Abstract
Calorie restriction (CR) improves health and extends life span in a variety of species. Despite many downstream molecules and physiological systems having been identified as being regulated by CR, the mechanism by which CR extends life span remains unclear. The Drosophila gene Indy (for I'm not dead yet), involved in the transport and storage of Krebs cycle intermediates in tissues important in fly metabolism, was proposed to regulate life span via an effect on metabolism that could overlap with CR. In this study, we report that CR down regulates Indy mRNA expression, and that CR and the level of Indy expression interact to affect longevity. Optimal life span extension is seen when Indy expression is decreased between 25 and 75% of normal. Indy long-lived flies show several phenotypes that are shared by long-lived CR flies, including decreased insulin-like signaling, lipid storage, weight gain, and resistance to starvation as well as an increase in spontaneous physical activity. We conclude that Indy and CR interact to affect longevity and that a decrease in Indy may induce a CR-like status that confers life span extension.
Keywords: Drosophila, insulin, physical activity, triglyceride
Aging is a complex biological process that causes deteriorative changes over time. It has been suggested that the interplay between environmental factors and genetic alterations may affect this near universal process. Calorie restriction (CR) is the most widely recognized life span-extending intervention, and it has been shown to extend lifespan in a variety of different organisms (1, 2). Progress has been made in identifying genes that regulate longevity, and many of them appear to belong to pathways related to nutrient sensing, metabolism or nutrient/metabolic signaling (3–7). The life span extending effects of a subset of these longevity genes has been shown to be associated with, and in some cases, causally related to CR life span extension (chico, Sir2, p53). Studies have suggested that alterations in the activity of these genes may mediate elements of the normal CR life span extending effect. Despite these advances little is understood about the molecular and genetic mechanisms underlying the healthy life span extension of CR.
In Drosophila melanogaster, mutations in the Indy gene dramatically extend life span (8). The INDY protein is a transmembrane transporter of Krebs cycle intermediates (citrate, succinate, fumarate, and alpha-ketoglutarate) predominantly found at the plasma membrane of cells in the midgut, fat body, and oenocytes, tissues important for the uptake, utilization, and storage of nutrients and the principal sites of intermediary metabolism in the fly (8–10). Several independently derived lines, each with a P-element in the non-coding region of the Indy locus leading to decreased expression of Indy mRNA, have been shown to extend life span. It has been reported that life span extension is seen even when the Indy mutation is crossed into different genetic backgrounds (e.g., Hyperkinetic, Shaker, drop dead, and the long- and short-lived laboratory selected outbred lines of Luckinbill) (8). In the nematode Caenorhabditis elegans, decreased expression of an Indy-like gene by RNAi has been shown to extend life span (11). An important feature of Indy life span extension is that it dramatically extends life span with very few physiological tradeoffs. For example, Indy flies show no reduction in resting metabolic rate and early or late life fecundity under normal laboratory rearing conditions, and no decrease in maximal flight velocity, negative geotaxis or 24-hour activity levels has been detected (12–14). It has been proposed that a decrease in Indy expression might extend life span by affecting intermediary metabolism and creating a metabolic state that overlaps with or mimics CR.
Insulin/IGF-like signaling (IIS) is one of the major pathways that respond to the energy and metabolic status of the body. Drosophila insulin-like peptides (dilps) signal through the insulin receptor (InR) and the InR substrate (chico in flies), which leads to activation of phosphoinositide-3-kinase (PI3K) and protein kinase B/Akt. This kinase cascade eventually phosphorylates the forkhead transcription factor dFOXO and causes dFOXO retention in the cytoplasm via binding to 14–3-3 proteins. Mutations that cause reduced activity of Insulin/IGF-like signaling have been shown to increase life span in nematodes, flies, and mice (15). However, in the nematode and to a lesser extent in the fly, experimental evidence suggests that Insulin/IGF-like signaling may not be essential for the life span extension of CR. In nematodes, long-lived daf-2/InR and daf-16/dFOXO mutants live even longer when subject to CR (16). In flies, knockdown of dilps using RNAi does not decrease the life span-extending effects of CR and dFoxO null mutations continue to have a CR responsive life span extension, suggesting that CR and IIS-mediated life span extension may be unrelated (17). However, CR fails to further increase the life span extension of long-lived chico (InR substrate) mutant flies, suggesting CR and IIS mediated life span extension may be related (18). Finally, recent studies showed that over-expression of dFOXO in flies may modulate the CR response (19), and CR does not further extend life span in mice having a targeted mutation of growth hormone receptor, leading to a suppressed level of insulin and IGF1 (20).
In this study, we investigate the relationship between Indy mutant life span extension and CR induced life span extension. We report here that Indy mutants induce an altered state of IIS and other CR-like phenotypes, including changes in starvation resistance, lipid storage, physical activity, and life span. We conclude that decreasing Indy induces a calorie restriction-like status that confers life span extension.
Results
Decreased Indy Expression Extends Life Span.
A recent report stated that a decrease in Indy expression is not associated with life span extension (21). This claim was based on an inability to detect a decrease in Indy transcription levels in the Indy302 allele using northern blot analysis; an inability to demonstrate life span extension in females for either the Indy206 or Indy302 alleles; and an inability to demonstrate life span extension in males or females in either the Indy206 or Indy302 alleles after backcrossing to w1118 or after treatment with tetracycline. To begin to understand these discrepancies, we examined the level of Indy transcription in our Indy206 and Indy302 stocks and in the Indy206 and Indy302 stocks provided by (21). Unlike the report by (21), which showed no transcriptional defect in the Indy302 stock using northern blot analyses, we found a 40–50% decrease in Indy transcript levels in the original Indy302 stock as well as in the Indy302 stock directly obtained from (21) using real-time quantitative PCR (Fig. S1 A and B).
We next examined the life span of the Indy206 allele on a series of different calorie foods after backcrossing to yw or w1118 and treating with tetracycline. We found that after Indy206 was backcrossed to a wild-type yw stock, allowing for meiotic recombination for 10 generations, and then treated with tetracycline for 3 generations to remove all Wolbachia (Fig. S2A), followed by culturing without tetracycline for several more generations, both male and female Indy206 heterozygotes continue to show a significant life span extension as compared to genetically matched controls when grown on normal rich food (Fig. 1 and Fig. S2B and C, 29% and 34% median lifespan extension in male and female, respectively, P < 0.0001). Interestingly, as shown by Toivenon et al. (21), there is no life span extension when Indy206 is backcrossed into the w1118 background (Fig. S3 A and B). Life span studies on the original Canton-S derived Indy206 stock, kept homozygous since 1989, continue to demonstrate a significant life span extension (Fig. S4 A and B).
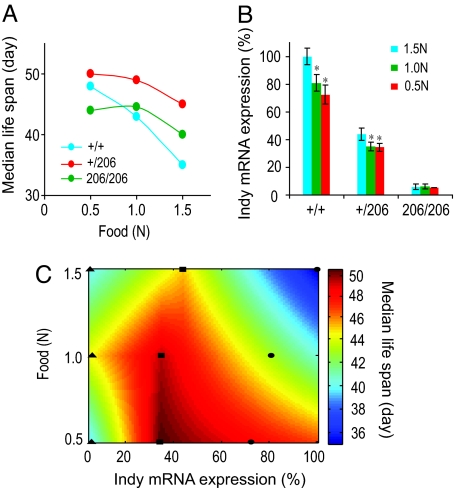
Fig. 1.
The interaction between CR and Indy affects life span. (A) Median life span of yw control flies (+/+, blue line), Indy206 heterozygous (+/206, red line), and Indy206 homozygous (206/206, green line) flies on 5% (0.5 N), 10% (1 N), and 15% (1.5 N) dextrose and yeast diets. (B) Diets regulate Indy mRNA expression in +/+ and +/206 flies. Data are presented as mean ± SD. Experiments were done in triplicate and each sample contained more than 20 heads and thoraxes of 20-day-old male flies. The * indicates P < 0.05 by t-test. (C) A heat map presentation showing correlations between diets, Indy mRNA expression and median lifespan of flies. Data were derived from A and B. ●, (+/+); ■, (+/206); ▴, (206/206).
Indy Life Span Extension and CR Life Span Extension Interact.
It has been hypothesized that a decrease in Indy activity extends life span through induction of metabolic/physiological changes that may be similar to calorie restriction (CR) (8, 9, 13). The food conditions used in (21), where Indy mutants showed no female life span extension and inconsistent male life span extension, is a low-calorie food. The food used in the studies by Toivonen (21) was a low-calorie food as it did not include the addition of live yeast, the standard procedure at the time of the initial publication on Indy life span extension (8), or the increase in addition of killed yeast to the base food that is in normal-calorie foods now. The report of a loss of Indy life span extension under these low calorie-food conditions (21) coupled with the hypothesis that Indy life span extension may be related to calorie restriction life span extension led us to further examine how food conditions might interact with Indy.
Life span studies were performed using a series of food calorie conditions on the 10 generation backcrossed yw;Indy206 stock. Consistent with the hypothesis that the Indy mutant may induce life span extension through a mechanism that overlaps with CR, we found a strong relationship between calorie content in the food and Indy mutant life span extension (Fig. 1A). At normal (1.0 N)- or high (1.5 N)-calorie conditions Indy206 heterozygote flies have a significant life span extension as compared to genetically matched controls (Fig. 1A and Table S1, 29% median lifespan extension, P < 0.0001). However, under low (0.5 N)-calorie conditions, the Indy heterozygote mutant flies show minimal life span extension over that already seen for controls on low calorie conditions (Fig. 1A and Table S1; 4% median lifespan extension, P < 0.03). These data suggest a relationship between Indy mutant life span extension and diet.
Decreases in the Calorie Content of Food Induce a Decrease in Indy Expression.
To explore whether low calorie food affects Indy expression, we measured Indy mRNA levels in control and Indy heterozygous mutant flies on high-, normal-, and low-calorie foods. Fig. 1B shows that for normal yw flies reduction of calorie content from high (1.5 N)- to normal (1.0 N)-calorie conditions results in an approximate 19% decrease in Indy mRNA. Reduction of calorie content from normal (1.0 N)- to low (0.5 N)-calorie conditions results in an additional 9% decrease in Indy mRNA. Reduction of calorie content from high (1.5 N)- to normal (1.0 N)-calorie conditions in yw;Indy heterozygous flies leads to a 20% reduction in Indy mRNA expression without an additional decrease upon further reduction to a low (0.5 N)-calorie food (Fig. 1B). The finding that the calorie content in food affects the expression of Indy mRNA lead us to further examine the relationship of Indy mRNA levels in CR related life span extension.
CR and Indy Interact to Determine Longevity.
The relationship between Indy mRNA expression, food calorie content, and life span is illustrated in the heat map in Fig. 1C. These data indicate that maximum life span extension is associated with Indy mRNA levels between 25–75% of normal. Outside of this range, when Indy mRNA levels are greater than 75% of normal or less than 25% of normal, the life span extension is diminished or largely lost. The correlation between the level of Indy mRNA and life span supports the hypothesis that the level of Indy expression plays an important role in life span determination.
Indy Long-Lived Flies Share Several Phenotypes with CR Long-Lived Flies.
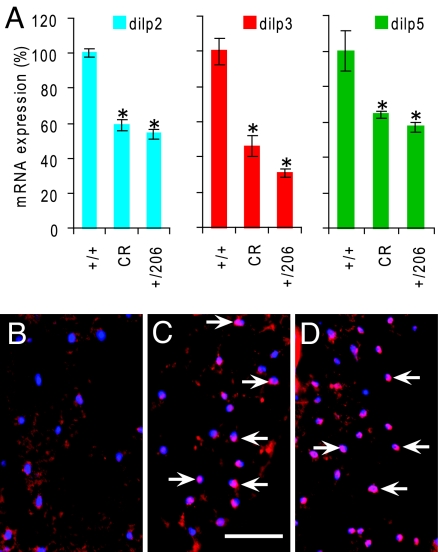
Long-lived Indy mutants on high-calorie food and normal flies on low-calorie food have reduced levels of dilp2, 3, and 5 mRNA expression. The state of insulin signaling provides one measure of the nutritional status of the fly. Under conditions of a nutrient challenge, such as CR, insulin signaling is decreased (17, 22). An indirect method for assessing the state of insulin signaling is the level of mRNA expression for the 3 Drosophila insulin-like peptides (Dilps) found in the neurosecretory insulin-producing cells (IPCs) of the brain: Dilp2, Dilp3, and Dilp5. Using qPCR, we confirmed that control flies on our low-calorie food (CR) have a 50–60% decrease in expression of Dilp2, Dilp3, and Dilp5 (Fig. 2A). We next found that Indy long-lived mutant flies on a high calorie diet also have a 50–60% decrease in levels of Dilp2, Dilp3, and Dilp5 (Fig. 2A). Thus Indy heterozygote long-lived flies on high calorie food show the same decrease in Dilp mRNA expresssion as normal control flies on low-calorie food.
Fig. 2.
CR and Indy alter insulin-like signaling. (A) Real-time PCR detection of mRNA expression levels of dilp2, dilp3, and dilp5 in yw control flies (+/+) on high-calorie food (1.5 N), yw control flies (+/+) on low-calorie food (0.5 N/CR), and Indy206 heterozygous (+/206) flies on high-calorie food (1.5 N). Data are presented as mean ± SD. Experiments were done in triplicate and each sample contained more than 20 heads and thoraxes of 20-day-old male flies. The * indicates P < 0.05 by t-test. (B–D) Immunohistochemical localization of dFOXO in the abdominal fat body of yw control (B and C) or Indy206 heterozygous (D) flies on high-calorie (1.5 N) food (B and D) or low-calorie (0.5 N/CR) food (C). Sections were visualized using a red fluorescent-conjugated secondary antibody for dFOXO and counter stained with DAPI for blue nuclear fluorescence. The white arrows indicate co-localization of dFOXO and DAPI in nuclei (pink). (Scale bar, 50 μm.)
Long-Lived Indy Mutants on High-Calorie Food and Normal Flies on Low-Calorie Food Have Increased Nuclear FoxO in Fat Body Cells.
Nuclear localization of FoxO in the insulin-responsive fat body cells is a more direct measure of the general state of insulin signaling in the fly (22). Under conditions of active insulin signaling, FoxO is phosphorylated and largely excluded from the nucleus. When insulin signaling is decreased, FoxO is not phosphorylated, and an increase in nuclear FoxO staining is seen (22). Normal control flies on high-calorie food have few anti-FoxO positive-staining nuclei in their fat body cells (22) (Fig. 2B). The number of anti-FoxO positive-staining nuclei in the fat body cells is greatly increased in normal flies under low-calorie food conditions (Fig. 2C). We found that long-lived Indy heterozygote flies on high-calorie foods have a similar high percentage of anti-FoxO-positive nuclei as normal flies on low-calorie food (Fig. 2C). Taken together, the decrease in Dilp2, 3, and 5 mRNA and increase in nuclear FoxO localization suggest that the long-lived Indy heterozygote flies on high- or normal-calorie food are in a decreased state of insulin signaling, similar to control flies on low-calorie food.
Long-Lived Indy Mutants on High-Calorie Food and Normal Flies on Low-Calorie Food Are Sensitive to Starvation.
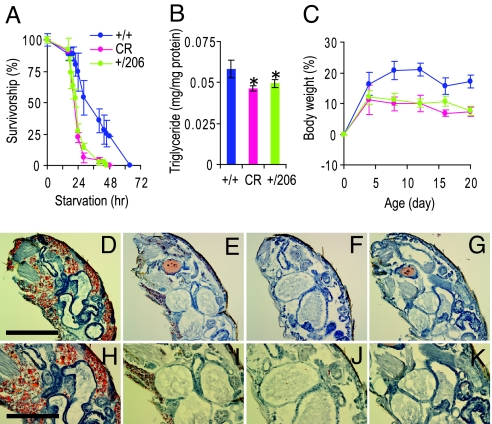
Under low-calorie food conditions, normal flies are unable to accumulate as much nutrient storage as on high-calorie foods. One manifestation of this is that normal flies that have been living on low-calorie food are sensitive to starvation stress as compared to normal flies that have been living on high-calorie food (Fig. 3A). Consistent with our hypothesis that Indy mutants alter nutrient storage in a manner that may overlap with calorie restriction, we found that long-lived Indy heterozygote flies living on high-calorie food are as sensitive to a starvation stress as normal flies on a low-calorie food (Fig. 3A).
Fig. 3.
CR and Indy induce reduced starvation resistance and body weight gain. (A) Twenty-day-old yw control flies (+/+) on high-calorie food (1.5 N, blue line) or low-calorie food (0.5 N/CR, red line), and Indy206 heterozygous flies (+/206) on high-calorie food (1.5 N, green line) were subjected to starvation challenge. Survivorship curves were collected from more than 100 flies and presented as mean ± SEM. Similar results were obtained from 2 other replicate experiments. (B) Total triglyceride measurements of flies on food conditions described above. Data are presented as mean ± SD. Experiments were done in triplicate and each sample contained more than 10 flies. The * indicates P < 0.05 by t test. (C) Change of body weight over a 20-day period was measured in flies on food conditions described above. Data are presented as mean ± SD. Experiments were done in triplicate, and more than 100 flies in each group were measured. (D–K) Oil Red O staining of yw control flies on 1.5 N food (D and H), 16-h fasted yw control flies on 1.5 N food (E and I), 16-h fasted yw control flies on 0.5 N/CR food (F and J), and 16-h fasted Indy heterozygous flies on 1.5 N food (G and K). Blue color is hemotoxylin counter stain. [Scale bars, 500 μm (D–G) and 250 μm (H–K).]
Long-Lived Indy Mutants on High-Calorie Food and Normal Flies on Low-Calorie Food Do Not Gain Weight.
Decreasing total caloric intake might be expected to be associated with a decrease in weight gain (23). As shown in Fig. 3C, normal flies on low-calorie food do not gain as much weight as their genetically identical cohorts on high-calorie food. Unlike control flies, Indy heterozygote long-lived flies on high-calorie food gain very little weight and thus have a similar lack of weight gain as normal control flies on low-calorie food (Fig. 3C).
Long-Lived Indy Mutants on High-Calorie Food and Normal Flies on Low-Calorie Food Have a Similar Decrease in Triglycerides and Fat Storage.
The finding that long-lived Indy heterozygotes living on a rich diet are as sensitive to starvation as normal flies living on a low-calorie food lead us to investigate the relationship between Indy long-lived flies and nutrient stores. We examined the level of trigylcerides as a measure of nutrient storage and ability to resist starvation. We found that normal flies on a low-calorie diet have a 20% (P = 0.001) decrease in total triglycerides, and Indy heterozygous flies on a high-calorie diet have a 15% (P < 0.001) decrease in total triglycerides compared with genetically matched controls (Fig. 3B). We further examined the relative amount of fat storage in long-lived normal flies on CR and long-lived Indy heterozygote flies on a high-calorie diet by examining the amount of Oil Red O lipid stain in fat body cells of adult fly sections after 16 h of fasting. After 16 h of fasting, normal flies that had been living on high-calorie food still had significant levels of Oil Red O staining (Fig. 3D, E, H, and I). However, within 16 h of fasting, normal flies that had been living on low-calorie food and Indy heterozygote flies living on high-calorie food had lost almost all Oil Red O staining in their fat body cells (Fig. 3F, G, J, and K). These observations suggest that both normal flies on low-calorie food and Indy heterozygote flies on high-calorie food store less lipids than normal flies living on high calorie-food conditions.
Long-Lived Indy Mutants on High-Calorie Food and Normal Flies on Low-Calorie Food Have a Higher Rate of Spontaneous Physical Activity.
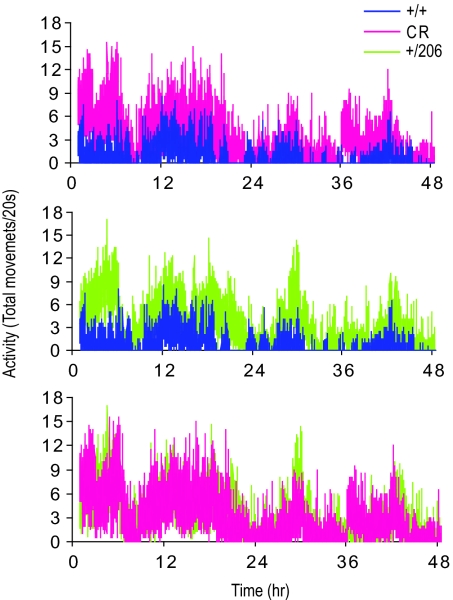
When mammals are placed on calorie-restricted diets, they increase their spontaneous physical activity (24). This is thought to be a normal component of the adaptive foraging behavior and has been observed in Drosophila (23). We found that normal control flies that have been living on low-calorie food show an increase in spontaneous physical activity. Interestingly, Indy long-lived heterozygote flies living on high-calorie food also show an increase in spontaneous physical activity that matches the increased physical activity seen with normal flies on low-calorie food (Fig. 4). Thus long-lived Indy heterozygote flies have the same higher rate of spontaneous physical activity that is normally associated with calorie restriction.
Fig. 4.
Indy and CR induce increased physical activity. Activities of 20-day-old yw control (+/+) flies on 1.5 N food (blue) or 0.5 N/CR food (red) and Indy heterozygous (+/206) flies on 1.5 N food (green) were recorded over a 48-h period using Drosophila activity monitor. Data were collected every 20 s and presented as total movement of 10 flies. Experiments were done in triplicate.
Long-Lived Indy Mutants Have Normal Intake of Food.
The life span extension seen in C. elegans eat mutants is thought to be due to a decrease in food intake related to a decrease in pharyngeal pumping. To test if the Indy heterozygous long-lived flies have a defect in food intake that could account for the similarity of phenotypes with CR, including life span extension, we examined food intake using a standard system of dye in their food. We found no difference in food intake between Indy long-lived heterozygote flies and matched normal controls on high-calorie food (Fig. S5). As noted previously, normal flies on low-calorie food had a modest increase in food intake (23). In fact, long-lived Indy heterozygous flies showed a similar modest increase in food intake when living on low-calorie food (Fig. S5). Not only do Indy long-lived heterozygous flies take in a normal amount of food, they increase this intake in response to lowering food calorie content in the food. The life span extension and other phenotypes shared between Indy long-lived heterozygous flies and normal CR flies is not due to a decrease in food intake.
Discussion
It is well recognized that in evaluating the effects of individual genes on complex biological phenomena such as aging the environmental context and genetic background of the organism needs to be taken into consideration. Genes that manifest their phenotype through alterations in metabolism are likely to lead to different effects based upon the environmental conditions (e.g., nutrition) in which they are examined. Given the central nature of metabolism and the large number of genes involved in setting the metabolic state of the organism, the effect of alterations in specific metabolically related genes on organismal function will be modulated by the specific genetic background of the organism.
The Indy gene product has been postulated to be involved in normal metabolism, as it is a plasma membrane transporter of Kreb's cycle intermediates found primarily in tissues responsible for uptake, utilization, and storage of nutrients in the fly (8). We found that the life span extension seen with the long-lived Indy mutation is sensitive to both food conditions and genetic background. When living on the typical Drosophila laboratory-culturing food conditions, similar to normal- or high-calorie food, reduction of Indy leads to significant life span extension. Under low-calorie food conditions however, the life span extending effects of calorie restriction mask the life span extending effect of Indy. The conclusion by Toivenon et al. (21) that the Indy mutation plays no role in life span extension is likely due to the use of low-calorie food conditions in their studies. Unlike the conclusions of Toivenon et al. (21) we directly show that reduction of Indy transcription, between a range of 25–75% of normal, has a strong positive effect on life span extension in high- and normal-calorie food conditions (Fig. 1C).
Genetic background is known to affect the calorie restriction life span-extension response in Drosophila. For example, the w1118 strain has a severely blunted life span-extension response to CR compared with other wild-type strains such as Canton-S (25). Similarly, life span extension induced by a decrease in Indy expression is also dependent upon genetic background. Toivenon et al. (21) backcrossed Indy into the w1118 strain, found a loss of Indy related life span extension, and interpreted this as demonstrating Indy expression is not involved in life span extension. We independently backcrossed Indy into w1118 and found that the w1118 does suppress the life span extending effect of Indy mutants (Fig. S3). However, when the Indy mutation is in a Canton-S background (the original strain it was isolated in) or yw background, a significant life span extension is seen (Figs. S2 and S4 and Fig. 1). The study by Toivenon et al. (21) rather than disproving that mutations in Indy are causally involved in life span extension provide evidence supporting a strong interaction between food conditions, genetic background, and Indy expression on longevity.
Our studies suggest an intimate relationship between food calorie content and Indy expression. Food calorie conditions directly affect the level of Indy transcription. A reduction of food calorie content, such as CR, causes a 20% or greater decrease in Indy mRNA expression in normal or heterozygous mutant Indy flies. Examination of life span, food conditions, and Indy mRNA expression, Fig. 1C demonstrates the strong interaction between Indy and food calorie content in the determination of longevity in flies. Our data support the hypothesis that the level of Indy expression plays an important role in life span determination regardless of whether the reduction in Indy mRNA expression is through the insertion of a P-element into the Indy region or a reduction of Indy mRNA via a change in food calorie content. These data suggest that the amount of Indy mRNA and food calorie content interact to achieve the significant life span extension seen.
The finding that food calorie content directly affects the level of Indy expression and that either CR or Indy expression can affect longevity suggested to us that there may be some overlap in the mechanisms by which CR and Indy mutations extend life span. In support of this we found that normal flies on low-calorie food (CR) and Indy heterozygous mutant flies on normal- or high-calorie food share several physiological and behavioral changes. Both normal animals on low-calorie conditions and Indy long-lived heterozygotes on normal-calorie conditions: (i) have a decrease in insulin signaling as measured by a decrease in transcription of Dilp2, 3, and 5 and an increase in dFOXO nuclear expression in fat body cells; (ii) a decrease in total triglycerides and total fat storage in fat body cells; (iii) do not gain weight; (iv) are sensitive to starvation; and (v) have a higher rate of spontaneous physical activity. The induction of these changes is not the result of a decrease in food intake in Indy long-lived heterozygotes that could secondarily cause CR since Indy long-lived heterozygotes take in as much food as normal flies, if not more (Fig. S5). This is in agreement with findings that functional knockdown of nac-2, a nematode transporter with sequence homology to Indy, has extended longevity, smaller body size, and decreased levels of fat (11).
The nature of the relationship between Indy life span extension and CR life span extension is not clear. Our data show that Indy long-lived heterozygote flies manifest a number of physiological and behavioral changes that occur when normal animals are placed under CR life span-extending conditions. The lack of an additive effect on life span extension when Indy long-lived heterozygote flies are placed on CR conditions, coupled with the finding that CR directly leads to a decrease in Indy transcription, provide genetic and molecular epistasis evidence, suggesting that CR and Indy interact to extend life span. The finding that CR reduces Indy expression suggests that a decrease in Indy may be one of the “downstream” components of the normal CR life span extending pathway in the fly.
The manipulation of food content, CR, and the level of Indy expression appear to interact to attain an optimal balance for achieving life span extension. The possibility that Indy is one of the downstream effectors of CR life span extension suggests that identification of the downstream physiological and molecular targets shared between these 2, CR and Indy, life span-extending interventions may provide further insights into the mechanisms of CR life span extension. A better understanding of the interaction between these 2 related interventions as well as other shared downstream elements could be of great benefit in developing interventions that can extend healthy life span and lead to other positive health benefits without the need for some of the unacceptable effects of severe CR. The realization that Indy and CR interact to extend life span suggests that a simultaneous modest modification of both could obviate the need for a more severe CR regime.
Methods
Fly Stocks and Life Span Assays.
Long-lived Indy mutants Indy206 were backcrossed into the yw or w1118 background. Female virgins from yw or w1118 stocks were first mated with Indy206 males to ensure the transfer of cytoplasmic constituents from yw or w1118 to progeny. Heterozygous mutant females were then backcrossed to yw or w1118 males for ten generations. To remove Wolbachia infection, these stocks were cultured on food containing 25 mg/mL tetracycline for 3 generations, followed by several generations in tetracycline-free media. The removal of Wolbachia was confirmed by PCR. For the life span assays, groups of 25 newly eclosed males and females were placed together in each vial with a total of at least 8 to 10 vials per assay. Flies were transferred to fresh food every other day, and the number of dead flies was scored. All flies were maintained in a humidified, temperature-controlled incubator with 12/12-h on/off light cycle at 25 °C in vials containing modified Sugar-Yeast food (26). The foods 0.5 N, 1 N, and 1.5 N contain 5%, 10%, and 15% (wt/vol) of dextrose and yeast, respectively. Two percent agar and 0.23% Tegosept (Apex) were added to all foods.
Starvation Stress, Oil Red O Stain, and Triglyceride Determination.
Male flies at 20 days of age were transferred to vials containing 2% agar. The number of dead flies in each vial was scored regularly. All starvation assays were performed using 100 to 200 flies per group. Flies were maintained under standard conditions (see above). For the Oil Red O stain, flies were starved for 16 h, then fixed in 4% paraformaldehyde/PBS for 20 min. Fixed samples were embedded in Tissue Freezing Medium (TFM, Triangle Biomedical), and sections were cut on a cryostat at a thickness of 10 μm. Sections were stained in 0.3% Oil Red O for 15 min and counter stained with haemotoxylin. To measure total triglyceride, ten 20-day-old flies were homogenized in 300 μL PBS containing 0.05% Tween 20 and centrifuged at 400 × g. Ten microliters of supernatant was then used to measure total triglyceride using the Triglyceride Determination Kit (Sigma).
Immunohistochemistry.
Twenty-day-old male flies were fixed, embedded, and sectioned as described above. Slides were incubated for 2 h at room temperature with an affinitiy-purified rabbit anti-dFoxo antibody (kind gift of Dr. Marc Tatar) at a dilution of 1:500 in PBS containing 5% normal goat serum and 0.1% Triton-X-100. After washing, slides were incubated for 1 h in Alexa 568-conjugated goat anti-rabbit IgG secondary antibody (Molecular Probes/Invitrogen), then washed, and mounted in antifade compound containing DAPI as a nuclear counterstain (Molecular Probes/Invitrogen). Images were collected on a Zeiss Axiovert microscope equipped with a cooled CCD camera, and running Axiovision 4.5 software.
Wolbachia DNA Detection.
Total DNA from more than 20 flies was isolated using a Maxwell 16 Instrument and tissue DNA purification kit (Promega). Wolbachia DNA was detected by PCR. The GADPH and Wolbachia primers (27) were mixed in the same PCR with denaturation at 95 °C for 15 s, annealing at 60 °C for 20 s, and extension at 72 °C for 30 s in a total of 30 cycles. The primer sequences were described in Table S2.
RNA Preparation, cDNA Synthesis, and Real-Time PCR.
Total RNA was prepared from more than 20 heads and thoraxes from 20-day-old male flies using the RNeasy Mini Kit (Qiagen). The RNA was treated with DNase and converted to cDNA using oligo-d(T)15 (Invitrogen) and SuperScript III reverse transcriptase (Invitrogen) as described previously (28). Real-time PCR reactions were performed using a 7500 Fast Real-Time PCR System (Applied Biosystems), SYBR Green Master Mix (Applied Biosystems), and gene-specific primers (Table S2). A 2-step PCR was carried out with denaturation at 95 °C for 15 s, annealing, and extension combined at 60 °C for 1 min in a total of 40 cycles. The uniqueness of amplicons was analyzed using dissociation.
Monitoring Activity Levels.
Glass vials containing 10 male flies and the appropriate food source were placed in locomotor recording chambers with circular rings of infrared beams at 3 different levels (TriKinetics). The data were recorded every 20 s in a 48-h period. Activity monitors were housed in incubators set at 25 °C on a 12/12-h on/off light cycle as was used throughout this research.
Body Weight and Feeding Assay.
Five cohorts of 20 to 25 male flies were anesthetized (CO2) and weighed immediately using a Mettler-Toledo analytical scale. The same groups of flies were measured longitudinally from day 1 and then every 4 days until day 20. For the feeding assay, 20-day-old flies maintained on 5% and 15% dextrose-yeast were transferred to the same foods with addition of 0.5% of FD & C no. 1 blue food dye. After 24 h, the flies were homogenized in PBS and the amount of dye ingested was determined by spectrophotometer for dye absorbance at 625 nm.
Statistical Analyses.
All statistical analyses were performed using JMP (version 5.1) software (SAS Institute). Life span data were analyzed by log-rank tests. Maximum life span was calculated as the median lifespan of the longest surviving 10% of the population. Student's t test was used for mRNA expression, triglyceride body weight change and feeding rate data analyses.
Supplementary Material
Acknowledgments.
We thank Mr. Will Lightfoot for fly food preparation. This work was supported by National Institutes on Aging (NIA) grants NIA K25AG028753 to NN, AG16667, AG24353 and AG25277 to SLH, and AG23088 to BR. SLH is an Ellison Medical Research Foundation Senior Investigator and recipient of a Glenn Award for Research in Biological Mechanisms of Aging.
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/cgi/content/full/0904115106/DCSupplemental.
References
- 1.Kenyon C. The plasticity of aging: Insights from long-lived mutants. Cell. 2005;120:449–460. doi: 10.1016/j.cell.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 2.Guarente L, Picard F. Calorie restriction– the SIR2 connection. Cell. 2005;120:473–482. doi: 10.1016/j.cell.2005.01.029. [DOI] [PubMed] [Google Scholar]
- 3.Lin S-J, Defossez P-A, Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. [DOI] [PubMed] [Google Scholar]
- 4.Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wood JG, et al. Sirtuin activators mimic caloric restriction and delay ageing in metazoans. Nature. 2004;430:686–689. doi: 10.1038/nature02789. [DOI] [PubMed] [Google Scholar]
- 6.Bishop NA, Guarente L. Two neurons mediate diet-restriction-induced longevity in C. elegans. Nature. 2007;447:545–549. doi: 10.1038/nature05904. [DOI] [PubMed] [Google Scholar]
- 7.Panowski SH, et al. PHA-4/Foxa mediates diet-restriction-induced longevity of C. elegans. Nature. 2007;447:550–555. doi: 10.1038/nature05837. [DOI] [PubMed] [Google Scholar]
- 8.Rogina B, et al. Extended Life-Span Conferred by Cotransporter Gene Mutations in Drosophila. Science. 2000;290:2137–2140. doi: 10.1126/science.290.5499.2137. [DOI] [PubMed] [Google Scholar]
- 9.Knauf F, et al. Functional characterization and immunolocalization of the transporter encoded by the life-extending gene Indy. Proc Natl Acad Sci USA. 2002;99:14315–14319. doi: 10.1073/pnas.222531899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Knauf F, et al. The life-extending gene Indy encodes an exchanger for Krebs-cycle intermediates. Biochem J. 2006;397:25–29. doi: 10.1042/BJ20060409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fei Y-J, et al. Relevance of NAC-2, an Na+-coupled citrate transporter, to life span, body size and fat content in Caenorhabditis elegans. Biochem J. 2004;379:191–198. doi: 10.1042/BJ20031807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gargano JW, et al. Rapid iterative negative geotaxis (RING): A new method for assessing age-related locomotor decline in Drosophila. Exp Gerontol. 2005;40:386–395. doi: 10.1016/j.exger.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 13.Marden JH, et al. Conditional tradeoffs between aging and organismal performance of Indy long-lived mutant flies. Proc Natl Acad Sci USA. 2003;100:3369–3373. doi: 10.1073/pnas.0634985100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Martin I, Grotewiel MS. Distinct genetic influences on locomotor senescence in Drosophila revealed by a series of metrical analyses. Exp Gerontol. 2006;41:877–881. doi: 10.1016/j.exger.2006.06.052. [DOI] [PubMed] [Google Scholar]
- 15.Tatar M, Bartke A, Antebi A. The endocrine regulation of aging by insulin-like signals. Science. 2003;299:1346–1351. doi: 10.1126/science.1081447. [DOI] [PubMed] [Google Scholar]
- 16.Houthoofd K, et al. Life extension via dietary restriction is independent of the Ins/IGF-1 signalling pathway in Caenorhabditis elegans. Exp Gerontol. 2003;38:947–954. doi: 10.1016/s0531-5565(03)00161-x. [DOI] [PubMed] [Google Scholar]
- 17.Min K-J, et al. Drosophila lifespan control by dietary restriction independent of insulin-like signaling. Aging Cell. 2008;7:199–206. doi: 10.1111/j.1474-9726.2008.00373.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Clancy DJ, et al. Dietary restriction in long-lived dwarf flies. Science. 2002;296:319. doi: 10.1126/science.1069366. [DOI] [PubMed] [Google Scholar]
- 19.Giannakou ME, Goss M, Partridge L. Role of dFOXO in lifespan extension by dietary restriction in Drosophila melanogaster: Not required, but its activity modulates the response. Aging Cell. 2008;7:187–198. doi: 10.1111/j.1474-9726.2007.00362.x. [DOI] [PubMed] [Google Scholar]
- 20.Bonkowski MS, et al. Targeted disruption of growth hormone receptor interferes with the beneficial actions of calorie restriction. Proc Natl Acad Sci USA. 2006;103:7901–7905. doi: 10.1073/pnas.0600161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Toivonen JM, et al. No influence of Indy on lifespan in Drosophila after correction for genetic and cytoplasmic background effects. PLoS Genet. 2007;3:e95. doi: 10.1371/journal.pgen.0030095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bauer JH, et al. Expression of dominant-negative Dmp53 in the adult fly brain inhibits insulin signaling. Proc Natl Acad Sci USA. 2007;104:13355–13360. doi: 10.1073/pnas.0706121104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bross TG, Rogina B, Helfand SL. Behavioral, physical, and demographic changes in Drosophila populations through dietary restriction. Aging Cell. 2005;4:309–317. doi: 10.1111/j.1474-9726.2005.00181.x. [DOI] [PubMed] [Google Scholar]
- 24.Chen D, et al. Increase in activity during calorie restriction requires Sirt1. Science. 2005;310:1641. doi: 10.1126/science.1118357. [DOI] [PubMed] [Google Scholar]
- 25.Libert S, et al. Regulation of Drosophila life span by olfaction and food-derived odors. Science. 2007;315:1133–1137. doi: 10.1126/science.1136610. [DOI] [PubMed] [Google Scholar]
- 26.Chapman T, Partridge L. Female fitness in Drosophila melanogaster: An interaction between the effect of nutrition and of encounter rate with males. Proc Biol Sci. 1996;263:755–759. doi: 10.1098/rspb.1996.0113. [DOI] [PubMed] [Google Scholar]
- 27.Zhou W, Rousset F, O'Neill S. Phylogeny and PCR-based classification of Wolbachia strains using wsp gene sequences. Proc Biol Sci. 1998;265:509–515. doi: 10.1098/rspb.1998.0324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang P-Y, et al. Mullerian Inhibiting Substance acts as a motor neuron survival factor in vitro. Proc Natl Acad Sci USA. 2005;102:16421–16425. doi: 10.1073/pnas.0508304102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.