
Northern Hybridization and Western Blot Analysis—Northern blot analysis was performed as described previously (19). For immunoblotting analysis, BMMs were stimulated with RANKL for the indicated periods of time. Cells were then washed with ice-cold phosphate-buffered saline and lysed in extraction buffer (50 mm Tris-HCl (pH 8.0), 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, and protease inhibitors). Cell lysates were subjected to SDS-PAGE and Western blotting.
Real-time PCR—Real-time PCRs were performed using the TaqMan universal PCR master mix and an ABI Prism 7000 sequence detection system (Applied Biosystems). TaqMan primers for the indicated genes were obtained from Applied Biosystems. For each reaction, 250 ng of cDNA template generated using SuperScript II (Invitrogen) was used.
RESULTS
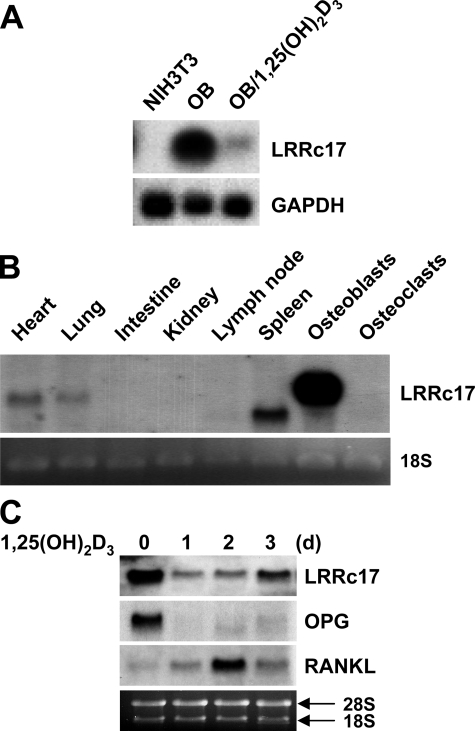
LRRc17 Is Predominantly Expressed in Osteoblasts and Is Down-regulated in Response to 1,25(OH)2D3—During a study comparing the mRNA expression profiles of osteoblasts stimulated with 1,25(OH)2D3 under osteoclastogenic conditions and NIH3T3 fibroblastic cells, which do not support osteoclastogenesis (see “Experimental Procedures”), we isolated LRRc17 as a gene with an expression profile characterized by higher mRNA levels in osteoblasts than in fibroblasts and significant suppression in response to 1,25(OH)2D3 treatment (Fig. 1A). The putative LRR-containing protein LRRc17, the function of which was previously unknown, was originally identified as p37NB using cDNA subtraction analysis of genes with higher expression levels in an S-type neuroblastoma cell line than in an N-type neuroblastoma cell line (20). The gene was also described in a large scale mRNA screen of the human pancreas (21) and as a result of the complete sequencing of human chromosome 7 (22). The full-length LRRc17 cDNA encodes a secreted protein that contains five putative LRR domains. The predicted mouse LRRc17 protein, which is composed of 443 amino acids, is 87% identical and 92% similar to the human LRRc17 protein.
FIGURE 1.
LRRc17 mRNA expression. A, Northern blot analysis of NIH3T3 cells, osteoblasts (OB), and osteoblasts treated with 1,25(OH)2D3 for 2 days (OB/1,25(OH)2D3). Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as a loading control. B, Northern blot analysis of LRRc17 in various mouse tissues, osteoblasts, and bone marrow-derived osteoclasts. C, Northern blot analysis was performed using total RNA from primary calvarial osteoblasts stimulated with 1,25(OH)2D3 for the indicated periods of time. Sequences specific for LRRc17, OPG, and RANKL were used as probes.
To further examine the distribution of LRRc17 mRNA, various mouse tissues were analyzed using Northern blots. Similar to humans, two isoforms of LRRc17 mRNA were detected, likely a result of alternative splicing (20-22). LRRc17 mRNA expression was weakly detected in heart and lung tissues, whereas a strong signal representing the smaller LRRc17 mRNA isoform was detected in the spleen (Fig. 1B). LRRc17 was strongly expressed in primary osteoblasts but not in osteoclasts. Upon stimulation with 1,25(OH)2D3, the expression levels of LRRc17 and OPG markedly decreased, whereas expression of RANKL, an osteoclast differentiation factor, was induced in primary osteoblasts (Fig. 1C) and UAMS32 cells, a stromal/osteoblastic cell line (supplemental Fig. S1).
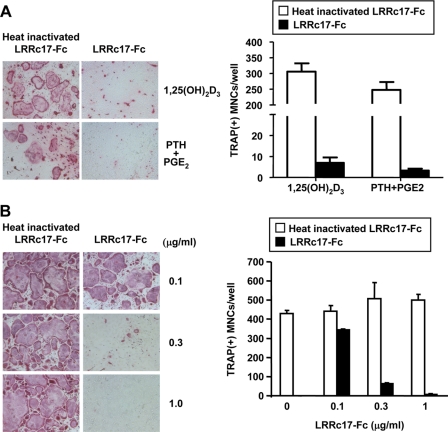
LRRc17 Inhibits Osteoclast Development—Because LRRc17 expression was detected in osteoblasts and was regulated by 1,25(OH)2D3, we hypothesized that LRRc17 contributes to the differentiation of osteoclast precursors into mature, multinucleated osteoclasts. To test this hypothesis, we constructed a recombinant form of LRRc17 by fusing LRRc17 to the Fc portion of human IgG1 (LRRc17-Fc). To determine the role of LRRc17 in osteoclastogenesis, LRRc17-Fc was added to cocultures containing bone marrow precursor cells and osteoblast/stromal cells in the presence of the osteotropic factor 1,25(OH)2D3. The addition of LRRc17-Fc significantly inhibited the formation of multinucleated TRAP+ osteoclasts (TRAP+ MNCs) in the cocultures containing 1,25(OH)2D3 (Fig. 2A). The inhibitory effect of LRRc17 on osteoclastogenesis was also observed in cocultures containing PTH and prostaglandin E2, two additional osteotropic factors (Fig. 2A). These results indicate that LRRc17 plays a negative regulatory role in development of mature osteoclasts from osteoblast-stimulated bone marrow precursor cells. The levels of M-CSF and RANKL expression induced by 1,25(OH)2D3 in osteoblasts, however, were not affected by treatment with LRRc17-Fc (supplemental Fig. S2).
FIGURE 2.
Role of LRRc17 in osteoclast differentiation. A, mouse bone marrow cells and primary calvarial osteoblasts were cocultured for 6 days with 1,25(OH)2D3 (1 × 10-8 m) or PTH (1 × 10-8 m)/prostaglandin E2 (PGE2, 5 × 10-8 m) in the presence of 3 μg/ml heat-inactivated recombinant murine LRRc17-Fc or native LRRc17-Fc. Cells were fixed and stained for TRAP (left). The TRAP+ MNCs were counted as osteoclasts (right). B, BMMs were cultured for 4 days with M-CSF (30 ng/ml) and RANKL (150 ng/ml) in the presence of various concentrations of heat-inactivated recombinant murine LRRc17-Fc or native LRRc17-Fc as indicated. Cells were fixed and stained for TRAP (left). TRAP+ MNCs were counted as osteoclasts (right).
LRRc17-Fc also inhibited osteoclastogenesis from BMMs treated with a combination of recombinant M-CSF and RANKL in the absence of osteoblasts/stromal cells (Fig. 2B), suggesting that LRRc17 may act directly on osteoclast precursors to inhibit osteoclast differentiation. We confirmed that recombinant LRRc17-Fc did not affect the proliferation or survival of BMMs using a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay (data not shown). Taken together, our results suggest that LRRc17 directly acts on osteoclast precursors and thereby inhibits RANKL-induced osteoclast differentiation.
Macrophages, osteoclasts, and dendritic cells originate from common myeloid lineage precursors (3, 4, 8, 23). Therefore, we determined whether LRRc17 affects the fates of monocyte lineage-derived cell types other than osteoclasts. Bone marrow cells were cultured in the presence of M-CSF alone to generate BMMs, the functions of which were measured by examining the phagocytosis of zymosan particles. The results showed that LRRc17-Fc did not inhibit the phagocytic activity of BMMs (Fig. 3A). On the other hand, we showed that RANKL treatment inhibited the phagocytic activity of BMMs (24). Because recombinant LRRc17-Fc protein inhibited osteoclastogenesis, we reasoned that BMMs treated with LRRc17-Fc might retain a phagocytic activity even in the presence of RANKL. When we cultured BMMs for 3 days with M-CSF and RANKL, BMMs treated with LRRC17-Fc were able to phagocytose the zymosan particles, whereas control IgG-treated BMMs lost their phagocytic activity under the same conditions.
FIGURE 3.
Effect of LRRc17 on dendritic cell differentiation and the phagocytic activity of macrophages. A, BMMs were cultured for 3 days with M-CSF alone or M-CSF and RANKL in the presence of 1 μg/ml murine LRRc17-Fc or control IgG. Cultured cells were incubated with fluorescein-conjugated zymosan particles for 1 h and washed with phosphate-buffered saline. Cells were fixed and observed with UV illumination under a microscope. Fluorescein-conjugated zymosan particles incorporated by the cells appear as bright dots in the dark field. B and C, BMMs were cultured for 4 days with GM-CSF to generate dendritic cells in the presence of 1 μg/ml murine LRRc17-Fc or control IgG. B, the cells were harvested and stained for FACS analysis with anti-CD11c antibodies (dotted lines) or control IgG (solid lines). C, LPS (1 μg/ml) was added to cultures to induce dendritic cell maturation. The cells were harvested the next day and stained for FACS analysis with anti-CD86 or anti-I-Ab antibodies (solid lines, without LPS; dotted lines, with LPS).
Previously, we demonstrated that BMMs developed into dendritic cells in response to stimulation with GM-CSF in vitro (24). Therefore, we tested whether LRRc17 affected the differentiation of BMMs into dendritic cells. After BMMs were cultured with GM-CSF for 4 days, immature dendritic cells were treated with LPS overnight in the presence of LRRc17-Fc or control IgG. The cells were then harvested and examined for dendritic cell markers, including CD11c, CD86, and I-Ab. As reported previously (24), BMM-derived dendritic cells expressed high levels of CD11c, CD86, and I-Ab, and LPS up-regulated the expression of CD86 and I-Ab. The addition of LRRc17-Fc to the cultures did not affect the differentiation of dendritic cells from BMMs or LPS-induced activation of dendritic cells (Fig. 3, B and C). Taken together, these data suggest that LRRc17 specifically regulates RANKL-induced osteoclast differentiation rather than affecting other cell types derived from the common precursor cells, dendritic maturation, or the phagocytic activity of macrophages.
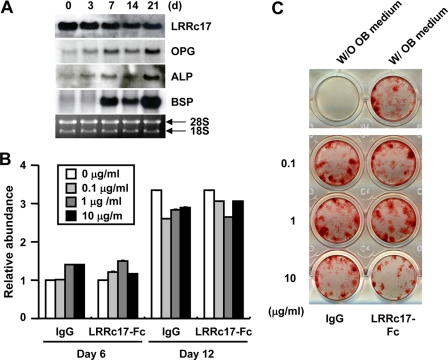
Effects of LRRc17 on Osteoblast Differentiation—It is well known that the destruction of the bone matrix by osteoclasts is coupled with the formation of new matrix by osteoblasts. Because LRRc17 affects osteoclast differentiation and osteoblasts abundantly express LRRc17, we investigated whether LRRc17 plays a role in osteoblast differentiation. To examine the expression profile of LRRc17 during osteoblast differentiation, Northern blot analysis was performed using total mRNA from primary osteoblasts cultured for the indicated periods of time in the presence of ascorbic acid and β-glycerophosphate (Fig. 4A). During osteoblast differentiation, the expression of ALP and bone sialoprotein (BSP), markers of osteoblast development, was strongly induced. Moreover, the expression of LRRc17 was slightly reduced during osteoblast differentiation, whereas OPG expression increased. We then tested whether LRRc17-Fc affected the proliferation and differentiation of osteoblasts. ALP activity and nodule formation were similar in control IgG- and LRRc17-Fc-treated osteoblasts (Fig. 4, B and C). Together, these results suggested that LRRc17 had no significant effect on osteoblast differentiation or proliferation.
FIGURE 4.
Effect of LRRc17 on osteoblast differentiation. A, Northern blot analysis was performed using total RNA from primary calvarial osteoblasts stimulated with ascorbic acid and β-glycerophosphate for the indicated periods of time. Sequences specific for LRRc17, OPG, ALP, and bone sialoprotein (BSP) were used as probes. B and C, stromal cells derived from bone marrow were cultured with ascorbic acid, β-glycerophosphate, and BMP-2 in the presence of various concentrations of murine LRRc17-Fc or control IgG as indicated. B, on days 6 and 12 of culture, ALP activity was measured at 570 nm. C, on day 12, cells were fixed and stained for nodules. W/O OB medium, without osteogenic medium; W/OB medium, with osteogenic medium.
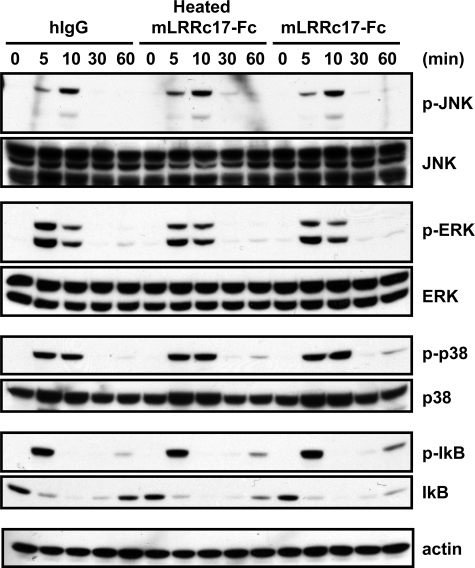
LRRc17 Inhibits RANKL-mediated NFATc1 Expression by Blocking PLCγ Signaling—RANKL, a critical osteoclastogenic factor, activates NF-κB, JNK, p38 MAP kinase, ERK, and Akt. To examine whether LRRc17 affects immediate early signaling pathways that are important for osteoclastogenesis, BMMs were treated with human IgG (hIgG), heat-inactivated LRRc17-Fc, or LRRc17-Fc and then were stimulated with RANKL for the indicated periods of time (Fig. 5). Consistent with previous results, RANKL activated JNK, p38, NF-κB, ERK, and Akt in control BMMs treated with human IgG or heat-inactivated LRRc17-Fc. Somewhat surprisingly, treatment with LRRc17 did not affect the RANKL-induced activation of NF-κB, JNK, p38 MAP kinase, ERK, or Akt. To investigate the potential inhibitory mechanism of LRRc17, we used real-time PCRs to examine the expression profiles of various genes that are important for RANKL-induced osteoclastogenesis (Fig. 6A). The expression levels of RANK, MITF, and c-Fos were similar in control and LRRc17-Fc-treated samples during RANKL-mediated osteoclast differentiation. However, the induction of NFATc1 expression, a key regulator of osteoclastogenesis, was strongly attenuated by LRRc17-Fc treatment. Consistent with the reduced levels of NFATc1, the expression levels of TRAP and OSCAR, two down-stream target genes of NFATc1, was also inhibited by LRRc17-Fc. Because costimulatory signaling via DAP12 and FcRγ is important for RANKL-induced osteoclastogenesis (11), we tested whether LRRc17 affected RANKL-mediated PLCγ activation. In preosteoclasts, treatment with LRRc17-Fc strongly attenuated the PLCγ2 activation induced by either RANKL stimulation or cross-linking of anti-OSCAR antibodies (Fig. 6B).
FIGURE 5.
LRRc17 does not affect immediate RANKL-induced signaling. BMMs were stimulated with 500 ng/ml RANKL for the indicated periods of time in the presence of 1 μg/ml human IgG (hIgG), heat-inactivated murine LRRc17-Fc, or murine LRRc17-Fc. Whole-cell extracts were subjected to Western blot analysis with specific antibodies as indicated. p-JNK, phospho-JNK,; p-ERK, phospho-ERK; p-p38, phospho-p38; p-IkB, phospho-IκB.
FIGURE 6.
LRRc17 blocks PLCγ2 signaling and the induction of NFATc1 expression during RANKL-mediated osteoclastogenesis. A, real-time quantitative PCR analysis of RANK, MITF, c-Fos, NFATc1, OSCAR, TRAP, and CSF1R. RNA was isolated on the indicated days after stimulation with M-CSF and RANKL. Day 0 indicates the day RANKL was added to the BMM cultures. B, BMMs were cultured for 2 days with M-CSF and RANKL. The cultured cells were starved in 0.5% FBS for 6 h and then stimulated with 500 ng/ml RANKL (left panel) or anti-OSCAR antibodies cross-linked with secondary anti-rat IgG antibodies (right panel) for the indicated period time in the presence of 1 μg/ml heat-inactivated murine LRRc17 (mLRRc17) or murine LRRc17. Whole-cell extracts were subjected to Western blot analysis with specific antibodies as indicated. The numbers below the lanes indicate the fold induction of PLCγ2 phosphorylation (pPLCγ2) relative to control samples.
Taken together, these results suggest that the inhibitory activity of LRRc17 functions through a down-regulation of NFATc1 expression. To test this directly, we examined whether overexpression of NFATc1 counteracted the inhibitory effect of LRRc17 on RANKL-mediated osteoclastogenesis. LRRc17-Fc significantly attenuated RANKL-induced osteoclast formation in control, vector-infected BMMs. Overexpression of constitutively active NFATc1, however, overcame the inhibitory effect of LRRc17 on osteoclastogenesis (Fig. 7). Therefore, these data indicate that LRRc17 attenuates RANKL-induced up-regulation of NFATc1 expression, which, in turn, inhibits osteoclast differentiation.
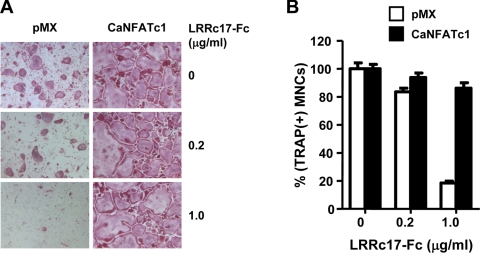
FIGURE 7.
Overexpression of NFATc1 overcomes the inhibitory effect of LRRc17 on RANKL-induced osteoclastogenesis. BMMs were transduced with pMX-IRES-EGFP (pMX, control) or retrovirus containing sequence for a constitutively active form of NFATc1 (CaNFATc1), where IRES indicates internal ribosomal entry site and EGFP indicates enhanced green fluorescent protein. BMMs were cultured for 4 days with M-CSF and RANKL in the presence of various concentrations of murine LRRc17-Fc or control IgG as indicated. A, cultured cells were fixed and stained for TRAP. B, TRAP+ MNCs with more than three nuclei were counted as osteoclasts.
DISCUSSION
Osteoblasts and osteoclasts are the principal cell types responsible for bone remodeling. Various factors and hormones continuously regulate these cells to maintain bone homeostasis. Osteoblasts produce bone matrix and regulate the differentiation and activity of osteoclasts by responding to osteotropic factors and producing such cytokines as M-CSF and RANKL. To identify proteins produced by osteoblasts that negatively regulate osteoclast formation, we compared the mRNA profiles of osteoblasts stimulated with 1,25(OH)2D3 with those of the NIH3T3 fibroblastic cell line, which does not support osteoclastogenesis even in the presence of osteotropic factors. As a result, we identified LRRc17, a secreted protein composed of LRR domains; expression of this protein is down-regulated in osteoblasts in response to proosteoclastogenic factors, resulting in the inhibition of osteoclastogenesis.
Using flow cytometry and an LRRc17-Fc fusion protein, we demonstrated that a putative LRRc17 receptor is expressed on the surface of osteoclast precursors, including BMMs and RAW264.7 cells (supplemental Fig. S3). We also found that LRRc17 attenuated RANKL-induced expression of NFATc1. Thus, our data suggest that LRRc17 acts directly on osteoclast precursors through a putative LRRc17 receptor. In addition to osteoclasts, osteoblasts also express a surface molecule that can interact with LRRc17 (supplemental Fig. S3). LRRc17, however, did not affect the osteoblastic expression of M-CSF and RANKL driven by such osteotropic factors as 1,25(OH)2D3 (supplemental Fig. S2). In addition, we did not observe any effects of recombinant LRRc17 on osteoblast differentiation and proliferation, suggesting that the effects of LRRc17 on bone remodeling are limited to osteoclasts. We, however, cannot rule out the possibility that LRRc17 may affect osteoblast lineage cells or other cell types in vivo. Detailed studies using LRRc17-deficient mice are needed to elucidate the physiological roles of LRRc17 beyond osteoclasts.
Macrophages, osteoclasts, and dendritic cells are derived from common precursors, and consequently, share a number of molecular signatures. Indeed, many factors that are critical for osteoclasts, such as RANKL, also regulate macrophages or dendritic cells (4, 8). Therefore, it is not unexpected that a putative LRRc17 receptor was also detected in macrophage/monocyte lineage cells. When we examined whether LRRc17 affected the differentiation of BMMs into each of these cell types, however, we found that LRRc17 does not appear to affect the phagocytic activity of macrophages or the differentiation of BMM into dendritic cells. These results suggest that the effects of LRRc17 may be limited to the fate of osteoclasts rather than such immunocytes as macrophages and dendritic cells.
The growing family of proteins containing LRR domains includes intracellular, extracellular, and cell-surface proteins that control a diverse range of physiological processes, including bone metabolism. For example, biglycan and decorin, two members of the small leucinerich repeat proteins and proteoglycans, are highly expressed in extracellular bone matrix, where they influence the differentiation and proliferative activity of bone cells (25). Small leucine-rich repeat proteins and proteoglycan-deficient mice develop an osteoporotic phenotype, characterized by a failure to achieve peak bone mass due to decreased bone formation (26). LRRc17, which is also abundantly expressed in osteoblast cells, appears to play a different role than biglycan and decorin. In fact, LRRc17 directly regulates the differentiation of osteoclasts rather than osteoblasts.
OPG is the best characterized inhibitor of osteoclast differentiation. When compared with LRRc17, however, OPG is more broadly expressed and has been detected in lung, liver, heart, and kidney (27). Nevertheless, the expression of both LRRc17 and OPG is suppressed in osteoblasts in response to the pro-osteoclastogenic factor 1,25(OH)2D3. Although both secreted molecules act as negative regulators of RANKL-induced osteoclastogenesis, the underlying inhibitory mechanisms are clearly different. OPG directly binds to RANKL, whereas LRRc17 does not (supplemental Fig. S4). Instead, LRRc17 binds to a putative receptor on osteoclast precursor cells and inhibits RANKL-induced NFATc1 expression by blocking PLCγ signaling. In general, when RANKL expression is up-regulated, OPG expression is relatively suppressed, although the published data are somewhat contradictory (28, 29). Laboratory studies have shown that the relative expression levels of OPG and RANKL are critical for the regulation of osteoclastic activity, and in turn, physiologic and pathologic bone turnover. Studies designed to assess the relationship between serum OPG and RANKL levels and bone metabolism/osteoporosis in postmenopausal women, however, have yielded conflicting results (28), suggesting that other factors, such as LRRc17, may be involved in pathogenic bone metabolism. Further examination of the LRRc17 expression levels in animal models and osteoporotic patients are needed to elucidate the roles of LRRc17 in bone metabolism and diseases.
In conclusion, our data strongly suggest that LRRc17 functions as an inhibitory molecule for osteoclastogenesis. In addition, the regulation of LRRc17 expression in osteoblasts by 1,25(OH)2D3 suggests that LRRc17 is produced by osteoblasts and contributes to the interactions between osteoblasts and osteoclasts, which are critical to ensuring proper regulation of bone metabolism in response to osteotropic factors. To our knowledge, LRRc17 is the first LRR superfamily member that has been shown to directly regulate osteoclastogenesis. Of note, in addition to RANKL, RANK, and OPG, which are critical for osteoclast differentiation, LRRc17 appears to be another important regulator of osteoclast development. Therefore, our work highlights an additional regulatory layer in bone homeostasis. Further studies examining the detailed mechanisms of this regulation will allow for a clearer understanding of the roles of LRRc17 and its potential as a therapeutic target to treat bone diseases, including osteoporosis and rheumatoid arthritis.
Supplementary Material
This work was supported, in whole or in part, by National Institutes of Health Grant AR053843 (to Y. C.) through the NIAMS. This work was also supported in part by the Korea Science and Engineering Foundation National Research Laboratory Program grant funded by the Korean government (MEST) (Grant R0A-2007-000-20025-0) (to N. K.) and Grant R13-2002-013-03001-0 from the Korea Science and Engineering foundation through the Medical Research Center for Gene Regulation at Chonnam National University.
The on-line version of this article (available at http://www.jbc.org) contains four supplemental figures.
Footnotes
The abbreviations used are: RANK, receptor activator of NF-κB; RANKL, RANK ligand; OPG, osteoprotegerin; LRR, leucine-rich repeat; LRRc17, leucinerich repeat-containing 17; PLCγ, phospholipase C-γ; JNK, c-Jun NH2-terminal kinase; ERK, extracellular signal-regulated kinase; MAP, mitogen-activated protein; MITF, microphthalmia transcription factor; 1,25(OH)2D3, 1,25-dihydroxyvitamin D3; PTH, parathyroid hormone; M-CSF, macrophage colony-stimulating factor; GM-CSF, granulocyte-macrophage colony-stimulating factor; OSCAR, osteoclast-associated receptor; α-MEM, α-minimum essential medium; FBS, fetal bovine serum; BMM, bone marrow-derived macrophage; TRAP, tartrate-resistant acid phosphatase; MNC, multinucleated cell; FACS, fluorescence-activated cell sorter; ALP, alkaline phosphatase; LPS, lipopolysaccharide.
References
- 1.Lacey, D. L., Timms, E., Tan, H. L., Kelley, M. J., Dunstan, C. R., Burgess, T., Elliott, R., Colombero, A., Elliott, G., Scully, S., Hsu, H., Sullivan, J., Hawkins, N., Davy, E., Capparelli, C., Eli, A., Qian, Y. X., Kaufman, S., Sarosi, I., Shalhoub, V., Senaldi, G., Guo, J., Delaney, J., and Boyle, W. J. (1998) Cell 93 165-176 [DOI] [PubMed] [Google Scholar]
- 2.Suda, T., Takahashi, N., Udagawa, N., Jimi, E., Gillespie, M. T., and Martin, T. J. (1999) Endocr. Rev. 20 345-357 [DOI] [PubMed] [Google Scholar]
- 3.Teitelbaum, S. L. (2000) Science 289 1504-1508 [DOI] [PubMed] [Google Scholar]
- 4.Walsh, M. C., Kim, N., Kadono, Y., Rho, J., Lee, S. Y., Lorenzo, J., and Choi, Y. (2006) Annu. Rev. Immunol. 24 33-63 [DOI] [PubMed] [Google Scholar]
- 5.Yasuda, H., Shima, N., Nakagawa, N., Yamaguchi, K., Kinosaki, M., Mochizuki, S., Tomoyasu, A., Yano, K., Goto, M., Murakami, A., Tsuda, E., Morinaga, T., Higashio, K., Udagawa, N., Takahashi, N., and Suda, T. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 3597-3602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boyle, W. J., Simonet, W. S., and Lacey, D. L. (2003) Nature 423 337-342 [DOI] [PubMed] [Google Scholar]
- 7.Lee, Z. H., and Kim, H. H. (2003) Biochem. Biophys. Res. Commun. 305 211-214 [DOI] [PubMed] [Google Scholar]
- 8.Takayanagi, H. (2005) J. Mol. Med. 83 170-179 [DOI] [PubMed] [Google Scholar]
- 9.Hirotani, H., Tuohy, N. A., Woo, J. T., Stern, P. H., and Clipstone, N. A. (2004) J. Biol. Chem. 279 13984-13992 [DOI] [PubMed] [Google Scholar]
- 10.Takayanagi, H., Kim, S., Koga, T., Nishina, H., Isshiki, M., Yoshida, H., Saiura, A., Isobe, M., Yokochi, T., Inoue, J., Wagner, E. F., Mak, T. W., Kodama, T., and Taniguchi, T. (2002) Dev. Cell 3 889-901 [DOI] [PubMed] [Google Scholar]
- 11.Koga, T., Inui, M., Inoue, K., Kim, S., Suematsu, A., Kobayashi, E., Iwata, T., Ohnishi, H., Matozaki, T., Kodama, T., Taniguchi, T., Takayanagi, H., and Takai, T. (2004) Nature 428 758-763 [DOI] [PubMed] [Google Scholar]
- 12.Mocsai, A., Humphrey, M. B., Van Ziffle, J. A., Hu, Y., Burghardt, A., Spusta, S. C., Majumdar, S., Lanier, L. L., Lowell, C. A., and Nakamura, M. C. (2004) Proc. Natl. Acad. Sci. U. S. A. 101 6158-6163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kim, N., Takami, M., Rho, J., Josien, R., and Choi, Y. (2002) J. Exp. Med. 195 201-209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suda, T., Jimi, E., Nakamura, I., and Takahashi, N. (1997) Methods Enzymol. 282 223-235 [DOI] [PubMed] [Google Scholar]
- 15.Wong, B. R., Rho, J., Arron, J., Robinson, E., Orlinick, J., Chao, M., Kalachikov, S., Cayani, E., Bartlett, F. S., III, Frankel, W. N., Lee, S. Y., and Choi, Y. (1997) J. Biol. Chem. 272 25190-25194 [DOI] [PubMed] [Google Scholar]
- 16.Kim, K., Kim, J. H., Lee, J., Jin, H. M., Kook, H., Kim, K. K., Lee, S. Y., and Kim, N. (2007) Blood 109 3253-3259 [DOI] [PubMed] [Google Scholar]
- 17.Kim, K., Lee, J., Kim, J. H., Jin, H. M., Zhou, B., Lee, S. Y., and Kim, N. (2007) J. Immunol. 178 5588-5594 [DOI] [PubMed] [Google Scholar]
- 18.Lee, J., Kim, K., Kim, J. H., Jin, H. M., Choi, H. K., Lee, S. H., Kook, H., Kim, K. K., Yokota, Y., Lee, S. Y., Choi, Y., and Kim, N. (2006) Blood 107 2686-2693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim, K., Kim, J. H., Lee, J., Jin, H. M., Lee, S. H., Fisher, D. E., Kook, H., Kim, K. K., Choi, Y., and Kim, N. (2005) J. Biol. Chem. 280 35209-35216 [DOI] [PubMed] [Google Scholar]
- 20.Kim, D., LaQuaglia, M. P., and Yang, S. Y. (1996) Biochim. Biophys. Acta 1309 183-188 [DOI] [PubMed] [Google Scholar]
- 21.Clark, H. F., Gurney, A. L., Abaya, E., Baker, K., Baldwin, D., Brush, J., Chen, J., Chow, B., Chui, C., Crowley, C., Currell, B., Deuel, B., Dowd, P., Eaton, D., Foster, J., Grimaldi, C., Gu, Q., Hass, P. E., Heldens, S., Huang, A., Kim, H. S., Klimowski, L., Jin, Y., Johnson, S., Lee, J., Lewis, L., Liao, D., Mark, M., Robbie, E., Sanchez, C., Schoenfeld, J., Seshagiri, S., Simmons, L., Singh, J., Smith, V., Stinson, J., Vagts, A., Vandlen, R., Watanabe, C., Wieand, D., Woods, K., Xie, M. H., Yansura, D., Yi, S., Yu, G., Yuan, J., Zhang, M., Zhang, Z., Goddard, A., Wood, W. I., Godowski, P., and Gray, A. (2003) Genome Res. 13 2265-2270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Curtiss, N. P., Bonifas, J. M., Lauchle, J. O., Balkman, J. D., Kratz, C. P., Emerling, B. M., Green, E. D., Le Beau, M. M., and Shannon, K. M. (2005) Genomics 85 600-607 [DOI] [PubMed] [Google Scholar]
- 23.Teitelbaum, S. L., and Ross, F. P. (2003) Nat. Rev. Genet. 4 638-649 [DOI] [PubMed] [Google Scholar]
- 24.Takami, M., Kim, N., Rho, J., and Choi, Y. (2002) J. Immunol. 169 1516-1523 [DOI] [PubMed] [Google Scholar]
- 25.Waddington, R. J., Roberts, H. C., Sugars, R. V., and Schonherr, E. (2003) Eur. Cells Mater. 6 12-21 [DOI] [PubMed] [Google Scholar]
- 26.Ameye, L., and Young, M. F. (2002) Glycobiology 12 107R-116R [DOI] [PubMed] [Google Scholar]
- 27.Simonet, W. S., Lacey, D. L., Dunstan, C. R., Kelley, M., Chang, M. S., Luthy, R., Nguyen, H. Q., Wooden, S., Bennett, L., Boone, T., Shimamoto, G., DeRose, M., Elliott, R., Colombero, A., Tan, H. L., Trail, G., Sullivan, J., Davy, E., Bucay, N., Renshaw-Gegg, L., Hughes, T. M., Hill, D., Pattison, W., Campbell, P., Sander, S., Van, G., Tarpley, J., Derby, P., Lee, R., and Boyle, W. J. (1997) Cell 89 309-319 [DOI] [PubMed] [Google Scholar]
- 28.Kearns, A. E., Khosla, S., and Kostenuik, P. J. (2008) Endocr. Rev. 29 155-192 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Theoleyre, S., Wittrant, Y., Tat, S. K., Fortun, Y., Redini, F., and Heymann, D. (2004) Cytokine Growth Factor Rev. 15 457-475 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.