Abstract
The tumor suppressor gene Lot1 is highly expressed during brain development. During cerebellar development, Lot1 is expressed by proliferating granule cells with a time course matching the expression of the pituitary adenylate cyclase-activating polypeptide (PACAP) receptor, a neuropeptide receptor that plays an important role in the regulation of granule cell proliferation/survival. Although it has become clear that Lot1 is a negative regulator of cell division in tumor cells, its role in neuronal proliferation is not understood. We previously demonstrated that in cerebellar granule cells Lot1 expression is regulated by the PACAP/cAMP system. The aim of this study was to investigate the role played by Lot1 in neuron proliferation/survival and to identify the molecular mechanisms underlying its actions. Using a Lot1-inducible expression system, we found that in PC12 cells Lot1 negatively regulates proliferation and favors differentiation by up-regulating the expression of the PACAP receptor. In cerebellar granule cells in culture, an increase in Lot1 expression was paralleled by inhibition of proliferation and up-regulation of the PACAP receptor, which in turn positively regulated Lot1 expression. Silencing of Lot1 leads to an increase in granule cell proliferation and a reduction in survival. Confirming the in vitro results, in vivo experiments showed that PACAP induced an increase in Lot1 expression that was paralleled by inhibition of cerebellar granule cell proliferation. These data show that Lot1 is a key element of the PACAP/cAMP pathway that negatively regulates neuronal precursor proliferation. The existence of a PACAP receptor/Lot1-positive feedback loop may powerfully regulate neural proliferation during critical phases of cerebellar development.
Lot1 (lost on transformation 1) (1) is a tumor suppressor gene member of a new family of zinc finger proteins, the PLAG (pleiomorphic adenoma gene) family, which includes Lot1 (PLAGL1), PLAGL, and PLAGL2 (2). The mouse ortholog of rat Lot1 was independently identified by Spengler et al. (3) and designated as Zac1, which is highly homologous to the human and rat Lot1 genes.
Lot1 was first considered as a tumor suppressor gene based on its ability to induce cell cycle arrest and apoptosis, when misexpressed in epithelial cell lines (3, 4). Its expression is frequently down-regulated in the ovarian and breast carcinoma cells (1, 5, 6), and it is localized on chromosome 6q24-25, a common site for loss of heterozygosity in many solid tumors (3, 7). In addition, Lot1 is maternally imprinted, a mode of epigenetic control of gene expression levels that is common to many genes involved in growth control (8, 9). Functional analysis of Lot1 demonstrated that it may cause growth suppression as a target of mitogenic signaling pathways (10). The growth inhibitory potential of Lot1, the patterns of its expression in cancer cells, and its chromosomal localization confirm the hypothesis that this gene is a suitable candidate as a tumor suppressor.
Interestingly, Lot1/Zac1 has been shown to be abundantly expressed in many proliferative areas during brain development (11-15). Comparative analysis of the expression profiles of the three PLAG family members showed that these genes are expressed in both unique and overlapping patterns both in the central and peripheral nervous systems (15), suggesting that these PLAG genes might control cell fate and proliferation decisions during brain development. In recent studies, we demonstrated that Lot1 was expressed by proliferating neuronal precursor cells during neurogenesis (13, 16). Lot1 expression was observed during the first postnatal week in the external granule cell layer of the cerebellum (13), composed primarily of proliferating cerebellar granule cell precursors. We also showed that primary cultures of cerebellar granule cells exhibit a temporal pattern of Lot1 expression resembling that of in vivo development, with mRNA and protein levels progressively decreasing with differentiation (13, 16). Furthermore, in cerebellar granule cells, Lot1 expression was found to be controlled by cAMP cascade activated by the pituitary adenylate cyclase-activating polypeptide (PACAP),2 which suggested that this gene may be an important element of the cAMP-mediated pathway that negatively regulates neuronal proliferation and favors differentiation (16). Interestingly, it has been recently demonstrated that Zac1 is a negative regulator of retinal size, controlling the absolute number of rod and amacrine cells generated during development (17).
PACAP is a neuropeptide belonging to the vasoactive intestinal peptide family and is highly expressed both in the central and peripheral nervous system. Several lines of evidence suggest that PACAP, besides having neurotrophic and neuroprotective actions on various neural cell types, enhances neuron differentiation (18-22) and acts as an anti-mitogenic signal in developing cerebral cortex (23). PACAP-38 binds with high affinity to three G-protein-coupled receptors, PAC1-R, VPAC1-R, and VPAC2. All three receptors activate adenylate cyclase through Gs coupling, whereas PAC1-R also activates phospholipase C through Gq coupling (24). It has been shown that in the PC12 cell line PACAP induces neurite generation (25, 26) through the cAMP pathway (27, 28). The finding that a correlation exists between the expression patterns of Lot1/Zac1 and PAC1-R in some brain areas during development (11, 13), that Lot1 expression is induced by the PACAP-cAMP system (16), and that Lot1 is capable of inducing transcription of the PAC1-R in different cell lines (29, 30), suggests that Lot1 may be a key element of the pathway that mediates the various PACAP actions.
Although it has become clear that Lot1 is a suppressor of cell division in tumor cells, its role in the regulation of neuronal proliferation during development is not understood. The aim of this study was to establish whether Lot1 expression affects proliferation/differentiation/survival of neuronal cells and to identify the molecular mechanisms that underlie its action. To this purpose we have used two neuronal culture systems as follows: (i) the pheochromocytoma PC12 cell line that represents one of the principal model systems that has been used to dissect the molecular details of neuronal proliferation/differentiation (31, 32), and (ii) primary cultures of cerebellar granule cell precursors that have been shown to express high levels of Lot1 after PACAP activation (16). In addition, to validate the results obtained in vitro, we used an in vivo approach to establish whether an increase in Lot1 expression induced by PACAP is accompanied by changes in granule cell proliferation.
EXPERIMENTAL PROCEDURES
Plasmids—The following reporter plasmids were used for this study: cmv-βGal, Tk-Luc, AP1-Luc, CRE-Luc (33), and Lot1-Luc (13). The inducible Lot1 reporter plasmid pIND-Lot1 was constructed by inserting the cDNA of rat Lot1 containing the HA epitope (hemagglutinin influenza, YPYDVPDYA) (13) into the HindIII/EcoRI sites of pIND1 vector (Invitrogen).
Cell Cultures and Treatments—Ecdysone-responsive (EcR) PC12 cells (34), established using the co-expression vector pVgRXR and Zeocin selection, were generously provided by Dr. H. Green (Harvard Medical School, Boston). Parental EcR PC12 cells and those stably transfected with pIND-Lot1 (EcR Lot1) were grown on polystyrene tissue culture plates or dishes, coated with poly-d-lysine (20 μm, Sigma) in Dulbecco's modified Eagle's medium (Sigma) supplemented with 10% horse serum (Sigma), 5% plasma-derived fetal bovine serum (Sigma), and 1% penicillin/streptomycin (Invitrogen) at 37 °C in a humidified atmosphere with 5% CO2. Cells were split at 1-week intervals by dissociation with trypsin/EDTA with a midweek change of medium. Starting 24 h after plating, medium was aspirated and replenished every other day. Ponasterone A (Invitrogen) at the concentrations indicated in figure legends (Figs. 1, 2, 3, 4), 10 μm forskolin (Sigma), 10 nm PACAP-38 (Sigma), 50 ng/ml mouse NGF (Sigma), 1 μm PACAP6-38 (Bachem), 10 μm H89 (PKA inhibitor, Calbiochem), or vehicle was added after each change of medium. Primary cultures of cerebellar granule cell precursors (GCP) were prepared from the cerebella of 7-day-old Wistar rat pups as described previously (35). Cells were plated on poly-d-lysine (20 μm, Sigma)-coated dishes at the density of 2 × 103 cells/mm2 and maintained in B27-supplemented neurobasal medium (Invitrogen) supplemented with 2 mm glutamine and 0.05 mg/ml gentamicin (Sigma). GCP were treated 2 h after plating with either 10 μm forskolin, 10 nm PACAP-38, or 1 μm PACAP6-38.
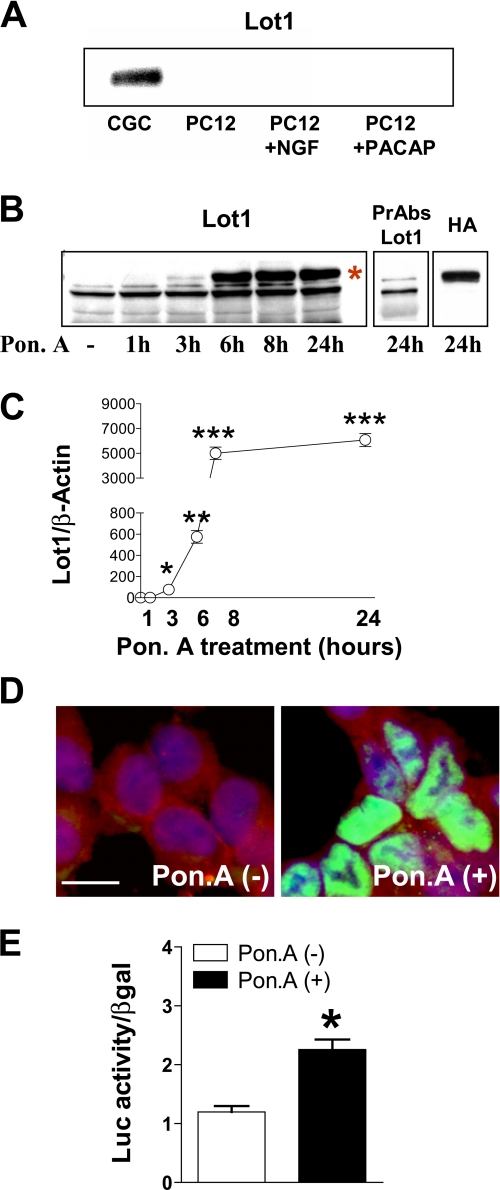
FIGURE 1.
Inducible expression of Lot1 in PC12 cells. A, expression of Lot1 protein was assayed by Western blot on nuclear extracts of cerebellar GCP at 1 day in vitro and of PC12 cells untreated and treated with NGF (50 ng/ml) or PACAP-38 (10 nm) for 5 days. B, expression of Lot1 protein was assayed by Western blot in total cell extracts of EcR Lot1 cells treated with ponasterone A (Pon. A) (10 μm) for the indicated times. The asterisk indicates the band corresponding to Lot1 protein. On the right are shown immunoblots of homogenates of EcR Lot1 cells, treated with ponasterone A (10 μm) for 24 h, and exposed to preabsorbed anti-Lot1 serum (PrAbs Lot1) and anti-HA antibody (HA) to check for the specificity of the band corresponding to Lot1 protein. C, Lot1 protein levels following treatment with ponasterone A were normalized to β-actin content. Bars are the mean ± S.E. of three experiments. *, p < 0.05; **, p < 0.01; ***, p < 0.001; Bonferroni's test after ANOVA. D, confocal microscope images of EcR Lot1 cells untreated (panel on the left) or stimulated with ponasterone A (10 μm)(panel on the right) and double-immunostained for Lot1 (green) and N-CAM (red). Cell nuclei were stained by Hoechst dye (blue). Scale bar, 20 μm. E, luciferase reporter analysis of Lot1 promoter. EcR Lot1 cells were transfected (as indicated under “Experimental Procedures”) with 1.5 μg of Lot1 luciferase reporter plasmid (Lot1-Luc) and 0.5 μg of the β-galactosidase reporter plasmid (pCMV-βGal). Twenty four hours after transfection cells were incubated with (black bars) or without (white bars) 10 μm ponasterone A for a further 24 h. Bars are the mean ± S.E. of three experiments. *, p < 0.01 (two-tailed t test).
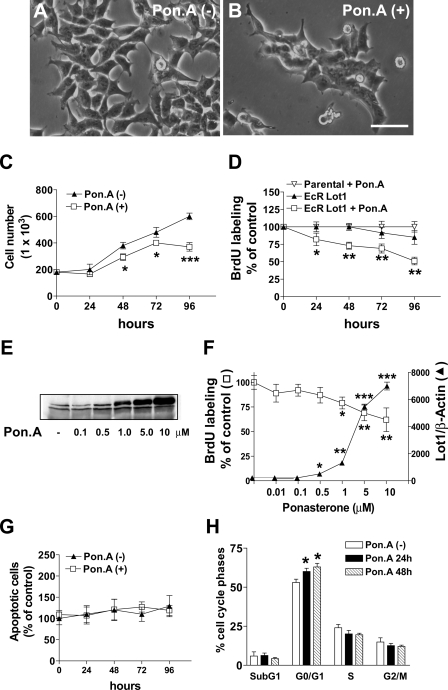
FIGURE 2.
Effect of Lot1 expression on proliferation, cell death, and cell cycle progression in PC12 cells. A and B, phase contrast images of EcR Lot1 cells cultured without (A) or with (B) ponasterone A (Pon. A) (10 μm) for 2 days. Scale bar, 50 μm. C, EcR Lot1 cells were cultured with or without ponasterone A (10 μm) for the indicated periods, after which cells were dissociated, stained by trypan blue, and counted on a hemocytometer. Data are expressed as the mean ± S.E. of three independent experiments. D, EcR Lot1 cells and parental PC12 cells were stimulated as indicated in C, and BrdUrd (10 μm) was added for the last 2 h, and thereafter cells were processed for a proliferation BrdUrd assay (see “Experimental Procedures”). Data, given as % of untreated conditions, are expressed as the mean ± S.E. of five independent experiments. E, ponasterone A dose-dependent expression of Lot 1 in EcR Lot1 cells. EcR Lot1 cells were grown with varying concentrations of ponasterone A for 48 h and then assayed for Lot1 expression by immunoblotting. F, relationship between Lot1 expression (solid triangle; normalized toβ-actin content) and cell proliferation (open squares; evaluated with a BrdUrd incorporation assay) at different concentrations of ponasterone A. Data of proliferation assay are given as % of untreated condition. Data are the mean ± S.E. of 3-4 experiments. G, EcR Lot1 cells were cultured without or with ponasterone A (10 μm) for the indicated periods, after which cells were processed for a cell death detection assay (see “Experimental Procedures”). Data, given as % of untreated conditions, are the mean ± S.E. of three independent experiments. H, flow cytometry analysis of DNA content in EcR Lot1 cells cultured without or with ponasterone A (10 μm) for 1 or 2 days. Bars are the mean ± S.E. of three experiments. The asterisks (C, D, and F-H) indicate a significant difference between the treated versus untreated condition; *, p < 0.05; **, p < 0.01; ***, p < 0.001; Bonferroni's test after ANOVA.
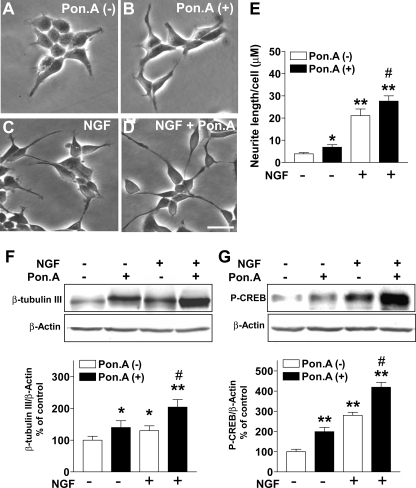
FIGURE 3.
Effect of Lot1 on NGF-induced neurite outgrowth in PC12 cells. A-D, phase contrast images showing the morphology of EcR Lot1 cells. EcR Lot1 cells were incubated for 48 h in the absence (A) or presence (B) of 10 μm ponasterone A (Pon. A), 50 ng/ml NGF (C), or 50 ng/ml NGF plus 10 μm ponasterone A (D) in serum-free medium. Scale bar, 40 μm. E, quantification of neurite outgrowth, shown as mean length (μm) per cell. Photomicrographs from six adjacent fields were taken for each cell line in each experiment. Data are expressed as mean ± S.E. of 3-6 independent experiments. *, p < 0.01; **, p < 0.001, treated versus untreated cells; #, p < 0.05 ponasterone A plus NGF versus NGF-only treated cells (Bonferroni's test after ANOVA). F and G, examples of immunoblotting with anti-β-tubulin III (F), anti-P-CREB (G), and anti-β-actin antibodies obtained from total cell extracts in the indicated different conditions. β-Tubulin III (F) and P-CREB (G) protein levels were normalized to β-actin content and expressed as percent of untreated condition. Bars are the mean ± S.E. of three independent experiments. *, p < 0.05; **, p < 0.01, treated versus untreated conditions; #, p < 0.01 ponasterone A plus NGF versus NGF-only treated cells (Bonferroni's test after ANOVA).
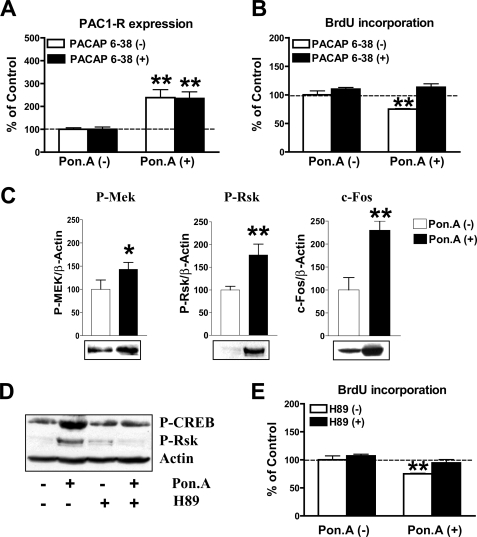
FIGURE 4.
Lot1 promotes PAC1-R expression and activation of PKA-dependent pathways in PC12 cells. A, PAC1-R mRNA expression, quantified by RT-qPCR, in EcR Lot1 cells cultured without or with ponasterone A (Pon. A) (10 μm) and in the presence or absence of the PAC1-R antagonist, PACAP6-38 (1 μm). Data are expressed as mean ± S.E. of three independent experiments. B, proliferation, evaluated with a BrdUrd assay, of EcR Lot1 cells exposed to the same conditions indicated in A. BrdUrd (10 μm) was added for the last 2 h in culture. Data are the mean ± S.E. of four independent experiments. C, Western blot analysis of Mek and Rsk phosphorylation and c-Fos expression in EcR-Lot1 cells cultured without or with ponasterone A (Pon. A) (10 μm), protein expressions were normalized to β-actin content. Bars are the mean ± S.E. of three independent experiments. Representative examples of Western blots are shown below the graphs. *, p < 0.05; **, p < 0.01; two-tailed t test. D, Western blot analysis of P-CREB, P-Rsk, and β-actin expression in EcR Lot1 cells cultured without or with ponasterone A (10 μm) and in the presence or absence of the PKA antagonist, H89 (10 μm). E, sister cultures were exposed to the same conditions as in D, and cell proliferation was evaluated with a BrdUrd assay. BrdUrd (10 μm) was added for the last 2 h in culture. Data, given as % of the untreated condition, are the means ± S.E. of four independent experiments. The asterisks (A, B, and E) indicate a significant difference between the treated versus untreated condition; *, p < 0.05; **, p < 0.01; Bonferroni's test after ANOVA.
Animals and Treatments—Wistar rats (Harlan, Italy) were kept in our animal facility under conditions of constant temperature and with 12-h light/12-h dark cycle. Pregnant mothers were isolated in single cages when pregnancy became evident. Day of birth was considered P0 and pups were treated at P7. The young rats were lightly anesthetized with ether and injected with 0.01 μg of PACAP-38 over 1 min under the skull by using a 25-μl microsyringe (Hamilton) connected to a needle (0.5-mm diameter) equipped with a guard limiting its penetration to 6 mm from the insertion point. The injection (15 μl) was made freehand at a point ∼6 mm behind the interaural line; the syringe was held horizontally so that the tip of the needle was positioned in the subarachnoid space between the skull and the surface of the cerebellum, as reported previously (36). Control experiments performed with trypan blue have shown that the needle did not penetrate into the neuroepithelium and that the dorsal part of the cerebellar hemispheres and vermis were completely bathed with the dye. After injection, the needle was left in situ for 1 min to avoid reflux on withdrawal. Twenty four hours after PACAP-38 injection, animals received a single subcutaneous injection (150 μg/g body weight) of 5-bromo-2-deoxyuridine (BrdUrd; Sigma), a marker of proliferating cells and their progeny (37) in 0.9% NaCl solution. Animals were sacrificed 2 h after the BrdUrd injection. Pups were killed by decapitation, and the cerebellum was rapidly dissected and fixed by immersion in Glyo-Fix (Thermo Electron Corp., Waltham, MA) for 24 h. Experiments were performed in accordance with the Italian and European Community Law for the use of experimental animals and were authorized by a local bioethical committee.
Transfection—PC12 cells were transfected with polyethyleneimine, 25 kDa (PEI-25, Sigma), as described previously (38, 39). Cells were plated 24 h before transfection on poly-d-lysine-coated plates or dishes. For stable transfection of the pIND plasmid, EcR PC12 cells were seeded ∼5 × 106 cells in 10-cm dishes and transfected with 10 μg of plasmid DNA. Cells expressing Lot1 (EcR Lot1) were selected by growth in Geneticin® (G418, 500 mg/ml; Invitrogen) and screened for expression by immunoblotting. For luciferase assays, cells were plated at 1 × 105 cells/24-well plate and transfected with 2 μg of plasmid DNA. 0.5 μg of β-galactosidase reporter plasmid (pCMV-βGal) was co-transfected together with 1.5 μg of Lot1 luciferase reporter plasmid (Lot1-Luc) (13) to normalize for transfection efficiency.
Luciferase Assay—Luciferase activity was measured as described previously (40) 48 h after transfection and normalized for β-galactosidase activity in the same sample (see below). Luciferase activity was measured with a TD 20/20 luminometer (Promega).
β-Galactosidase Assay—Cell lysates (20 μl) were added to 100 μl of β-galactosidase assay buffer (Na2HPO4 60 mm, NaH2PO4 40 mm, KCl 10 mm, MgCl2 1 mm, β-mercaptoethanol 0.34%, pH 7.5) containing 20 mg/ml o-nitrophenyl β-d-galactopyranoside (Sigma) and incubated for 1 h at 37 °C. Reaction was stopped with 50 μl of Na2CO3 1 m, and absorbance at 420 nm was measured on a Benchmark multiplate reader (Bio-Rad).
Immunofluorescence—For immunofluorescence studies in PC12 cells, the following antibodies were used: anti-N-CAM (monoclonal antibody at 1:200; Sigma) and anti-Lot1 (polyclonal antibody at 1:100; see Ref. 13). Cells, plated on poly-d-lysine-coated coverslips at 5 × 104 cells/well (24-well plates), were fixed for 30 min in 4% paraformaldehyde, 4% sucrose in 120 mm sodium phosphate buffer, pH 7.4, and then rinsed three times with PBS. Coverslips were incubated overnight at 4 °C with appropriate dilutions of the primary antibody in the following blocking buffer: 1.5% goat serum, 0.1% Triton X-100 in PBS, pH 7.4. Cells were then incubated with Cy3-conjugated goat anti-mouse secondary antibody or with fluorescein isothiocyanate-conjugated goat anti-rabbit secondary antibody (dilution 1:200; Sigma) for 1-2 h at room temperature. After all incubations, specimens were extensively washed with PBS containing 0.1% Triton X-100. Coverslips were mounted on glass slides in PBS containing 70% glycerol and Hoechst 33342 (2 μg/ml). Phase contrast and fluorescence images were taken with an Eclipse TE 2000-S microscope (Nikon, Tokyo, Japan) equipped with an AxioCam MRm digital camera (Zeiss, Oberkochen, Germany).
Cell Proliferation Assay—EcR PC12 parental and EcR Lot1 cells were plated on poly-d-lysine-coated 96-well plates at 1 × 104 cells/well. At the end of the different treatments (described in detail under “Results” and Figs. 2 and 4 legends), cells were incubated with 10 μm BrdUrd for an additional 2 h. BrdUrd incorporation was measured in triplicate with the cell proliferation ELISA kit (Roche Applied Science) according to the manufacturer's instructions. Data were normalized versus total protein level and evaluated with the Lowry protein assay (41).
Cell Count—EcRLot1 cells ± ponasterone A at various time points were dissociated, gently centrifuged, and resuspended in 5 ml of PBS, pH 7.4. An aliquot containing 0.4% trypan blue was incubated at room temperature for 5 min, and cells were counted on a hemocytometer. Only cells that excluded the blue dye and had a well defined cellular outline were scored as live and were counted.
Real Time Reverse Transcriptase Quantitative PCR (RT-qPCR)—Total RNA was prepared from EcRLot1 cells, GCP, or cerebella with TRI Reagent® (Sigma) according to the manufacturer's instructions. cDNA synthesis was achieved with 1 μg of total RNA using the iScript™ cDNA synthesis kit according to manufacturer's instructions. The primer sequences used are as follows: (i) Lot1 (NM_012760), forward 5′-TTAGCTGCGTAGTTGCGTGTTA-3′ and reverse 5′-CGGGTCCCTGAAAAGAACACA-3′; (ii) glyceraldehyde-3-phosphate dehydrogenase (NM_008084), forward 5′-GAACATCATCCCTGCATCCA-3′ and reverse 5′-CCAGTGAGCTTCCCGTTCA-3′; (iii) PACAP receptor (PAC1-R) (NM_133511), forward 5′-CCCTCGCCACCCTCACTAC-3′ and reverse 5′-CCTTGATGAAGACGGAGATAGCC-3′; and (iv) cyclin-dependent kinase inhibitor 1C (p57Kip2) (NM_001033757), forward 5′-CTGACTTTCGCCAAGC-3′ and reverse 5′-AACTAACTCATCGCAGACG-3′. Real time PCR was performed using iQ™SYBR GREEN supermix (Bio-Rad), according to the manufacturer's instructions, on an iCycler iQ real time PCR detection system (Bio Rad). For each experiment the reactions were done in triplicate. Fluorescence was determined at each step of every cycle. Real time PCR assay was done under the following universal conditions: 2 min at 50 °C, 10 min at 95 °C, 50 cycles of denaturation at 95 °C for 15 s, and annealing/extension at 60 °C for 1 min. In all cases, a single product of the appropriate size was detected by gel electrophoresis, and the analysis of the melting curve excluded the presence of dimerized primers (data not shown). Relative quantification was performed using the comparative CT method, in which arithmetic formulas are used to obtain the same result as the one yielded by the relative standard curve method. The comparative CT method was used because the target gene and the reference control gene (glyceraldehyde-3-phosphate dehydrogenase) had similar amplification efficiency.
Western Blotting—The following antibodies were used: antiphosphorylated Erk1/2 (polyclonal antibody at 1:1000), anti-phosphorylated-RSK (polyclonal antibody at 1:1000), and anti-phosphorylated MEK1/2 (polyclonal antibody at 1:1000) (Cell Signaling Technologies, Beverly, MA); anti-phosphorylated CREB (polyclonal antibody at 1:2000) (Upstate Biotechnology, Inc., Lake Placid, NY); anti-HA (monoclonal antibody, at 1:500) (Santa Cruz Biotechnology); anti-Lot1 (polyclonal antibody at 1:1000) (13); anti-c-Fos (polyclonal antibody at 1:20,000), anti-β-tubulin III (polyclonal antibody at 1:1000), and anti-β-actin (polyclonal antibody at 1:2000) (Sigma). The specificity of the Lot1 immunohistochemical staining was confirmed by replacing the primary anti-Lot1 serum with the antibody pre-absorbed as described previously (13). For the preparation of total cell extracts, cells were lysed in lysis buffer (Tris-HCl 10 mm, SDS 2%, dithiothreitol 10 mm, protease and phosphatase inhibitor mixtures 1%). Cell extracts were immediately processed by Western blot or kept frozen (-80 °C) until assayed. Protein concentration of samples was estimated by the Lowry method (41). Equivalent (15 μg) amounts of proteins per sample were subjected to electrophoresis on a 10% SDS-polyacrylamide gel. The gel was then blotted onto a nitrocellulose membrane, and equal loading of protein in each lane was assessed by brief staining of the blot with 0.1% Ponceau S. Blotted membranes were blocked for 1 h in 5% milk in TBS (Tris-HCl 10 mm, NaCl 150 mm, pH 8.0) plus Tween 20 0.1% and then incubated overnight at 4 °C with primary antibodies. Membranes were washed and incubated for 1 h at room temperature with peroxidase-conjugated anti-rabbit IgG (dilution 1:1000; Amersham Biosciences). Specific reactions were revealed with the ECL Western blotting detection reagent (Amersham Biosciences).
Fluorescence Cell Sorter—A flow cytometric method was used to assess the cell cycle. PC12 cells were washed with PBS and harvested after treatment with ponasterone A for either 24 or 48 h. The pellet was incubated in ice (8-10 min) and then resuspended in 50 μg/ml propidium iodide containing Triton 0.1%, tri-sodium citrate 0.1%, and 0.1 mg/ml RNase (Sigma) at 4 °C. Cell cycle analysis was performed by using a FACSCalibur (BD Biosciences) to determine the percentage of cells in the G0-G1,S,G2 + M phases of the cell cycle.
Analysis of Neurite Outgrowth—PC12 cells differentiation was assessed by plating cells on poly-d-lysine-coated 6-well plates (35 × 104 cells/well), switched to serum-free medium for 1 h, and followed by stimulations as indicated in the legend of Fig. 3. Phase contrast photographs of the cultures were taken at various time intervals with an Eclipse TE 2000-S microscope (Nikon, Tokyo, Japan) equipped with an AxioCam MRm (Zeiss, Oberkochen, Germany) digital camera. Ten different areas were randomly selected, and neurite outgrowth was measured using the image analysis system Image Pro Plus (Media Cybernetics, Silver Spring, MD). Only cells with neurites longer than one cell body diameter were considered as neurite-bearing cells. All experiments were performed at least three times. The total length of neurites was divided for the total number of cells counted in the areas.
Cell Death—The Cell Death Detection ElisaPlus kit (Roche Applied Sciences) was used to determine apoptosis using the protocol provided by the manufacturer. This kit is based on a quantitative sandwich enzyme immunoassay principle, using mouse monoclonal antibodies directed against DNA and histones, respectively. This allows the apoptosis-specific detection and relative quantification of mononucleosomes and oligonucleosomes that are released into the cytoplasm of cells dying from apoptosis. 1 × 104 cells/well were plated on poly-d-lysine-coated 96-well plates. Nucleosomes were photometrically detected at 405 nm, and the final absorbance was obtained by subtracting the observed absorbance of the negative control and the background.
Immunofluorescence and Determination of the Labeling Index in GCP Cultures—GCP, plated on poly-d-lysine-coated coverslips, were cultured for 18 h, then treated with 10 μm BrdUrd for an additional 6 h, fixed, and processed for BrdUrd and β-tubulin III (an early neuronal marker) double-fluorescence immunohistochemistry, and counterstained with Hoechst 33342, as described previously (16). Fluorescence images, taken from random microscopic fields (10-12 for each coverslip), were superimposed and used to determine the labeling index (LI), defined as percent of cells co-labeled with BrdUrd and β-tubulin III over total cell number in three independent experiments in duplicate. Pyknotic cells were excluded from the count of total cell number.
Antisense Experiment—To silence the expression of Lot1, experiments were performed by using an antisense oligonucleotide (5′-tcgTGGGTCTGGAGGtga-3′) specific for the 5′-coding region of rat Lot1 (RNA structure version 4.5; CLC RNA Workbench) and, as a control, with the same sequence with mixed basis (5′-cggTCAGAGTGAGTAGGgac-3′), a “scrambled” sequence that does not recognize any eukaryotic coding region (Basic Local Alignment Search Tool from NCBI (blast.ncbi.nlm.nih)). Both oligonucleotides were phosphorothioated to make them more resistant to RNase attack. Treatments were performed in GCP switched after 2 h in culture to serum-free medium for 30 min and exposed to different concentrations of the oligonucleotides for 24 h. All chemicals and oligonucleotides were from Sigma.
BrdUrd Immunohistochemistry and Cell Count—Cerebella from rat pups were embedded in paraffin and cut with a microtome in 8-μm-thick sections. One of eight sections was processed for BrdUrd immunohistochemistry as described previously (42). Cell count was done in the external granular layer (Fig. 8, C and D), the site of cerebellar granule cell production at this age, in three lobuli (II, III, and VI) of each sampled section. A random area in the external granule cell layer of each lobule was first traced, and all immunostained cells present within the traced area were then counted. The volume of the sampled region was obtained by multiplying the area by the section thickness. Cell number was expressed as cells/mm3.
FIGURE 8.
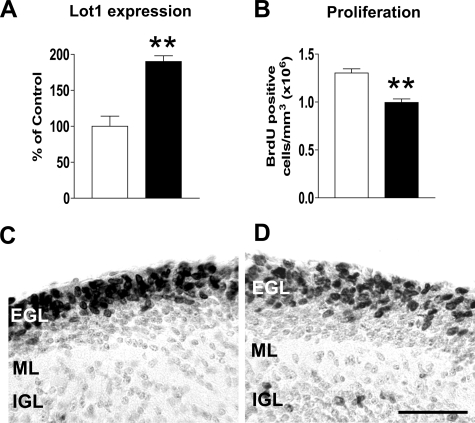
Effect of PACAP on Lot1 expression and GCP proliferation in vivo. A, relative quantification by RT-qPCR of Lot1 expression in cerebellar extract from P7 rat pups untreated (n = 4) or treated (n = 4) with PACAP-38 (0.01 μg). Animals were scarified 6 h after a PACAP-38 intracranial injection. Data, given as % of control condition, are expressed as the mean ± S.E. B, quantification of BrdUrd-positive cells in the cerebellum of untreated (n = 4) and PACAP-treated rat pups (n = 4). These animals received one intraperitoneal BrdUrd injection 22 h after a PACAP-38 intracranial injection and were sacrificed after 2 h. BrdUrd-positive cells were counted in the EGL and were expressed as BrdUrd positive cells/mm3. Values are means ± S.E.; **, p < 0.01 (two-tailed t test). C and D, photomicrographs of sagittal sections across the cerebellum of a P7 untreated (C) and PACAP-38 (0.01 μg) treated rat (D). Sections were taken at the level of the midline. These animals received one BrdUrd injection 22 h after a PACAP-38 intracranial injection and were sacrificed after 2 h. Sections were immunostained for BrdUrd and counterstained with toluidine blue. Scale bar, 100 μm. IGL, inner granular layer; ML, molecular layer.
Statistics—Data are expressed as means ± S.E., and statistical significance was assessed by one-way analysis of variance (ANOVA) followed by Bonferroni's post hoc tests. The two-tailed Student's t test was used when comparing two groups. A probability level of p < 0.05 was considered to be statistically significant. Statistical analysis was performed using GraphPad Prism 3.00 (GraphPad Software, San Diego).
RESULTS
Establishment of a Lot1-inducible Expression System in PC12 Cell Lines—The PC12 cell line is a model system widely used for peripheral neuronal proliferation/differentiation studies (32). We first examined whether PC12 cells exhibit a basal expression of Lot1, similarly to cerebellar GCP (13, 16). We found that, unlike GCP, PC12 cells did not exhibit a basal expression of Lot1 (Fig. 1A). This is in agreement with recent data suggesting loss of Lot1 expression in human pheochromocytomas (60). To establish whether Lot1 may be induced in differentiated PC12 cells, we exposed PC12 cells to either PACAP (PACAP-38) or NGF for 5 days. We found that Lot1 was also not expressed in differentiated PC12 cells (Fig. 1A). We took advantage of such a cellular model, where Lot1 is not endogenously expressed, to create PC12 cell lines in which Lot1 expression could be induced through the EcR-based gene regulation system. We used ponasterone A to activate the system and induce Lot1 expression in PC12 clones (EcR Lot1 clones).
The level of Lot1 expression, assessed at the protein level, was different in different EcR Lot1 cell clones (data not shown). For this study, we selected the EcR Lot1 clone that showed the highest induction of Lot1 expression. The time course of Lot1 induction by ponasterone A in EcR Lot1 cells was evaluated by Western blotting. The addition of ponasterone A (10 μm) to the EcR Lot1 cells rapidly stimulated (within 3 h) the expression of the Lot1 gene product (Fig. 1, B and C), with maximum induction at 8 h (Fig. 1, B and C). The pre-absorbed anti-Lot1 antibody specifically abolished the Lot1 band (labeled in Fig. 1B with an asterisk), and the anti-HA antibody recognized the HA epitope included in the EcR Lot1 plasmid (Fig. 1B), indicating the specificity of the Western blot band for Lot1. We used immunocytochemistry to verify the subcellular localization of Lot1 in EcR Lot1 cells stimulated with ponasterone A for 48 h. In agreement with previous evidence (3), we confirmed the nuclear localization of the induced protein (Fig. 1D).
We transiently transfected EcR Lot1 cells with a Lot-luciferase reporter plasmid (16). EcR Lot1 cells were then stimulated with ponasterone A (10 μm) for 24 h. Expression of Lot1 increased the activity of the Lot-luciferase reporter (Fig. 1E), thus demonstrating the ability of the inducibly expressed Lot1 to act as transcription factor in PC12 cells.
Effect of Lot1 on Cell Proliferation in PC12 Cell Lines—After 2 days of stimulation with ponasterone A, the number of EcR Lot1 cells underwent a reduction (Fig. 2B) as compared with unstimulated cells (Fig. 2A). We quantified the effects of treatment with ponasterone A (10 μm) on cellular proliferation by the following: (i) directly counting the number of cells (Fig. 2C) and (ii) BrdUrd incorporation (Fig. 2D). Lot1-expressing cells showed a significant reduction in the growth capacity after 24-48 h of ponasterone A stimulation. Maximum reduction occurred after 4 days and attained a value of -50% with respect to untreated EcR Lot1 cells (Fig. 2, C and D). Parental PC12 cells did not show any decrease in cell proliferation at each time point, irrespective of the presence of ponasterone A (Fig. 2D), indicating that ponasterone A per se did not alter PC12 cell proliferation.
Treatment of the EcR Lot1 cells with ponasterone A resulted in a concentration-dependent induction of Lot1 expression (Fig. 2E). Lot1 expression increased with the doses of ponaster-oneAupto10 μm (Fig. 2, E and F) during 48 h of exposure. Higher doses of ponasterone A did not further increase the amounts of produced Lot1 (data not shown). Dose-response curves revealed an inverse relationship between Lot1 expression and cell proliferation (Fig. 2F), suggesting that Lot1 was responsible for inhibition of cell proliferation. On the basis of these results, all subsequent experiments in EcR Lot1 cells were performed in the presence of 10 μm ponasterone A, the dose that induced maximum effects on proliferation.
Effect of Lot1 on Cell Death and Cell Cycle Progression in PC12 Cell Lines—Lot1 overexpression appears to exert apoptotic effects and induce cell cycle arrest in G0/G1 in different cell lines (3). We first assessed whether the reduced number of cells observed following stimulation of Lot1 expression was because of an increase in apoptotic cell death. Evaluation of apoptosis using a cell death enzyme-linked immunosorbent assay method showed no differences between cells that did and did not express Lot1 after 96 h of culture (Fig. 2G). This result was confirmed by flow cytometry analysis of the sub-G1 peak, which is considered to indicate the proportion of apoptotic cells over total (43). We found that the percentage of apoptotic cells was similar irrespective of Lot1 expression (Fig. 2H). These findings show that in this cellular system Lot1 expression did not induce cell death.
Flow cytometry analysis of DNA content (Fig. 2H) showed that following induction of Lot1 expression a significantly larger fraction of cells was in G0/G1 phase of the cell cycle (12% after 24 h and 20% at 48 h), compared with unstimulated cells. We found that a smaller fraction of cells was in S and G2 + M phases, although this difference was not statistically significant (Fig. 2H). Although flow cytometry analysis does not allow discrimination of the percentage of cells in G0 and G1, due to the similar DNA content in these phases, these data strongly suggest that the reduced proliferation observed following induction of Lot1 expression was because of cell cycle arrest in the G0/G1 phase.
Effect of Lot1 on Differentiation of PC12 Cell Lines—During development, cessation of neuronal proliferation is closely coupled with differentiation. Differentiation of PC12 cells, toward sympathetic and chromaffin-like phenotypes, can be induced by different trophic factors, including NGF and PACAP (44). We investigated whether Lot1 expression alone or by virtue of synergy with other agents could promote neuronal differentiation in PC12 cell lines. We found that Lot1 was able to induce a beginning of neuronal morphology in PC12 cells, as assessed by evaluation of neurite length (Fig. 3, A and E). Although untreated EcR Lot1 cells exhibited a polygonal morphology with occasional short fine processes (Fig. 3A), treatment with ponasterone A for 48 h induced a neurite length increase by +70% (Fig. 3, B and E). Although the effect of Lot1 was smaller than that induced by NGF (see below), these data clearly show that Lot1 expression was able to promote initial differentiation even in the absence of pro-differentiative stimuli. To analyze the effects of Lot1 expression on NGF-induced differentiation, EcR Lot1 cells were cultivated for 2 days in the presence of NGF ± ponasterone A (Fig. 3, C and D). After NGF treatment, PC12 cells exhibited short neurites within 24 h (data not shown) and more elongated neurites within 48 h (Fig. 3C). Co-treatment with NGF + ponasterone A induced a greater length increase (+700% versus untreated cells) (Fig. 3, D and E) than treatment with NGF alone (+500% versus untreated cells) (Fig. 3E).
The growth-associated protein β-tubulin type III is frequently used as a marker for differentiation in PC12 cells (45). EcR Lot1 cells stimulated with ponasterone A for 48 h exhibited a higher expression of β-tubulin III than untreated cells (Fig. 3F), which is in agreement with the appearance of neurites (see above). EcR Lot1 cells stimulated with NGF + ponasterone A expressed a higher level of β-tubulin III when compared with cells stimulated with either ponasterone A or NGF alone (Fig. 3F). CREB is a transcription factor that is a key target of the NGF-stimulated signaling pathway (46). Upon exposure of PC12 cells to NGF, CREB becomes phosphorylated (P-CREB) (46). and this phosphorylation promotes pro-differentiative gene expression. Exposure of EcR Lot1 cells to ponasterone A for 48 h induced a 2-fold increase in P-CREB (Fig. 3G), whereas exposure to NGF + ponasterone A induced a 4-fold increase (Fig. 3G). These data suggest that Lot1 alone is able to induce a partial differentiation of PC12 cells and to enhance the differentiation induced by NGF. The increase in P-CREB after induction of Lot1 expression raises the possibility that Lot1 might act by enhancing the signaling pathways downstream to NGF receptors.
Effect of Lot1 on PAC1-R Gene Expression and Activation of the PKA/CREB and the MEK/ERK Pathways in PC12 Cell Lines—In several cell lines, including PC12 cells, transfection of Lot1 induces the expression of type I PACAP receptor (PAC1-R) by transcriptional mechanisms (30). PACAP, a key signal neuropeptide, has been proposed as an autocrine regulatory factor that plays a role in the differentiation of neural cortical precursor cells (47) and neurite outgrowth in PC12 cells (25, 27, 48). In addition, PACAP has been shown to play a negative role in neuronal proliferation (22, 47, 49, 50).
To verify whether the effect of Lot1 expression on cell proliferation/differentiation in EcR Lot1 cells was mediated by an increased expression of PAC1-R, we first evaluated, by RT-qPCR, the mRNA level of PAC1-R 24 h after ponasterone A stimulation. As shown by Fig. 4A, Lot1 induced an increase (2.5-fold) of PAC1-R mRNA in EcR Lot1 cells. Lot1-induced PAC1-R expression was unchanged in the presence of a PAC1-R receptor antagonist, PACAP6-38 (Fig. 4A), indicating a direct transcriptional mechanism of Lot1 on PAC1-R expression. To establish whether the anti-proliferative and pro-differentiative actions of Lot1 required activation of PAC1-R, we investigated cell proliferation and differentiation in the presence of PACAP6-38. We found that the anti-proliferative effect of Lot1 (Fig. 4B; see also Fig. 2D) was reverted by co-incubation with PACAP6-38 (Fig. 4B), confirming that the anti-proliferative action of Lot1 on PC12 cells requires PAC1-R activation. We also found that the pro-differentiative effect of Lot1 was reverted by co-incubation with PACAP6-38 (data not shown).
Although PAC1-R is known to be positively coupled with the adenylate cyclase, phospholipase C, and calcium pathways (27), there is now evidence that the effects of its activation on cell proliferation and neurite outgrowth in PC12 cells are mainly mediated by an increase in cAMP levels (51). MEK/ERK phosphorylation, dependent on cAMP elevation (27), mediates the effect of PACAP on neurite outgrowth (26, 52). We used Western blot analysis to determine whether expression of Lot1/PAC1-R in EcR Lot1 cells induced activation of pathways downstream to the cAMP/PKA system. We found that EcR Lot1 cells have a low basal phosphorylation of CREB (Fig. 3G) and MEK (Fig. 4C). After 2 days of treatment with ponasterone A, there was a notably increased phosphorylation of CREB (Fig. 3G) and MEK (Fig. 4C), compared with unstimulated cultures. Because the activity of MEK has been demonstrated to cause activation of RSK (alternatively named 90-kDa ribosomal S6 kinase (p90rsk)) (53), as well as expression of the immediate early gene c-fos (54, 55), we examined the levels of phosphorylated RSK and of c-Fos. Fig. 4C shows that 2 days of treatment with ponasterone A caused a strong phosphorylation of RSK and induction of c-Fos (Fig. 4C).
We then investigated the role of PKA in the effects exerted by Lot1 on cell proliferation. EcR Lot1 cells were treated with ponasterone A, or left unstimulated, either in the presence or absence of the H89, a strong inhibitor of PKA. As indicated in Fig. 4E, in the presence of H89, Lot1 expression was not accompanied by a decrease in cell proliferation (Fig. 4E). In parallel, upon treatment with H89, the increase in phosphorylation of CREB as well as RSK induced by Lot1 was also abolished (Fig. 4D). These results suggested that the activation of PKA was essential for the anti-proliferative effects exerted by Lot1.
Relations between Lot1 and PAC1-R Expression and Cerebellar Granule Cell Precursor Proliferation—We have recently shown that GCP exhibit high levels of Lot1 expression, which progressively decrease with granule cell differentiation (13, 16), and that Lot1 expression is controlled by the PACAP/cAMP cascade (16).
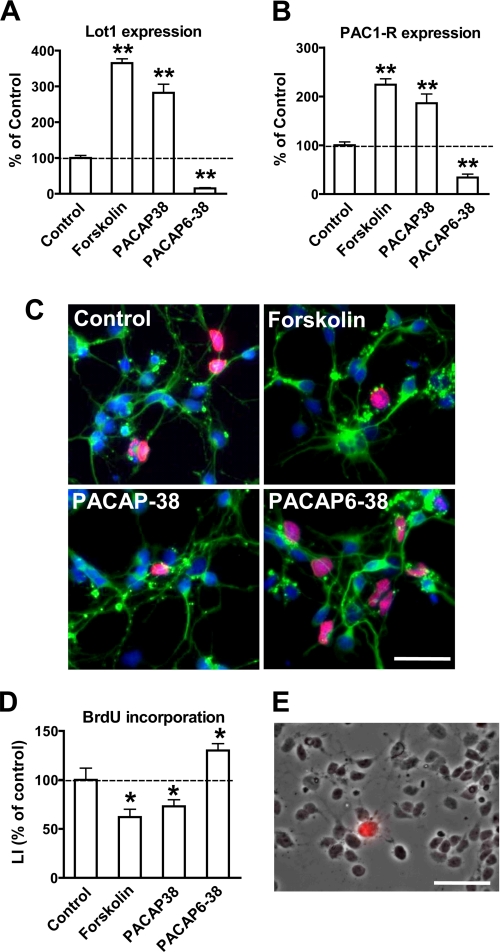
Confirming previous evidence (16), exposure of GCP in culture to either forskolin, a potent activator of adenylyl cyclase, or PACAP induced a notable increase in Lot1 mRNA levels (Fig. 5A). Conversely, exposure to an antagonist of PAC1-R, PACAP6-38, caused a dramatic decrease of Lot1 expression (Fig. 5A). We next examined the PAC1-R mRNA levels and found a close relationship between Lot1 and PAC1-R expression (Fig. 5B). Analysis of GCP proliferation in cultures exposed to forskolin, PACAP, or PACAP6-38 showed that proliferation was significantly inhibited by forskolin and PACAP (-40 and -30%, respectively) (Fig. 5, C and D), as reported previously (16). Conversely, exposure to PACAP6-38 induced a significant increase in GCP proliferation (Fig. 5, C and D). The pro-proliferative action exerted by the block of PAC1-R indicates that in the culture there was a basal activation of the PAC1-R, because of the presence of PACAP in the culture medium. Relative quantification by RT-qPCR of PACAP in control cultures showed a small amount of the PACAP transcript (data not shown). In the developing and adult rat cerebellum PACAP is not expressed by granule cells but by Purkinje cells (56, 57). Immunohistochemistry for calbindin-28 kDa, a marker of Purkinje cells, revealed the presence of around 1% of Purkinje cells (Fig. 5E), which accounts for the presence of PACAP in the culture medium. Taken together, these results correlate GCP proliferation with Lot1 and PAC1-R expression.
FIGURE 5.
Effect of PACAP-38 and PACAP6-38 on proliferation of GCP in culture. A and B, Lot1 (A) and PAC1-R (B) mRNA expression were quantified by RT-qPCR in GCP cultures treated with forskolin (10 μm), PACAP-38 (10 nm), or PACAP6-38 (1 μm) for 20 h. Data, given as % of untreated conditions, are expressed as the mean ± S.E. of three independent experiments. C, double immunofluorescence for β-tubulin III and BrdUrd of GCP at 1 day in vitro. One hour after plating GCP cultures were stimulated with forskolin (10 μm), PACAP-38 (10 nm), or PACAP6-38 (1 μm) for 20 h. BrdUrd (10 μm) was added for the last 6 h, and thereafter cells were processed for double immunofluorescence with anti-BrdUrd (red) and anti-β-tubulin III (green) antibodies. Cell nuclei were stained by Hoechst dye (blue). Scale bar, 50 μm. D, LI, defined as percentage of BrdUrd-positive cells over total cell number, was determined for GCP treated as reported in C. Data are expressed as the means ± S.E. of three independent experiments. E, phase-contrast image of GCP after 1 day in culture, merged with an immunofluorescence image showing a Purkinje cell immunostained for anti-calbindin-28 kDa (red). Scale bar, 40 μm. The asterisks (B, D, and E) indicate a significant difference between the treated versus untreated condition; *, p < 0.05; **, p < 0.01; Bonferroni's test after ANOVA.
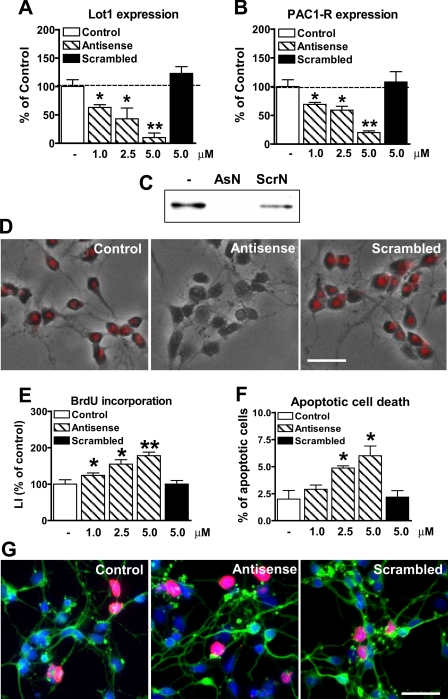
Effect of Inhibition of Lot1 Expression on Cerebellar Granule Cell Precursor Proliferation/Survival—To define whether Lot1 is an essential gene for GCP proliferation, in cultures of GCP we suppressed its expression with antisense oligonucleotides (AsN), using scrambled oligonucleotides (ScrN) as control. To test the efficacy of treatment with AsN, we checked Lot1 expression at the mRNA level by RT-qPCR and at the protein level by Western blot and immunohistochemistry (Fig. 6, A, C and D). After a 12-h treatment, AsN but not ScrN markedly decreased the expression of Lot1 in a dose-dependent manner (Fig. 6, A, C, and D). Analysis of PAC1-R expression by RT-qPCR showed that it decreased in parallel with that of Lot1 (Fig. 6, A and B), which is in line with a direct transcriptional action of Lot1 on PAC1-R expression.
FIGURE 6.
Dose-dependent effect of Lot1 antisense on GCP proliferation/survival. A and B, relative quantification by RT-qPCR of Lot1 (A) and PAC1-R (B) expression in GCP at 1 day in vitro after 12 h of exposure to different concentrations of Lot1 antisense or to a scrambled oligonucleotide. Data, given as % of control condition, are expressed as the means ± S.E. of three independent experiments. C, examples of immunoblotting with anti-Lot1 antibody obtained from GCP extracts from cultures untreated, treated with AsN (5 μm) or treated with ScrN (5 μm). D, immunohistochemistry for Lot1 (red) in cultures of GCP-untreated, treated with antisense (5 μm), or scrambled oligonucleotides (5 μm). Final pictures were obtained by merging phase contrast images with fluorescence images. Scale bar, 50 μm. E, LI, defined as percentage of BrdUrd-positive cells over total cell number, was determined for GCP treated as reported in D. F, percentage of apoptotic GCP treated as reported in D, evaluated by counting pyknotic nuclei evidenced by Hoechst staining. Data in E and F are expressed as the mean ± S.E. of four independent experiments. G, double immunofluorescence for β-tubulin III (green) and BrdUrd (red) of GCP at 1 day in vitro untreated, treated with antisense (5 μm) or treated with a scrambled oligonucleotide (5 μm). Scale bar, 50 μm. The asterisks in A, B, E, and F indicate a significant difference between the treated versus untreated condition; *, p < 0.05; **, p < 0.01; Bonferroni's test after ANOVA.
We then evaluated neuronal proliferation after 24 h of treatment with different concentrations of AsN or ScrN. Treatment with AsN, but not with ScrN, resulted in a dose-dependent increase in neuronal proliferation (Fig. 6, E and G). We also evaluated cell death by counting Hoechst-stained pyknotic nuclei after treatment with AsN or ScrN to establish whether Lot1 is involved in this process. Inhibition of Lot1 expression with AsN resulted in a dose-dependent increase in apoptotic cell death (Fig. 6F).
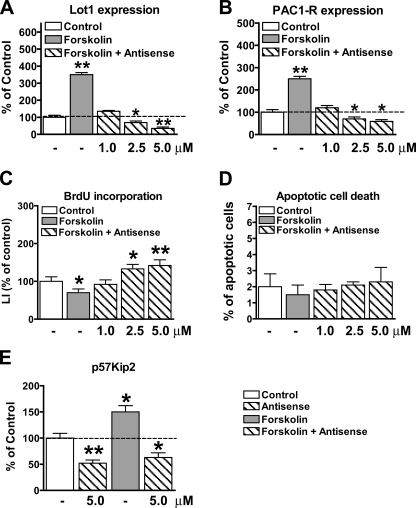
Mechanisms Underlying Lot1 Actions—The experiments reported above demonstrate that Lot1 negatively regulates GCP proliferation, positively regulates GCP survival, and increases the expression of PAC1-R. Lot1 may exert its anti-proliferative and anti-apoptotic actions by increasing the transcription of PAC1-R, with consequent enhancement of the PACAP/cAMP pathway, and/or by promoting the transcription of genes other than PAC1-R. To clear this issue, we investigated the effect of the PACAP/cAMP pathway on proliferation/survival in cultures in which Lot1 expression was inhibited with AsN. The PACAP/cAMP pathway was activated downstream to PAC1-R, through forskolin, because inhibition of Lot1 expression leads to a reduced expression of PAC1-R (see Fig. 6B). Although in control cultures, forskolin induced a robust increase in Lot1 and PAC1-R expression (Fig. 7, A and B), in cultures treated with AsN, Lot1 and PAC1-R expression was reduced in a dose-dependent manner (Fig. 7, A and B), demonstrating that AsN was able to inhibit Lot1/PAC1-R expression also in a condition leading to stimulation of Lot1/PAC1-R transcription. Evaluation of cell proliferation in cultures treated with forskolin + AsN showed an increase in cell proliferation that closely matched the decreased expression of Lot1. This clearly shows that Lot1 by itself is necessary to inhibit cell proliferation. Evaluation of cell death in parallel cultures showed that a reduction in Lot1 expression by AsN did not change cell death, when cultures were stimulated with forskolin (Fig. 7D). This suggests that the increase in apoptotic cell death, observed when Lot1 expression was inhibited (Fig. 6G), was not mediated by Lot1 per se, but was due to under-activation of the PACAP/cAMP pathway because of the reduced expression of PAC1-R.
FIGURE 7.
Concomitant effects of Lot1 silencing and activation of the PACAP/cAMP pathway on granule cell proliferation/survival. A and B, relative quantification by RT-qPCR of Lot1 (A) and PAC1-R (B) expression in GCP at 1 day in vitro after 12 h of treatment with forskolin (10 μm) or forskolin plus different concentrations of Lot1 antisense. Data, given as % of control condition, are expressed as the mean ± S.E. of three independent experiments. C, LI, defined as percentage of BrdUrd-positive cells over total cell number, was determined for GCP treated as reported in A. D, percentage of apoptotic GCP evaluated by counting pyknotic nuclei evidenced by Hoechst staining. Data in C and D are expressed as the mean ± S.E. of three independent experiments. E, relative quantification by RT-qPCR of p57Kip2 expression in GCP at 1 day in vitro after 12 h of treatment with forskolin (10 μm), antisense oligonucleotides (5 μm), or forskolin plus antisense oligonucleotides (5 μm). Data, given as % of control condition, are expressed as the mean ± S.E. of three independent experiments. The asterisks in A-E indicate a significant difference between the treated versus untreated condition; *, p < 0.05; **, p < 0.01; Bonferroni's test after ANOVA.
In an attempt to identify the mechanism by which Lot1 inhibits cell proliferation, we investigated the expression of p57Kip2, a cyclin-dependent kinase inhibitor, in cultures treated with AsN, forskolin or forskolin + AsN. We focused on p57Kip2 because this gene appears to control the transition from proliferation to differentiation during brain development (58) and is a direct transcriptional target of Lot1 (59). We found that inhibition of Lot1 expression with AsN led to a parallel decrease of p57Kip2 expression (Fig. 7E). Conversely, up-regulation of Lot1 expression by forskolin led to a corresponding increase of p57Kip2 expression (Fig. 7E). Importantly, in cultures treated with forskolin but in the presence of AsN there was no increase in p57Kip2 expression, indicating that Lot1 is essential for transcription of p57Kip2.
From these experiments it can be concluded that the anti-proliferative and anti-apoptotic effects of the PACAP/cAMP pathway are mediated through different mechanisms. Although the anti-proliferative effects are mediated by Lot1, most likely through p57Kip2, the anti-apoptotic effects are mediated by other components of the pathway.
Effect of PACAP on Lot1 Expression and GCP Proliferation in Vivo—To establish whether the effect of Lot1 on GCP proliferation observed in vitro reflects the in vivo condition, neonate (P7) rats received one intracranial injection of PACAP at the level of the cerebellum to activate the PACAP/cAMP pathway. Results showed that at 6 h after PACAP injection, the cerebellum exhibited a large increase of Lot1 expression, as evaluated at the mRNA level (Fig. 8A). Evaluation of the effect of PACAP on GCP proliferation 24 h after the injection, through BrdUrd labeling, showed that there was a significant decrease in the number of proliferating cells in the EGL, where the dividing precursors of granule neurons are present (Fig. 8, B-D). These data indicate that the effects of PACAP in vivo mimic those observed in vitro.
DISCUSSION
During brain development the generation of an appropriate neuron number requires a fine-tuning between cell proliferation and cell death. This study provides novel evidence that the tumor suppressor gene Lot1 is a key element of the PACAP/cAMP pathway that regulates proliferation/survival of neuronal cells. In particular, in the PC12 cell line, Lot1 appears to negatively regulate proliferation and favor differentiation, and in cerebellar granule cell precursors Lot1 inhibits proliferation and promotes survival.
Lot1 Inhibits Proliferation of PC12 Cell Lines—We provide here evidence that PC12 cells cloned from a rat adrenal pheochromocytoma (32) do not express Lot1. We took advantage of the PC12 cell line to create a cellular model where Lot1 expression could be induced by activating the ecdysone system. We found that in this cellular model induction of Lot1 expression caused a strong inhibition of cell proliferation and promoted a partial neuronal differentiation. In agreement with other studies (30), we demonstrated that Lot1 transcriptionally induced an increase in PAC1-R expression. By using a PAC1-R antagonist, we additionally demonstrated that the effect of Lot1 on PC12 cell proliferation and differentiation was dependent on the PAC1-R.
The signaling cascades downstream to PAC1-R/cAMP system have been shown to include the MEK pathway (61), the protein kinase A (PKA), and the cAMP-binding guanine nucleotide exchange factors Epac1 and -2 (62). By using a PKA inhibitor, we found that Lot1-induced inhibition of cell proliferation required activation of PKA. We additionally found that the PAC1-R/cAMP system, up-regulated by Lot1, induced activation of the MEK pathway, with a consequent increase of c-Fos expression. According to recent studies, PACAP induces PC12 cell neurite outgrowth through either the MEK and/or the Epac pathway, independently of PKA (26, 63, 64). Thus, although in PC12 cells PACAP-induced neuritogenesis appears to be PKA-independent, according to our study the anti-proliferative action of PACAP is PKA-dependent.
PC12 cells can produce PACAP which, binding to PAC1-R, stimulates PACAP gene expression (48), indicating that PACAP may act as an autocrine regulatory factor. The fact that PC12 cells exhibit continuous proliferation implies that they express low levels of PAC1-R and/or PACAP. Our observation that the increased expression of PAC1-R induced by Lot1 was accompanied by inhibition of cell proliferation indicates that this increase was sufficient to drive the cAMP cascade and to inhibit cell proliferation. Preliminary data from our laboratory suggest that an increase in the expression of Lot1 can induce an increase in the production of PACAP also. We are currently investigating whether the increase in PACAP expression is directly mediated by Lot1 or indirectly through the PAC1-R/cAMP cascade.
Lot1 and PAC1-R have been shown to be highly expressed in the human adrenal gland (65, 66). Accordingly, loss of Lot1 expression in pheochromocytomas (60) may underlie dysregulation of cell proliferation in this tumor. In this study, by inducing Lot1 expression, we could demonstrate that Lot1 exerts anti-proliferative effects in PC12 cells and the underlying mechanisms. Lot1 effects in the PC12 cells are likely to replicate those physiologically present in adrenal gland cells.
Lot1 Inhibits Proliferation and Is Involved in the Survival of Cerebellar Granule Cell Precursors—In the EGL, granule cells proliferate actively and then stop dividing, to migrate through the different layers of the cerebellar cortex (67). Although the different stages of cerebellar development are well described, the molecular mechanisms that shift granule neurons from proliferation to differentiation remain largely unknown.
PACAP and PAC1-R are actively expressed during cerebellar development (68-70). Specifically, the expression of the PAC1-R reaches a maximum in the EGL between P4 and P12 (71), a period that corresponds to intense neurogenesis (67), with a time window temporally matching Lot1 expression (13). In vitro experiments have shown that PAC1-R is expressed by granule cell precursors (72) and that PACAP stimulates differentiation of these neurons by inhibiting cell proliferation and stimulating neurite outgrowth (50, 72, 73). Recent studies have shown that disruption of the PACAP gene in mutant mice induces altered cerebellar development, for a delay of granule cell differentiation and an increase in granule cell death (22, 50).
We provide here novel evidence that acute, in vivo administration of PACAP increases the expression of Lot1 and exerts an anti-proliferative effect on GCP, confirming the key role of the PACAP/cAMP system in the ontogenesis of the cerebellum. Concerning the mechanisms by which this system regulates cell proliferation, our in vitro results demonstrate that Lot1 is a key effector of this pathway because it directly inhibits GCP proliferation and up-regulates the transcription of PAC1-R. Conversely, inhibition of either Lot1 expression or PAC1-R activity (by PACAP6-38) increases GCP proliferation. These data demonstrate that expression of Lot1 and PAC1-R signaling are closely regulated and are required to inhibit GCP proliferation.
In vivo and in vitro experiments have shown that PACAP has an important survival-promoting effect on cerebellar granule cells (36, 73, 74). This study confirms the survivalpromoting effect of the PACAP/cAMP system on cerebellar granule cells. It additionally shows that inhibition of Lot1 expression was followed by down-regulation of PAC1-R expression and an increase in cell death, suggesting that Lot1 participates in the survival-promoting effect of the PACAP/cAMP system by enhancing PAC1-R expression (see also below).
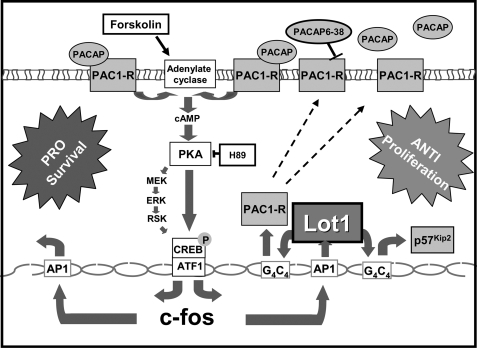
Potential Mechanisms by which the PACAP/cAMP/Lot1 System Modulates Cerebellar Granule Cell Proliferation and Survival—According to a recent study, Lot1 appears to inhibit cell proliferation by up-regulating the expression if the cyclin-dependent kinase inhibitor p57Kip2 (59). In that study (59) it was shown that mouse Lot1 and p57Kip2 have a strikingly similar expression pattern and that LOT1 binds within the CpG island of LIT1 (KCNQ1OT1), which is a paternally expressed antisense RNA thought to negatively regulate p57Kip2 in cis. Moreover, it has been demonstrated that PACAP rapidly elicits increased levels of p57Kip2, to block cell cycle progression of cerebral cortical precursors (75). By silencing Lot1 expression, we could show that Lot1 is essential for transcription of p57Kip2 in GCP. These data suggest that PACAP inhibits GCP proliferation through Lot1, which, by increasing p57Kip2 expression, accelerates GCP exit from the cell cycle (Fig. 9).
FIGURE 9.
Putative signaling pathways involved in the anti-proliferative/prosurvival actions of Lot1 in cerebellar granule cell precursors. Lot1 expression in GCP increases the transcription of PAC1-R through a G4C4 response element. The larger number of PAC1-R increases the efficacy of the PACAP-prosurvival effect and, by a positive feedback, the transcription of Lot1. Lot1 increases p57Kip2 expression, which may inhibit GCP proliferation.
Our data show that the PACAP/cAMP system even in the absence of Lot1 expression was able to promote GCP survival, indicating that this effect was mediated by a Lot1-independent mechanism. Although this mechanism is largely unknown so far, in light of recent data c-fos may be a good candidate for the survival promoting activity exerted by the PACAP/cAMP system (76, 77) on GCP (Fig. 9). It should be noted that although Lot1 does not directly promote cell survival, it indirectly favors cell survival by increasing the expression of PAC1-R, thereby enhancing the efficacy of the PACAP/cAMP system.
In conclusion, our data clearly show that the PACAP/cAMP system exerts both anti-proliferative and survival-promoting effects on cerebellar granule cells and that Lot1 is involved in both these effects. Although the anti-proliferative effects are most likely directly mediated by Lot1, through inhibition of the cell cycle machinery, the survival-promoting effects are indirectly mediated through a Lot1-induced increase in PAC1-R expression (Fig. 9).
Lot1-PAC1-R Interactions—This study shows that Lot1 is upregulated in GCP by the PACAP/cAMP system and that Lot1 up-regulates the expression of PAC1-R. Thus, Lot1 and PAC1-R appear to reciprocally regulate their expression through a positive feedback mechanism. Feedback pathways exist in diverse neuronal systems, including Drosophila miRNA9a, which regulates sensory organ precursor number by down-regulating Senseless expression (78) or Branchless and Hedgehog pathways that form a positive feedback loop to regulate the onset of neuroblast division (79). In the mammalian brain, nitric oxide acts in a positive feedback loop with brain-derived neurotrophic factor to regulate neural progenitor cell proliferation and differentiation (80).
Our findings that Lot1 and PAC1-R reciprocally regulate their expression provide a basis for the formulation of a model for the role played by Lot1 in the regulation of GCP proliferation and survival (Fig. 9). In this model, Lot1 expressed by newly generated GCP provides a positive feedback signal, which by increasing PAC1-R further enhances Lot1 expression. This amplifies the anti-proliferative and survival-promoting effects of the PACAP/cAMP pathway.
We recently demonstrated that Lot1 is expressed by proliferating GCP in the EGL, but is absent from mature cerebellar granule cells in the IGL (13), though PAC1-R is still highly expressed by the latter. This suggests that at the end of cerebellar development, additional events must take place that silence Lot1 expression in differentiated granule cells and regulate PAC1-R expression independently on Lot1. At this stage of development PAC1-R is no longer involved in the regulation of proliferation/differentiation but plays a role in the neuromodulatory/neurotrophic action exerted by PACAP.
Conclusions—This study shows that Lot1 is responsible for the growth-inhibitory effect exerted by PACAP on proliferating granule cells and suggests that a Lot1-PAC1-R-positive feedback loop may represent a key mechanism for regulation of neuron proliferation.
It has been demonstrated recently that PACAP exerts a powerful inhibitory action on the induction, growth, or survival of Hedgehog pathway-associated medulloblastoma, a highly aggressive tumor of the cerebellum in infants (81). Down-regulation of the expression of LOT1 has been linked to several types of tumors (1, 5, 60, 82, 83). From our results it may be speculated that mutations in either LOT1 and/or PAC1-R takes place in human medulloblastoma tumors. If so, it would be of relevance to determine whether down-regulation of LOT1 expression underlies the pathogenesis of this tumor.
Acknowledgments
We are grateful to Dr. H. Green for kindly supplying the Ecdysone-responsive (EcR) PC12 cells used in this work.
This work was supported by the University of Bologna through funding for basic research (to E. C.).
Footnotes
The abbreviations used are: PACAP, pituitary adenylate cyclase-activating polypeptide; BrdUrd, 5-bromo-2-deoxyuridine; PBS, phosphate-buffered saline; RT-qPCR, reverse transcriptase-quantitative PCR; LI, labeling index; ANOVA, analysis of variance; GCP, granule cell precursor; EcR, ecdysone-responsive; ERK, extracellular signal-regulated kinase; MEK, mitogen-activated protein kinase/extracellular signal-regulated kinase kinase; EGL, external granular layer; NGF, nerve growth factor; CREB, cAMP-response element-binding protein; PKA, cAMP-dependent protein kinase; AsN, antisense oligonucleotide; ScrN, scrambled oligonucleotide; HA, hemagglutinin; PAC1-R, PACAP type 1 receptor.
References
- 1.Abdollahi, A., Godwin, A. K., Miller, P. D., Getts, L. A., Schultz, D. C., Taguchi, T., Testa, J. R., and Hamilton, T. C. (1997) Cancer Res. 57 2029-2034 [PubMed] [Google Scholar]
- 2.Kas, K., Voz, M. L., Hensen, K., Meyen, E., and Van de Ven, W. J. (1998) J. Biol. Chem. 273 23026-23032 [DOI] [PubMed] [Google Scholar]
- 3.Spengler, D., Villalba, M., Hoffmann, A., Pantaloni, C., Houssami, S., Bockaert, J., and Journot, L. (1997) EMBO J. 16 2814-2825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bilanges, B., Varrault, A., Mazumdar, A., Pantaloni, C., Hoffmann, A., Bockaert, J., Spengler, D., and Journot, L. (2001) Oncogene 20 1246-1253 [DOI] [PubMed] [Google Scholar]
- 5.Abdollahi, A., Roberts, D., Godwin, A. K., Schultz, D. C., Sonoda, G., Testa, J. R., and Hamilton, T. C. (1997) Oncogene 14 1973-1979 [DOI] [PubMed] [Google Scholar]
- 6.Bilanges, B., Varrault, A., Basyuk, E., Rodriguez, C., Mazumdar, A., Pantaloni, C., Bockaert, J., Theillet, C., Spengler, D., and Journot, L. (1999) Oncogene 18 3979-3988 [DOI] [PubMed] [Google Scholar]
- 7.Colitti, C. V., Rodabaugh, K. J., Welch, W. R., Berkowitz, R. S., and Mok, S. C. (1998) Oncogene 16 555-559 [DOI] [PubMed] [Google Scholar]
- 8.Piras, G., El Kharroubi, A., Kozlov, S., Escalante-Alcalde, D., Hernandez, L., Copeland, N. G., Gilbert, D. J., Jenkins, N. A., and Stewart, C. L. (2000) Mol. Cell. Biol. 20 3308-3315 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Smith, R. J., Arnaud, P., Konfortova, G., Dean, W. L., Beechey, C. V., and Kelsey, G. (2002) Gene 292 101-112 [DOI] [PubMed] [Google Scholar]
- 10.Abdollahi, A., Bao, R., and Hamilton, T. C. (1999) Oncogene 18 6477-6487 [DOI] [PubMed] [Google Scholar]
- 11.Valente, T., and Auladell, C. (2001) Mech. Dev. 108 207-211 [DOI] [PubMed] [Google Scholar]
- 12.Valente, T., Junyent, F., and Auladell, C. (2005) Dev. Dyn. 233 667-679 [DOI] [PubMed] [Google Scholar]
- 13.Ciani, E., Frenquelli, M., and Contestabile, A. (2003) Brain Res. Dev. Brain Res. 142 193-202 [DOI] [PubMed] [Google Scholar]
- 14.Tsuda, T., Markova, D., Wang, H., Evangelisti, L., Pan, T. C., and Chu, M. L. (2004) Dev. Dyn. 229 340-348 [DOI] [PubMed] [Google Scholar]
- 15.Alam, S., Zinyk, D., Ma, L., and Schuurmans, C. (2005) Dev. Dyn. 234 772-782 [DOI] [PubMed] [Google Scholar]
- 16.Contestabile, A., Fila, T., Bartesaghi, R., and Ciani, E. (2005) J. Biol. Chem. 280 33541-33551 [DOI] [PubMed] [Google Scholar]
- 17.Ma, L., Cantrup, R., Varrault, A., Colak, D., Klenin, N., Götz, M., Mc-Farlane, S., Journot, L., and Schuurmans, C. (2007) Neural Dev. 2 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Vaudry, D., Gonzalez, B. J., Basille, M., Pamantung, T. F., Fournier, A., and Vaudry, H. (2000) Ann. N.Y. Acad. Sci. 921 293-299 [DOI] [PubMed] [Google Scholar]
- 19.Vaudry, D., Gonzalez, B. J., Basille, M., Pamantung, T. F., Fontaine, M., Fournier, A., and Vaudry, H. (2000) Proc. Natl. Acad. Sci. U. S. A. 97 13390-13395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arimura, A. (1998) Jpn. J. Physiol. 48 301-331 [DOI] [PubMed] [Google Scholar]
- 21.Somogyvári-Vigh, A., and Reglodi, D. (2004) Curr. Pharm. Des. 10 2861-2889 [DOI] [PubMed] [Google Scholar]
- 22.Allais, A., Burel, D., Isaac, E. R., Gray, S. L., Basille, M., Ravni, A., Sherwood, N. M., Vaudry, H., and Gonzalez, B. J. (2007) Eur. J. Neurosci. 25 2604-2618 [DOI] [PubMed] [Google Scholar]
- 23.Suh, J., Lu, N., Nicot, A., Tatsuno, I., and DiCicco-Bloom, E. (2001) Nat. Neurosci. 4 123-124 [DOI] [PubMed] [Google Scholar]
- 24.McCulloch, D. A., MacKenzie, C. J., Johnson, M. S., Robertson, D. N., Holland, P. J., Ronaldson, E., Lutz, E. M., and Mitchell, R. (2002) Biochem. Soc. Trans. 30 441-446 [DOI] [PubMed] [Google Scholar]
- 25.Deutsch, P. J., and Sun, Y. (1992) J. Biol. Chem. 267 5108-5113 [PubMed] [Google Scholar]
- 26.Vaudry, D., Stork, P. J., Lazarovici, P., and Eiden, L. E. (2002) Science 296 1648-1649 [DOI] [PubMed] [Google Scholar]
- 27.Hernandez, A., Kimball, B., Romanchuk, G., and Mulholland, M. W. (1995) Peptides 16 927-932 [DOI] [PubMed] [Google Scholar]
- 28.Hansen, T. O., Rehfeld, J. F., and Nielsen, F. C. (2000) J. Neurochem. 75 1870-1877 [DOI] [PubMed] [Google Scholar]
- 29.Ciani, E., Hoffmann, A., Schmidt, P., Journot, L., and Spengler, D. (1999) Brain Res. Mol. Brain Res. 69 290-294 [DOI] [PubMed] [Google Scholar]
- 30.Hoffmann, A., Ciani, E., Houssami, S., Brabet, P., Journot, L., and Spengler, D. (1998) Ann. N.Y. Acad. Sci. 865 49-58 [DOI] [PubMed] [Google Scholar]
- 31.Tischler, A. S., and Greene, L. A. (1978) Lab. Investig. 39 77-89 [PubMed] [Google Scholar]
- 32.Greene, L. A., and Tischler, A. S. (1976) Proc. Natl. Acad. Sci. U. S. A. 73 2424-2428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ciani, E., Guidi, S., Della Valle, G., Perini, G., Bartesaghi, R., and Contestabile, A. (2002) J. Biol. Chem. 277 49896-49902 [DOI] [PubMed] [Google Scholar]
- 34.Foehr, E. D., Lin, X., O'Mahony, A., Geleziunas, R., Bradshaw, R. A., and Greene, W. C. (2000) J. Neurosci. 20 7556-7563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ciani, E., and Paulsen, R. E. (1995) J. Mol. Neurosci. 6 131-139 [DOI] [PubMed] [Google Scholar]
- 36.Vaudry, D., Gonzalez, B. J., Basille, M., Fournier, A., and Vaudry, H. (1999) Proc. Natl. Acad. Sci. U. S. A. 96 9415-9420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nowakowski, R. S., Lewin, S. B., and Miller, M. W. (1989) J. Neurocytol. 18 311-318 [DOI] [PubMed] [Google Scholar]
- 38.Pennuto, M., Dunlap, D., Contestabile, A., Benfenati, F., and Valtorta, F. (2002) Mol. Biol. Cell 13 2706-2717 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boussif, O., Lezoualc'h, F., Zanta, M. A., Mergny, M. D., Scherman, D., Demeneix, B., and Behr, J. P. (1995) Proc. Natl. Acad. Sci. U. S. A. 92 7297-7301 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.de Wet, J. R., Wood, K. V., DeLuca, M., Helinski, D. R., and Subramani, S. (1987) Mol. Cell. Biol. 7 725-737 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lowry, O. H., Rosebrough, N. J., Farr, A. L., and Randall, R. J. (1951) J. Biol. Chem. 193 265-275 [PubMed] [Google Scholar]
- 42.Contestabile, A., Fila, T., Bartesaghi, R., and Ciani, E. (2008) Brain Pathol. 9 224-237 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Godard, T., Deslandes, E., Lebailly, P., Vigreux, C., Poulain, L., Sichel, F., Poul, J. M., and Gauduchon, P. (1999) Cytometry 36 117-122 [DOI] [PubMed] [Google Scholar]
- 44.Vaudry, D., Chen, Y., Hsu, C. M., and Eiden, L. E. (2002) Ann. N.Y. Acad. Sci. 971 491-496 [DOI] [PubMed] [Google Scholar]
- 45.Ng, Y. P., He, W., and Ip, N. Y. (2003) J. Biol. Chem. 278 38731-38739 [DOI] [PubMed] [Google Scholar]
- 46.Riccio, A., Pierchala, B. A., Ciarallo, C. L., and Ginty, D. D. (1997) Science 277 1097-1100 [DOI] [PubMed] [Google Scholar]
- 47.Lu, N., and DiCicco-Bloom, E. (1997) Proc. Natl. Acad. Sci. U. S. A. 94 3357-3362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hashimoto, H., Hagihara, N., Koga, K., Yamamoto, K., Shintani, N., Tomimoto, S., Mori, W., Koyama, Y., Matsuda, T., and Baba, A. (2000) J. Neurochem. 74 501-507 [DOI] [PubMed] [Google Scholar]
- 49.Dicicco-Bloom, E., Lu, N., Pintar, J. E., and Zhang, J. (1998) Ann. N.Y. Acad. Sci. 865 274-289 [DOI] [PubMed] [Google Scholar]
- 50.Botia, B., Basille, M., Allais, A., Raoult, E., Falluel-Morel, A., Galas, L., Jolivel, V., Wurtz, O., Komuro, H., Fournier, A., Vaudry, H., Burel, D., Gonzalez, B. J., and Vaudry, D. (2007) Peptides 28 1746-1752 [DOI] [PubMed] [Google Scholar]
- 51.Gunning, P. W., Landreth, G. E., Bothwell, M. A., and Shooter, E. M. (1981) J. Cell Biol. 89 240-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lazarovici, P., Jiang, H., and Fink, D., Jr. (1998) Mol. Pharmacol. 54 547-558 [DOI] [PubMed] [Google Scholar]
- 53.Hauge, C., and Frödin, M. (2006) J. Cell Sci. 119 3021-3023 [DOI] [PubMed] [Google Scholar]
- 54.Janknecht, R., Cahill, M. A., and Nordheim, A. (1995) Carcinogenesis 16 443-450 [DOI] [PubMed] [Google Scholar]
- 55.Whitmarsh, A. J., Shore, P., Sharrocks, A. D., and Davis, R. J. (1995) Science 269 403-407 [DOI] [PubMed] [Google Scholar]
- 56.Skoglösa, Y., Patrone, C., and Lindholm, D. (1999) Neurosci. Lett. 265 207-210 [DOI] [PubMed] [Google Scholar]
- 57.Nielsen, H. S., Hannibal, J., and Fahrenkrug, J. (1998) Neuroreport 9 2639-2642 [DOI] [PubMed] [Google Scholar]
- 58.Ye, W., Mairet-Coello, G., Pasoreck, E., and Dicicco-Bloom, E. (2009) Dev. Neurobiol. 69 1-21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Arima, T., Kamikihara, T., Hayashida, T., Kato, K., Inoue, T., Shirayoshi, Y., Oshimura, M., Soejima, H., Mukai, T., and Wake, N. (2005) Nucleic Acids Res. 33 2650-2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lemeta, S., Salmenkivi, K., Pylkkänen, L., Sainio, M., Saarikoski, S. T., Arola, J., Heikkilä, P., Haglund, C., Husgafvel-Pursiainen, K., and Böhling, T. (2006) Hum. Pathol. 37 749-754 [DOI] [PubMed] [Google Scholar]
- 61.Stork, P. J. (2003) Trends Biochem. Sci. 28 267-275 [DOI] [PubMed] [Google Scholar]
- 62.Shi, G. X., Rehmann, H., and Andres, D. A. (2006) Mol. Cell. Biol. 26 9136-9147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ravni, A., Vaudry, D., Gerdin, M. J., Eiden, M. V., Falluel-Morel, A., Gonzalez, B. J., Vaudry, H., and Eiden, L. E. (2008) Mol. Pharmacol. 73 1688-1708 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kiermayer, S., Biondi, R. M., Imig, J., Plotz, G., Haupenthal, J., Zeuzem, S., and Piiper, A. (2005) Mol. Biol. Cell 16 5639-5648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Conconi, M. T., Spinazzi, R., and Nussdorfer, G. G. (2006) Int. Rev. Cytol. 249 1-51 [DOI] [PubMed] [Google Scholar]
- 66.Varrault, A., Ciani, E., Apiou, F., Bilanges, B., Hoffmann, A., Pantaloni, C., Bockaert, J., Spengler, D., and Journot, L. (1998) Proc. Natl. Acad. Sci. U. S. A. 95 8835-8840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Altman, J. (1982) Exp. Brain Res. 6 (suppl.) 8-49 [Google Scholar]
- 68.Basille, M., Falluel-Morel, A., Vaudry, D., Aubert, N., Fournier, A., Fréger, P., Gallo-Payet, N., Vaudry, H., and Gonzalez, B. (2006) Regul. Pept. 137 27-33 [DOI] [PubMed] [Google Scholar]
- 69.Basille, M., Vaudry, D., Coulouarn, Y., Jegou, S., Lihrmann, I., Fournier, A., Vaudry, H., and Gonzalez, B. (2000) J. Comp. Neurol. 425 495-509 [PubMed] [Google Scholar]
- 70.Basille, M., Vaudry, D., Coulouarn, Y., Jégou, S., Lihrmann, I., Fournier, A., Vaudry, H., and Gonzalez, B. J. (2000) Ann. N.Y. Acad. Sci. 921 304-307 [DOI] [PubMed] [Google Scholar]
- 71.Basille, M., Gonzalez, B. J., Leroux, P., Jeandel, L., Fournier, A., and Vaudry, H. (1993) Neuroscience 57 329-338 [DOI] [PubMed] [Google Scholar]
- 72.Nicot, A., Lelièvre, V., Tam, J., Waschek, J. A., and DiCicco-Bloom, E. (2002) J. Neurosci. 22 9244-9254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Gonzalez, B. J., Basille, M., Vaudry, D., Fournier, A., and Vaudry, H. (1997) Neuroscience 78 419-430 [DOI] [PubMed] [Google Scholar]
- 74.Cavallaro, S., Copani, A., D'Agata, V., Musco, S., Petralia, S., Ventra, C., Stivala, F., Travali, S., and Canonico, P. L. (1996) Mol. Pharmacol. 50 60-66 [PubMed] [Google Scholar]
- 75.Carey, R. G., Li, B., and DiCicco-Bloom, E. (2002) J. Neurosci. 22 1583-1591 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Falluel-Morel, A., Aubert, N., Vaudry, D., Desfeux, A., Allais, A., Burel, D., Basille, M., Vaudry, H., Laudenbach, V., and Gonzalez, B. J. (2008) J. Mol. Neurosci. 36 8-15 [DOI] [PubMed] [Google Scholar]
- 77.Vaudry, D., Basille, M., Anouar, Y., Fournier, A., Vaudry, H., and Gonzalez, B. J. (1998) Ann. N.Y. Acad. Sci. 865 92-99 [DOI] [PubMed] [Google Scholar]
- 78.Li, Y., Wang, F., Lee, J. A., and Gao, F. B. (2006) Genes Dev. 20 2793-2805 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Barrett, A. L., Krueger, S., and Datta, S. (2008) Dev. Biol. 317 234-245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Cheng, A., Wang, S., Cai, J., Rao, M. S., and Mattson, M. P. (2003) Dev. Biol. 258 319-333 [DOI] [PubMed] [Google Scholar]
- 81.Lelievre, V., Seksenyan, A., Nobuta, H., Yong, W. H., Chhith, S., Niewiadomski, P., Cohen, J. R., Dong, H., Flores, A., Liau, L. M., Kornblum, H. I., Scott, M. P., and Waschek, J. A. (2008) Dev. Biol. 313 359-370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pagotto, U., Arzberger, T., Theodoropoulou, M., Grübler, Y., Pantaloni, C., Saeger, W., Losa, M., Journot, L., Stalla, G. K., and Spengler, D. (2000) Cancer Res. 60 6794-6799 [PubMed] [Google Scholar]
- 83.Singhal, S., Amin, K. M., Kruklitis, R., DeLong, P., Friscia, M. E., Litzky, L. A., Putt, M. E., Kaiser, L. R., and Albelda, S. M. (2003) Cancer Biol. Ther. 2 291-298 [DOI] [PubMed] [Google Scholar]