Abstract
Transposable elements (TEs) are major sources of new exons in higher eukaryotes. Almost half of the human genome is derived from TEs, and many types of TEs have the potential to exonize. In this work, we conducted a large-scale analysis of human exons derived from mammalian-wide interspersed repeats (MIRs), a class of old TEs which was active prior to the radiation of placental mammals. Using exon array data of 328 MIR-derived exons and RT–PCR analysis of 39 exons in 10 tissues, we identified 15 constitutively spliced MIR exons, and 15 MIR exons with tissue-specific shift in splicing patterns. Analysis of RNAs from multiple species suggests that the splicing events of many strongly included MIR exons have been established before the divergence of primates and rodents, while a small percentage result from recent exonization during primate evolution. Interestingly, exon array data suggest substantially higher splicing activities of MIR exons when compared with exons derived from Alu elements, a class of primate-specific retrotransposons. This appears to be a universal difference between exons derived from young and old TEs, as it is also observed when comparing Alu exons to exons derived from LINE1 and LINE2, two other groups of old TEs. Together, this study significantly expands current knowledge about exonization of TEs. Our data imply that with sufficient evolutionary time, numerous new exons could evolve beyond the evolutionary intermediate state and contribute functional novelties to modern mammalian genomes.
INTRODUCTION
One of the most intriguing questions in evolutionary biology is the origin of evolutionary novelties. Evolution can create new gene functions via different processes, many of which have been investigated in detail [e.g. the creation of new genes (1), gene duplication (2–4), divergence of protein-coding sequences (5) and evolution of transcriptional regulation (6,7)]. In recent years, comparative analyses of exon–intron structures of orthologous genes from multiple species have revealed frequent creation of new exons during the evolution of higher eukaryotes [reviewed by (8,9)]. A variety of molecular mechanisms, such as exaptation of transposable elements (TE; 10,11), exon duplication (12,13) and de novo exonization from intronic regions (14), can add new exons to evolutionarily ancient genes. Lee and colleagues (14) analyzed the multiple alignment of 17 vertebrate genomes and identified thousands of human exons that were created during primate evolution. Other studies also reported high incidences of exon creation events in primate and rodent genomes (10,15). The vast majority of newly created exons are alternatively spliced with low transcript inclusion levels in expressed sequence tag (EST) sequences, suggesting that they are non-functional evolutionary intermediates (8,9). Nevertheless, certain new exons may have acquired functions during evolution. A well-known example is the new exon (exon 8) of human gene ADAR2 (adenosine deaminase, RNA-specific, B1). This exon was derived from a primate-specific Alu retrotransposon. It inserts a 40-amino acid peptide segment into the catalytic domain of ADAR2, altering the enzymatic activity of the protein product (16). However, despite a growing list of anecdotal reports for regulatory and functional roles of new exons (9), genome-wide analyses of new exons were mostly based on EST data (10,11,14,15,17) and few identified exonization events have been subjected to detailed experimental characterization. Thus, the evolution and impact of new exons remain poorly understood.
TEs are major sources of new exons in higher eukaryotes (9). Almost half of the human genome is derived from TEs (18), and many types of TEs have the potential to exonize (11). For example, exonization of Alu elements has been studied extensively (19–21). Alu is a primate-specific TE that belongs to the SINE (Short Interspersed Nuclear Element) family (22). It was created from the fusion of two 7SL RNA Alu monomers approximately 60 million years ago. It is the most abundant class of TEs in the human genome, with over one million copies occupying approximately 10% of the genomic DNA (18). As Alu contains several sites that resemble consensus splice site signals, intronic Alus have the potential to be recruited into transcripts of their host genes as new exons (19). Prior EST-based studies indicate that all Alu exons are alternatively spliced, and most of them have low transcript inclusion levels (20). We recently performed a large-scale analysis of Alu exons, using high-density exon array data of 330 exons and detailed RT–PCR analysis of 38 exons in 10 human tissues (23). Our study revealed surprisingly diverse splicing patterns of Alu exons in human tissues. We identified a limited number of Alu exons that were constitutively spliced in a broad range of tissues, as well as Alu exons that were spliced in a tissue-specific manner, suggesting that certain Alu exons may have acquired functional and regulatory roles during primate evolution. Strikingly, of the 26 Alu exons with high transcript inclusion levels according to RT–PCR data, there was 4-fold enrichment in exons derived from AluJ, the most ancient Alu subfamily in the human genome (23). This observation suggests that evolutionary time may be a major factor in the functional establishment of new exons. In fact, in one gene (p75TNFR) of which an Alu exonization event was characterized in detail over the primate phylogeny, a series of nucleotide changes occurred during a period of approximately 33 million years between the initial Alu insertion event and the emergence of a functional alternative exon (24).
In this work, we analyzed human exons derived from a much older class of TEs, mammalian-wide interspersed repeats (MIRs). MIRs were actively transposed prior to the radiation of placental mammals (approximately 130 million years ago) (25). The human genome has approximately 368 000 copies of MIRs, some of which overlap with exons of protein-coding genes (11,18,26). Previous studies generated contradictory results on the exonization level of MIR elements in the human genome. Krull and colleagues (26,27) conducted phylogenetic analyses of five MIR exons and four Alu exons using DNAs and RNAs from various mammalian and primate species. Their results suggest that the splicing signals of Alu exons acquired in ancestral primate genomes are often lost in descendent primate lineages (27). In contrast, the splicing signals of MIR exons tend to be conserved over a long period of evolutionary time, suggesting stable exonization and potential acquisition of functional properties by MIR exons (26). However, since the studies of Krull et al. focused on a small number of exons, it is unclear whether their observations can be generalized to the entire classes of Alu- and MIR-derived exons. On the other hand, in a genome-wide analysis of exonized TEs in the human genome, Sela and colleagues (11) found that exonization of intronic Alus was far more frequent than exonization of intronic MIRs. One thousand and sixty intronic Alus (0.2% of all intronic Alus) overlap with human exons, when compared with 181 intronic MIRs (0.08% of all intronic MIRs). However, the analysis of Sela et al. was based on exon–intron structures inferred from sequence data (i.e. mRNAs and ESTs). It was difficult to distinguish functional exonization events from exons that were incorporated into transcripts owing to rare errors of the splicing machinery (28,29). To better understand the evolutionary and functional significance of exonized MIR elements, in the present study, we performed a large-scale analysis of MIR-derived exons in primate genomes. Here, we report our results from exon array analysis of 328 MIR exons, RT–PCR analysis of 39 MIR exons in 10 human tissues, and phylogenetic analysis of nine MIR exons in RNAs from primates and rodents.
RESULTS
Splicing signals and exon array profiles of MIR exons in human tissues
We collected a list of 328 MIR-derived exons in the human genome, using annotations from the UCSC Genome Browser database (30) and Affymetrix human Exon 1.0 array (see details in Materials and Methods). For the purpose of comparison, we also collected 330 Alu-derived exons [the list of Alu exons analyzed in (23)], as well as 13 103 constitutive exons in the human genome.
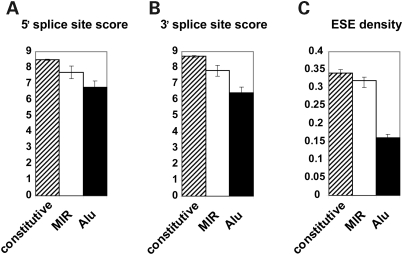
We compared the splicing signals of MIR exons, Alu exons and constitutive exons. For each exon, we scored its 5′ and 3′ splice sites using consensus splice site models in MAXENT (31). The average 5′ splice site score of MIR-derived exons was 7.71, lower than constitutive exons (8.49; P = 0.001, Wilcoxon test) but significantly higher than Alu-derived exons (6.78; P = 4.7e−7; Fig. 1A). The same trend was also observed for 3′ splice site. The average 3′ splice site scores of MIR exons, Alu exons and constitutive exons were 7.82, 6.43 and 8.70, respectively (Fig. 1B). This trend is consistent with the observation of Sela et al. (11) on an independent data set of TE-derived exons. We also calculated the density of exonic splicing enhancers (ESEs) on these three classes of exons. ESEs are short RNA sequence motifs that promote exon recognition during splicing (32). Using a set of 238 ESE hexamers from Fairbrother et al. (33), we calculated the average ESE density in different classes of exons as the proportion of total exon length covered by ESEs. The average ESE density was 0.32 in MIR exons, which was similar to constitutive exons (0.34, P = 0.15) and significantly higher than Alu exons (0.16, P < 2.2e−16; Fig. 1C).
Figure 1.
Splicing signals of constitutive exons, MIR-derived exons and Alu-derived exons. (A) 5′ splice site score. (B) 3′ splice site score. (C) Density of exonic splicing enhancers (ESEs). Error bars indicate 95% confidence interval.
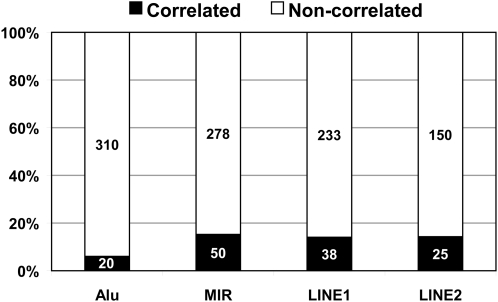
To investigate whether the difference in splicing signals between MIR exons and Alu exons is correlated with differences in their splicing profiles in human tissues, we analyzed a public Affymetrix Exon 1.0 array data set of 11 human tissues (breast, cerebellum, heart, kidney, liver, muscle, pancreas, prostate, spleen, testes, thyroid, with three replicates per tissue) (34). The Affymetrix human Exon 1.0 array is a high-density exon array platform designed for genome-wide analysis of pre-mRNA splicing, with approximately four probes per exon for well-annotated and predicted exons in the human genome (35,36). In our previous study of Alu-derived exons, we developed an exon-to-gene correlation metric to identify ‘correlated exons’ whose exon array probe intensities correlated strongly with the estimated expression levels of their host genes across a broad range of tissues [see details in Materials and Methods and (23)]. A strong correlation between exon signals and gene expression levels suggests stable inclusion of the exon in the transcript products. In this study, using the same correlation metric we identified 50 ‘correlated exons’ from a total of 328 MIR exons (15.2%). In contrast, of the 330 Alu exons analyzed in our previous work, 20 were ‘correlated exons’ (6.1%; Fig. 2). The percentage of correlated MIR exons was significantly higher than the percentage of correlated Alu exons (15.2% versus 6.1%, P-value = 1.3e−4; two-sided Fisher exact test). It should be noted that 15.2% is expected to be an underestimate for MIR exons with stable transcript inclusion, as noise in exon array probe intensities could even obscure the exon-to-gene correlation of constitutive exons (37). Nevertheless, as we applied the same analysis to MIR exons and Alu exons, these results indicate that a significantly larger fraction of MIR exons are spliced at substantial levels in human tissues.
Figure 2.
The percentage of ‘correlated exons’ among exons derived from young transposable elements (Alu) and old transposable elements (MIR, LINE1, LINE2).
RT–PCR analysis of 39 MIR exons in 10 human tissues
Exon array data provide valuable information of an exon's splicing profiles, but cannot reveal its absolute transcript inclusion levels in individual tissues. For example, owing to variations in microarray probe affinity (38,39), we cannot predict whether a correlated exon is constitutively spliced, or alternatively spliced with similar transcript inclusion levels in various tissues. The existence of microarray noise also makes it difficult to identify the tissue-specific splicing patterns of certain correlated exons.
In order to uncover the detailed splicing patterns of correlated MIR exons, we analyzed 39 exons in 10 human tissues by RT–PCR. In selecting exons for RT–PCR analysis, we skipped exons for which RT–PCR primer design was difficult, e.g. MIR exons adjacent to very short terminal exons or MIR exons with complex alternative splicing events at adjacent exons. For each exon tested by RT–PCR, we first compared the observed sizes of PCR products to the expected sizes of all potential exon inclusion and skipping isoforms. In case of any ambiguity, all potential exon inclusion and skipping PCR products were confirmed by sequencing.
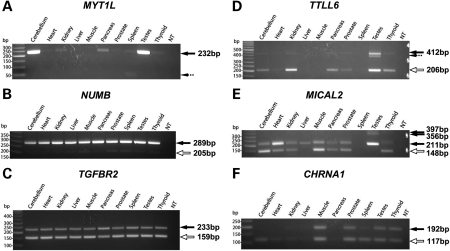
Our RT–PCR analysis identified MIR exons with a broad range of splicing patterns in human tissues (see Table 1, Supplementary Material, Fig. S1 for gel pictures of all 39 MIR exons analyzed). Prior to our analysis, two constitutively spliced MIR exons were reported in the literature (11,26). For example, a MIR-derived exon in MYT1L was shown to be constitutively spliced in a human neuronal cell line and mouse brain (11). In our work, we found this exon to be constitutively spliced in all tissues where its host gene was expressed (Fig. 3A). We identified a total of 15 MIR exons as constitutively spliced in all surveyed tissues. These data indicate that numerous MIR exons have acquired strong splicing signals that result in 100% transcript inclusion. It should be noted that in six of the 15 constitutive MIR exons, the MIR exon was present in multiple PCR products (see Supplementary Material, Fig. S1), owing to alternative splicing of flanking exon(s) as revealed by sequencing of PCR products. In addition to these constitutive exons, we identified three alternatively spliced MIR exons that were included in the major isoform products in all tissues (see Fig. 3B for an example of NUMB), and two exons that were included at medium levels in all tissues (see Fig. 3C for an example of TGFBR2).
Table 1.
RT–PCR analysis of 39 mammalian-wide interspersed repeat (MIR) exons in 10 human tissues
| Gene symbol | Transcript cluster ID | Probeset ID | MIR exon location (hg18) | MIR strand/mRNA | PCR skipping (bp) | PCR inclusion (bp) | Splicing pattern | Impact on mRNA/protein | Gene name | GO processes/known features |
|---|---|---|---|---|---|---|---|---|---|---|
| CSF3R | 2406783 | 2406822 | chr1:36719666-36719725 | Antisense | None | 236 | *Constitutive inclusion | 5′-UTR | colony stimulating factor 3 receptor (granulocyte) | Cell adhesion, cytokine receptor activity |
| CUL2 | 3284882 | 3284888 | chr10:35340782-35340898 | Sense | None | 268 | *Constitutive inclusion | Coding | cullin 2 | G1/S transition of mitotic cell cycle |
| FXYD4 | 3244008 | 3244011 | chr10:43188181-43188226 | Antisense | None | 115/251 | *Constitutive inclusion | 5′-UTR | FXYD domain containing ion transport regulator 4 | Ion transport |
| IL6ST | 2857416 | 2857457 | chr5:55314309-55314396 | Sense | None | 204 | *Constitutive inclusion | 5′-UTR | interleukin 6 signal transducer (gp130, oncostatin M receptor) | Cytokine-mediated signaling pathway |
| KLC1 | 3553872 | 3553899 | chr14:103221076-103221126 | Antisense | None | 221/248 | *Constitutive inclusion | Coding with ALT stop | kinesin light chain 1 | Microtubule motor activity |
| MTA2 | 3375862 | 3375867 | chr11:62118307-62118455 | Antisense | None | 373 | *Constitutive inclusion | Coding | metastasis associated 1 family, member 2 | Transcription factor |
| MTIF2 (2553751) | 2553730 | 2553751 | chr2:55348209-55348275 | Antisense | None | 313/451 | *Constitutive inclusion | 5′-UTR | mitochondrial translational initiation factor 2 | Regulation of translational initiation |
| MTIF2 (2553754) | 2553730 | 2553754 | chr2:55349218-55349379 | Sense | None | 251/224/296 | *Constitutive inclusion | 5′-UTR | mitochondrial translational initiation factor 2 | Regulation of translational initiation |
| MTX1 | 2360773 | 2360784 | chr1:153449652-153449806 | Antisense | None | 309 | *Constitutive inclusion | Coding | metaxin 1 | Protein transport |
| MYT1L | 2466822 | 2466898 | chr2:1979970-1980126 | Antisense | None | 232 | *Constitutive inclusion | 5′-UTR | myelin transcription factor 1-like | Cell differentiation, regulation of transcription, DNA-dependent |
| SLC6A12 | 3439510 | 3439537 | chr12:191453-191537 | Antisense | None | 329 | *Constitutive inclusion | 5′-UTR | solute carrier family 6 (neurotransmitter transporter, betaine/GABA), member 12 | Neurotransmitter transport |
| SRRM2 | 3645253 | 3645260 | chr16:2747783-2747947 | Sense | None | 321 | *Constitutive inclusion | Coding | serine/arginine repetitive matrix 2 | Pre-mRNA splicing factor activity |
| ST3GAL1 | 3154398 | 3154432 | chr8:134580560-134580614 | Antisense | None | 226/320/351 | *Constitutive inclusion | 5′-UTR | ST3 beta-galactoside alpha-2,3-sialyltransferase 1 | Protein amino acid glycosylation |
| TFIP11 | 3955875 | 3955908 | chr22:25238065-25238141 | Sense | None | 358/435 | *Constitutive inclusion | 5′-UTR | tuftelin interacting protein 11 | RNA splicing |
| TYK2 | 3850278 | 3850327 | chr19:10351291-10351455 | Antisense | None | 410 | *Constitutive inclusion | 5′-UTR | tyrosine kinase 2 | Growth hormone receptor binding, protein amino acid phosphorylation |
| AY070437 | 2649710 | 2649716 | chr3:159942513-159942627 | Antisense | 116 | 231/458 | *ALT major | Non-coding transcript | Homo sapiens cytokine receptor CRL2 precursor, mRNA, complete cds | Receptor activity |
| C12orf52 | 3432641 | 3432648 | chr12:112108373-112108504 | Sense | 342 | 474 | *ALT major to constitutive | 5′-UTR | chromosome 12 open reading frame 52 | Unknown |
| NUMB | 3571347 | 3571378 | chr14:72903358-72903442 | Antisense | 205 | 289 | *ALT major to constitutive | 5′-UTR | numb homolog (Drosophila) | Nervous system development |
| ARNTL | 3321150 | 3321174 | chr11:13304536-13304590 | Antisense | 121 | 176 | ALT medium | 5′-UTR | aryl hydrocarbon receptor nuclear translocator-like | Positive regulation of transcription from RNA polymerase II promoter |
| TGFBR2 | 2615360 | 2615384 | chr3:30639695-30639769 | Antisense | 159 | 233 | ALT medium | Coding | transforming growth factor, beta receptor II (70/80 kDa) | Protein kinase activation, organ development |
| EIF4G3 | 2400373 | 2400483 | chr1:21310403-21310463 | Antisense | 97 | 158 | ALT minor | 5′-UTR | eukaryotic translation initiation factor 4 gamma, 3 | Regulation of translational initiation |
| FGF14 | 3523499 | 3523565 | chr13:101844109-101844182 | Antisense | 148 | 222 | ALT minor | Exon in a non-coding transcript | fibroblast growth factor 14 | Cell–cell signaling |
| MYO18A | 3751323 | 3751412 | chr17:24488160-24488228 | Sense | 234/270 | None | No inclusion detected | 5′-UTR | myosin XVIIIA | ATP binding, apoptosis inhibitor activity |
| VWF | 3441685 | 3441800 | chr12:6102152-6102393 | Antisense | 156 | None | No inclusion detected | Coding with ALT start | von Willebrand factor | Cell–substrate adhesion, physiological response to wounding |
| ANKRD9 | 3580357 | 3580370 | chr14:102044731-102044936 | Sense | 90/124/159 | 365 | Tissue-specific ALT medium form in muscle and minor form in testes | 5′-UTR | ankyrin repeat domain 9 | Unknown |
| CHRNA1 | 2587937 | 2587951 | chr2:175330920-175330994 | Sense | 117 | 192 | Tissue-specific ALT minor form in cerebellum and spleen, medium form in all the other tissues | Coding | cholinergic receptor, nicotinic, alpha 1 (muscle) | Neuromuscular process, signal transduction |
| CPA5 | 3023912 | 3023916 | chr7:129772915-129772972 | Antisense | 245 | 289/303 | *Tissue-specific ALT major form in cerebellum, liver, pancreas and testes, medium form in spleen | 5′-UTR | carboxypeptidase A5 | Metallocarboxypeptidase activity |
| HORMAD1 | 2434546 | 2434564 | chr1:148953715-148953778 | Antisense | 171 | 235 | *Tissue-specific ALT major form in testes, medium form in cerebellum, spleen and thyroid | Coding, skipping causing PTC of downstream exon | HORMA domain containing 1 | Mitosis |
| IL16 | 3604287 | 3604317 | chr15:79369300-79369441 | Antisense | 212 | 354 | Tissue-specific ALT minor form in spleen and testes, no inclusion in the other tissues | Coding with PTC | interleukin 16 (lymphocyte chemoattractant factor) | Leukocyte chemotaxis |
| INPP5J | 3942766 | 3942773 | chr22:29850128-29850249 | Antisense | 225 | 346 | *Tissue-specific ALT major form in testes and kidney, ALT medium form in heart, muscle, pancreas, prostate and thyroid | 5′-UTR result in ALT start from a downstream exon | inositol polyphosphate-5-phosphatase J | Inositol/phosphatidylinositol phosphatase activity |
| MICAL2 | 3320717 | 3320794 | chr11:12227307-12227369 | Sense | 148 | 211/356/397 | *Tissue-specific ALT splicing to all degrees | Coding | microtubule associated monoxygenase, calponin and LIM domain containing 2 | Metabolic process resulting in cell growth |
| MUC15 | 3366903 | 3366911 | chr11:26545175-26545262 | Sense | 178 | 266 | *Tissue-specific ALT major form in heart, pancreas and thyroid, constitutive inclusion in the other tissues | Coding with ALT start or 5′-UTR | mucin 15, cell surface associated | Membrane protein |
| NEK11 | 2642441 | 2642445 | chr3:132229464-132229536 | Sense | 202 | 275 | *Tissue-specific ALT major form in heart and muscle, medium form in the other tissues | 5′-UTR | NIMA (never in mitosis gene a)- related kinase 11 | Cell cycle |
| RGAG1 | 3987148 | 3987152 | chrX:109575037-109575152 | Sense | 196 | 312/342/419/445/483 | *Tissue-specific ALT major form in heart, muscle, spleen and thyroid, constitutive inclusion in the other tissues | 5′-UTR | retrotransposon gag domain containing 1 | Nuclear protein |
| SCRG1 | 2794006 | 2794022 | chr4:174561800-174561919 | Sense | 82 | 202 | *Tissue-specific ALT major form in cerebellum and testes, minor form in the other tissues | 5′-UTR | scrapie responsive protein 1 | Nervous system development |
| TTLL6 | 3761551 | 3761579 | chr17:44232433-44232639 | Antisense | 206 | 412 | Tissue-specific ALT minor form in testes, higher level overall expression in testes | 5′-UTR result in ALT start from a downstream exon | tubulin tyrosine ligase-like family, member 6 | Protein modification process |
| TUSC3 | 3087167 | 3087215 | chr8:15659671-15659735 | Sense | 277/368 | 432 | Tissue-specific ALT medium form in cerebellum, prostate and spleen, minor form in the other tissues. | Coding with ALT stop | tumor suppressor candidate 3 | Protein amino acid N-linked glycosylation via asparagine |
| WSCD2 | 3430620 | 3430687 | chr12:107162429-107162488 | Antisense | 173 | 233/451 | ALT minor form in all tissues, but higher inclusion level in testes | Coding | WSC domain containing 2 | Membrane protein |
| ZFAND5 | 3209623 | 3209643 | chr9:74168206-74168342 | Antisense | 218 | 364/527 | Tissue specific alternative splicing to all degrees | 5′-UTR | zinc finger, AN1-type domain 5 | DNA binding |
ALT, Alternative; PTC, premature termination codon.
*Strongly included.
Figure 3.
Examples of ‘correlated’ mammalian-wide interspersed repeat (MIR) exons from exon array analysis, semi-quantitative RT–PCR and sequencing. In each gel figure, solid arrows show sequencing confirmed MIR exon inclusion forms. Hollow arrows show sequencing confirmed MIR exon skipping forms. Dashed arrows show sequencing confirmed non-specific PCR products.
We also identified 15 MIR exons showing tissue-specific shift in their transcript inclusion levels (see Supplementary Material, Fig. S1). In three genes (TTLL6, WSCD2, IL16), the MIR exons were skipped almost completely in most tissues, but appeared to have modest tissue-specific increase of their transcript inclusion levels in restricted tissues. For example, TTLL6 (tubulin tyrosine ligase-like family, member 6) is an apoptosis-related gene with preferential expression in testis. Previously, analyses of TTLL6 coding sequences from representative non-human primate species and worldwide human populations indicate strong positive selection on TTLL6 during recent primate and human evolution (40). Chen and colleagues (40) suggested that TTLL6 might contribute to the adaptive evolution of human male reproduction. In our RT–PCR analysis, an MIR-derived exon in TTLL6 had a detectable exon inclusion isoform as the minor transcript product in testis (Fig. 3D). Real-time qPCR analysis of this exon using isoform-specific primers indicated an inclusion level of approximately 20% in testis RNA (data not shown). Moreover, the splice sites of this exon were probably created during recent human evolution. Alignment of the human exon to its orthologous regions from non-human primates indicated that the ‘GT’ donor splice site downstream of the human exon was created by a human-specific nucleotide substitution, which converted an ‘AT’ dinucleotide in the common ancestor of human and chimpanzee to the ‘GT’ consensus splice site in the human gene. In the future, it would be interesting to examine the potential functional significance of this human-specific MIR exonization event in TTLL6.
Several exons showed strong switch between exon inclusion and skipping forms across various tissues. An example is the tissue-specific MIR exon in MICAL2 (microtubule-associated monoxygenase, calponin, and LIM domain containing 2), a gene involved in the regulation of cytoskeleton and repulsive neuronal guidance (41). The MIR exon is in the coding region of MICAL2 and encodes an in-frame 21-amino acid peptide within its protein product. RT–PCR analysis identified tissue-specific transcript inclusion levels—the exon was spliced as a constitutive exon in liver and testis, as the major splice form in heart, and as the minor splice form in cerebellum, muscle and thyroid (Fig. 3E). In HORMAD1 (HORMA domain containing 1), the MIR exon had comparable amount of inclusion and skipping forms in cerebellum and spleen, while in testes, the exon inclusion form was the predominant isoform (Supplementary Material, Fig. S1). In CHRNA1 (cholinergic receptor, nicotinic, alpha 1), the MIR exon was spliced at medium levels in muscle, prostate and testis, but had a much higher level of exon-skipping in cerebellum and spleen (Fig. 3F). The skipping isoform of this exon encodes a functional subunit of the acetylcholine receptor (AchR), while the exon inclusion isoform encodes a non-functional protein (42). Thus, the increased skipping of this MIR exon in cerebellum is expected to result in increased ratio of functional to non-functional AchR subunits. Although the physiological consequence of this tissue-specific splicing event is unknown, it could be a regulated process. In fact, a recent study shows that skipping of this MIR exon is mediated by binding of splicing regulator hnRNP H to an intronic UGGG motif upstream of the exon (43). A mutation within this UGGG motif, which significantly enhanced exon inclusion, was identified in a patient with congenital myasthenic syndrome (43). Together, these 15 exons represent the first examples of experimentally validated tissue-specific MIR exons in the human genome. As tissue-specific regulation of alternative splicing is a strong indicator of functional alternative splicing events (44), it is reasonable to speculate that these MIR exons may be involved in the fine tuning of mRNA level or protein function in a tissue-specific manner.
Of 39 correlated MIR exons that we analyzed experimentally, 12 (30.8%) were coding-region exons without introducing premature termination codons (PTCs; Table 1). This percentage is nearly twice as much as the percentage of coding-region Alu exons from our previous study (3/19, 15.8%) (23). Gotea and colleagues (45) hypothesized that exons created from old TEs are more likely to develop coding potentials and contribute to the proteome, while the roles of exons created from young TEs (e.g. Alus) are mostly regulatory [e.g. regulation of steady-state mRNA levels by induction of mRNA nonsense-mediated decay; also see (46)]. Despite the lack of statistical power in our analysis owing to the small number of Alu and MIR exons, the trend observed in our data set is consistent with the hypothesis of Gotea et al.
There is a significant enrichment of MIR exons in 5′-UTR when compared with 3′-UTR. This trend was consistent with the observation by Zhang et al. (10) in a genome-wide analysis of species-specific exons, as well as the observation in our recent study of Alu-derived exons (23). Zhang et al. (10) hypothesized that creations of new internal exons in 3′-UTR may be strongly selected against, because of the high likelihood of inducing mRNA nonsense-mediated decay. It should also be noted that these studies focused on creation of internal spliced exons from TEs. The impact of TEs on 3′ terminal exons was not investigated in our work. In fact, a recent study by Lee et al. (47) suggested a role of TEs in creating new polyadenylation sites at 3′-end of mammalian genes.
Splicing activities of exons derived from old and young transposable elements
Our data on MIR exons and Alu exons suggest that a larger percentage of exonized MIR elements have high transcript inclusion levels. There are two possible explanations for this observation. It could be that the consensus sequence of MIR elements contains strong splicing signals, making MIR elements more prone to exonization. It could also be owing to the fact that MIR is a much older class of TEs, so MIR-derived exons had more evolutionary time to accumulate additional splicing regulatory elements that strengthened their transcript inclusion levels.
To assess the potential contribution from putative splice sites within MIR elements, we scanned the consensus sequences of MIR, AluJo (the oldest Alu subfamily) and AluSx (the most abundant Alu subfamily in the human genome) for existence of splice site signals. The consensus sequences of MIR, AluJo and AluSx were downloaded from Repbase (48). It has been demonstrated that both MIR and Alu tend to exonize from antisense orientation (11,26). For example, of the 39 ‘correlated exons’ analyzed by RT–PCR, 24 (61.5%) were exonized from the antisense strand of MIR. Therefore, we performed sliding window scans of antisense strands of MIR, AluJo and AluSx to identify high-scoring putative 5′ and 3′ splice sites. In each window, we calculated splice site scores using consensus splice site models in MAXENT (31). We found that the antisense consensus sequence of MIR had much stronger putative splice sites when compared with AluJo and AluSx (see Supplementary Material, Fig. S2). The antisense consensus sequence of MIR had two extremely strong 5′ splice sites scored at 9.65 and 8.78, respectively. Likewise, we found a high-scoring 3′ splice site on antisense MIR with a MAXENT score of 9.95. In contrast, the potential splice sites on antisense AluJo and AluSx were much weaker. The strongest putative 5′ and 3′ splice sites were scored at 2.44 and 6.53 on antisense AluJo, and at 4.28 and 5.92 on antisense AluSx. The existence of strong putative splice sites on antisense MIR could make MIR elements more favorable substrates for exonization.
However, most intronic MIRs do not exonize, and the splice sites of many exonized MIRs are not derived from high-scoring putative splice sites within the MIR consensus sequence. Twenty-four of the ‘correlated exons’ analyzed by RT–PCR were from antisense strand of MIRs, among which 15 were spliced as constitutive or major alternative splice form in at least certain tissues. For each of these 15 ‘strongly included’ exons from antisense MIR, we used the alignment of the exon sequence to the antisense consensus MIR to determine the origin of splice sites. In three constitutive exons (ST3GAL1, FXYD4, CSF3R), both 5′ and 3′ splice sites were derived from the high-scoring putative splice sites within antisense MIR. In six exons, either 5′ or 3′ splice site (but not both) was derived from the high-scoring putative splice sites. In the remaining six exons, including three constitutive exons (TYK2, MTX1, MTA2), none of the splice sites was derived from the high-scoring putative splice sites. Taken together, of the 30 splice sites in 15 ‘strongly included’ exons derived from antisense MIR, 12 (40%) were from high-scoring putative splice sites within the consensus MIR. These data imply that the strong putative splice site signals within consensus MIR are not the sole contributor to the strong splicing activities of exonized MIR elements. It is likely that the older age of MIR also plays an important role, as it provides more opportunities for newly exonized MIRs to strengthen splicing signals and/or acquire valid open reading frames through a series of subsequent evolutionary changes.
To investigate whether exons derived from other old TEs also have stronger splicing activities when compared with Alu exons, we analyzed exon array data of human exons derived from LINE1 and LINE2 elements (see Materials and Methods). Of 271 LINE1-derived exons, 38 (14.0%) were correlated with gene expression levels. Similarly, of 175 LINE2-derived exons, 25 (14.3%) were correlated with gene expression levels. These percentages were similar to the percentage of ‘correlated exons’ among MIR-derived exons (15.2%) but significantly higher than the percentage among Alu-derived exons (6.1%; Fig. 2).
Phylogenetic conservation of MIR exon splicing
Based on the RT–PCR results in 10 tissues, we divided the 39 experimentally analyzed MIR exons into two distinct sub-categories. The first category consisted of 26 ‘strongly included’ exons that were used as constitutive or major alternative splice forms in at least certain tissues. The second category consisted of 13 ‘weakly included’ exons that were always used as medium or minor alternative splice forms or had no detectable exon inclusion isoform in any tissue.
We found that the splice sites of ‘strongly included’ MIR exons were much more conserved across species than the splice sites of ‘weakly included’ MIR exons. Of the 52 splice sites in 26 ‘strongly included’ exons, 32 were conserved between human and mouse genomes according to the human–mouse pairwise genome alignment. In contrast, of the 26 splice sites in 13 ‘weakly included’ exons, only eight were conserved between human and mouse, a statistically significant difference (32/52 versus 8/26; P = 0.016, two-sided Fisher exact test). Moreover, the splice sites of these ‘strongly included’ exons were much stronger. The average 5′ splice site score of the 26 ‘strongly included’ exons was 9.16, compared with 6.55 for the 13 ‘weakly included’ exons (P = 0.002, two-sided Wilcoxon test). Similarly, the average 3′ splice site scores of ‘strongly included’ exons and ‘weakly included’ exons were 8.58 and 5.60, respectively, although the difference was not statistically significant (P = 0.12).
To directly assess the evolutionary conservation of MIR exon splicing, we selected nine MIR exons (Table 2) and analyzed the splicing patterns of their orthologous regions in fibroblast cell lines of three non-human primates (chimpanzee, rhesus macaque and marmoset) and mouse kidney. To ensure that the fibroblast cell line is representative of other tissues, in this analysis we focused on MIR exons with consistent splicing patterns across human tissues. Also, to simplify the interpretation of PCR products from non-human primates, we restricted our analysis to MIR exons flanked by constitutive exons at both 5′ and 3′ ends in human genes. If RT–PCR of fibroblast cells yielded no PCR products, we would also attempt RT–PCR of kidney RNAs of non-human primates.
Table 2.
RT–PCR analysis of nine MIR exons in humans, chimpanzees, rhesus macaques, marmosets and mice
| Human | Chimpanzee | Macaque | Marmoset | Mouse | |
|---|---|---|---|---|---|
| IL6ST | Constitutive | Constitutive | Constitutive | ALT major | Constitutive |
| SRRM2 | Constitutive | Constitutive | Constitutive | Constitutive | Constitutive |
| CUL2 | Constitutive | Constitutive | Constitutive | Constitutive | Constitutive |
| MTX1 | Constitutive | Constitutive | Constitutive | Constitutive | Constitutive |
| MTA2 | Constitutive | Constitutive | Constitutive | Constitutive | Constitutive |
| TYK2 | Constitutive | Constitutive | Constitutive (with ALT upstream splice sites) | Constitutive | Constitutive |
| NUMB | ALT major | ALT major | ALT major | ALT medium | ALT medium |
| ARNTL | ALT medium | ALT medium | ALT medium | ALT minor | No inclusion |
| TGFBR2 | ALT medium | ALT medium | ALT minor | ALT medium | ALT medium |
ALT, Alternative.
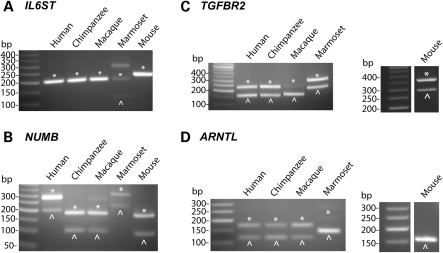
In six constitutively spliced MIR exons we analyzed, the splicing pattern of the human exon was highly conserved across species (Table 2). For example, the MIR exons of five genes (SRRM2, CUL2, MTX1, MTA2, TYK2) were constitutively spliced in all species (Supplementary Material, Fig. S3). In IL6ST (interleukin 6 signal transducer), the MIR exon was constitutively spliced in human, chimpanzee, rhesus fibroblasts and mouse kidney (Fig. 4A), suggesting that the exon was constitutively spliced before the divergence of primates and rodents (approximately 90 million years ago). In marmoset, we could not design RT–PCR primers because the sequences of flanking exons were unavailable in the low-coverage marmoset genome assembly. Nevertheless, our RT–PCR analysis using primers designed for human exons detected both exon inclusion and skipping forms in marmoset RNA, with the exon inclusion form being the predominant splice form (Fig. 4A).
Figure 4.
Splicing patterns of MIR exons in fibroblast cell lines of humans, chimpanzees, rhesus macaques, marmosets and mouse kidney. In each gel figure, exon inclusion forms and exon skipping forms are denoted as asterisks and arrow heads, respectively.
The other three exons we analyzed were alternatively spliced, with high or medium transcript inclusion levels in human tissues. These exons showed various patterns of evolutionary conservation of their splicing profiles. The MIR exon of NUMB had conserved alternative splicing profiles, with at least medium inclusion levels in all species we analyzed (Fig. 4B). In TGFBR2, the splicing pattern of the MIR exon was conserved in chimpanzee, marmoset and mouse, while the exon inclusion form was almost completely lost in rhesus macaque (Fig. 4C). In ARNTL, phylogenetic analysis suggested a gradual increase in the inclusion level of the MIR exon during primate evolution—the MIR exon was completely skipped in mice, included at a low level in marmosets and included at a medium level in rhesus macaques, chimpanzees and humans (Fig. 4D).
Taken together, these data suggest that for the majority of ‘strongly included’ MIR exons in the human genome, their splicing profiles in human tissues were present in the common ancestor of primates and rodents. The strong conservation of their splicing patterns in individual primate lineages implies that these exons are functionally important and that their splicing patterns have been subject to negative selection during primate evolution, as suggested by Krull and colleagues (26).
DISCUSSION
In this manuscript, we describe a large-scale analysis of MIR-derived exons in primate genomes. Our results significantly expand current knowledge about exonization of MIR elements. Prior to our study two constitutively spliced MIR exons were reported in the literature (11,26). In this work, we discovered and confirmed constitutive splicing of 15 MIR exons in a broad range of human tissues. We further analyzed a subset of these constitutive MIR exons in other species, and found that their constitutive splicing events were highly conserved between primates and rodents (Table 2). We also identified 15 MIR exons with tissue-specific shift in splicing patterns. The diverse splicing profiles of exonized MIR elements suggest that these exons can influence the regulation of gene expression or protein function through many ways—either constitutively or in a tissue-specific manner. Together, these experimental results provide many candidates for detailed functional studies of TE-derived exons.
We expect that the complete set of MIR exons with functional and/or regulatory roles is considerably larger than what we have identified in this study. Some strongly included MIR exons may be missed by our analysis owing to noise and artifacts in their exon array probe signals. It is also possible that some functional MIR exons are preferentially spliced in tissues or developmental states that are not investigated in this work.
Our phylogenetic analysis of MIR exons in DNA and RNA of primate and rodent species indicates that exonization can occur at any evolutionary time point. For the majority of constitutively spliced MIR exons, the exonization processes were likely completed prior to the divergence of primates and rodents (Table 2). In ARNTL (Fig. 4D), the MIR element was exonized during primate evolution. In TTLL6, the splice sites of a testis-specific MIR exon were not present in any non-human primates, suggesting that its splicing signal was acquired very recently during human evolution. It is possible that such newly created MIR exons are still evolving towards specified functions.
The comparison between MIR exons and Alu exons raises an interesting question of why exonized MIR elements tend to have stronger splicing signals and higher transcript inclusion levels. While several high-scoring putative splice sites are present in the consensus sequence of MIR, they cannot fully account for the difference in splicing levels of MIR and Alu exons, as only a minor fraction of splice sites of ‘strongly included’ MIR exons are derived from high-scoring putative splice sites within the consensus MIR. Moreover, splice sites alone are insufficient for proper exon recognition, as other splicing regulatory motifs (such as exonic/intronic splicing enhancers) also play important roles in pre-mRNA splicing (32). A key distinction between MIR and Alu is their evolutionary age. MIR was active prior to the radiation of placental mammals (approximately 130 million years ago), while Alu was created during primate evolution (approximately 60 million years ago). Previous phylogenetic studies of several TE-derived exons revealed that a series of independent evolutionary events were typically required for an intronic TE to evolve into a functional exon (24,27). For example, Singer and colleagues reconstructed the entire evolutionary history of the creation and functional establishment of an Alu-derived exon, which serves as the alternative first exon of tumor-necrosis factor receptor type 2 (p75TNFR) (24). Sequencing and phylogenetic analysis of orthologous DNAs of 13 primates suggests that the Alu element was inserted approximately 40–58 million years ago. Shortly after the Alu insertion, an A-to-G substitution created a start codon. However, a functional coding exon was not created until approximately 25 million years ago, when a C-to-T substitution created a splice donor site and a 7-bp deletion within the exonic region created a valid open reading frame. Considering the sequence changes required for turning an intronic TE into a functional exon, the older evolutionary age of MIR provides more opportunities for such sequence changes to occur, which could eventually lead to stronger splicing activities and higher protein-coding potentials of MIR-derived exons. In fact, exons derived from other classes of old TEs (such as LINE1 and LINE2) also had higher correlation with host genes' expression levels when compared with Alu-derived exons (Fig. 2). This further argues for the importance of evolutionary time in successful exonization of TEs.
Our work has important implications for understanding the evolution of new exons. The initial creation of new exons in functional genes (e.g. via exonization of TE, or de novo exonization from intronic regions) is generally considered detrimental, as new exons could disrupt functional elements within the protein products or cause frame-shifting and/or mRNA nonsense-mediated decay (9,49). A number of studies have shown that the majority of newly created exons in mammalian genomes are alternatively spliced, and most have low transcript inclusion levels according to EST and microarray data (10,15,50,51). Based on these observations, Modrek and Lee proposed that alternative splicing plays a major role in facilitating the creation and establishment of new exons. It allows functional genes to test new spliced isoforms (often produced at low levels), while keeping the majority of the original gene products intact (51). This evolutionary model describes the initial step of exon creation (i.e. relaxation of negative selection against exon creation via alternative splicing of the new exon). However, the subsequent evolution and eventual fate of new exons are far less understood. It is likely that most new exons are evolutionary intermediates dispensable for the organism, so the weak splicing signals of new exons can be lost again. On the other hand, over time a small percentage of new exons may acquire additional mutations that lead to stronger splicing regulatory signals and acquisition of functional properties. Our large-scale analysis of MIR exons in human tissues suggests that with sufficient evolutionary time, numerous new exons could evolve beyond the evolutionary intermediate state to acquire functional or regulatory roles. The phylogenetic conservation in the splice site signal and RNA splicing pattern of ‘strongly included’ MIR exons indicate strong functional constraints on these exons during recent evolution. These functional new exons could experience distinct modes of selection pressure over their entire evolutionary history—from weak negative selection or even positive selection after the initial creation of the new exon, to strong negative selection against loss of its splicing pattern after successful establishment of functional novelties.
MATERIALS AND METHODS
Compilation of Affymetrix exon array data on MIR-derived exons
We downloaded a public Affymetrix Exon 1.0 array data set on 11 human tissues (breast, cerebellum, heart, kidney, liver, muscle, pancreas, prostate, spleen, testes, thyroid) (34), with three replicates per tissue (http://www.affymetrix.com/support/technical/sample_data/exon_array_data.affx).
We compiled a list of Exon array probesets targeting exonized MIR elements. The locations of MIR elements in the human genome were downloaded from RepeatMasker annotation of the UCSC Genome Browser database (30). The locations of internal exons (i.e. exons flanked by both 5′ and 3′ exons) in human genes were taken from the UCSC KnownGenes database (30). This database combines transcript annotations from multiple sequence databases (30). To eliminate long exonic regions likely resulting from intron retention events, we removed probesets whose probe selection regions were over 250 bp as in Lee et al. (52). We then defined an exon as MIR-derived if the MIR element covered at least 25 bp of the exon and over 50% of the total length of the exon. We collected 363 exon array probesets targeting such MIR-derived exons. Since microarray probes targeting MIR repeats may cross-hybridize to off-target transcripts, we used a conservative approach to identify and remove individual probes showing abnormal intensities (see ‘Analysis of Exon array data’ below). After probe filtering, we collected a final list of 328 exon array probesets, with at least three reliable probes in each probeset to infer the splicing profiles of MIR-derived exons.
We used the same procedure to collect 330 Alu-derived exons, 271 LINE1-derived exons and 175 LINE2-derived exons. We also collected 13 103 high-confidence constitutive exons in the human genome from Lin et al. (23).
Analysis of exon splicing signals
For each exon, we scored its 5′ and 3′ splice sites using consensus splice site models in MAXENT (31). For 5′ splice site, we analyzed three nucleotides in exons and six nucleotides in introns. For 3′ splice sites, we analyzed three nucleotides in exons and 20 nucleotides in introns. We also calculated the density of ESEs using 238 ESEs from Fairbrother et al. (33). For each exon, the ESE density was calculated as the number of nucleotides covered by ESEs, divided by the total length of the exon.
Analysis of exon array data
Briefly, we first predicted the background intensities of individual exon array probes, using a sequence-specific linear model (53,54) trained from ‘genomic’ and ‘anti-genomic’ background probes on the Exon 1.0 array (55). For every probe, the predicted background intensity was an estimate for the amount of non-specific hybridization to the probe. This background intensity was subtracted from the observed probe intensity before downstream analyses (54). Second, for each gene we used a correlation-based iterative probe selection algorithm to construct robust estimates of overall gene expression levels, independent of splicing patterns of individual exons (56). Third, as oligonucleotide probes for TE-derived exons may be more likely to cross-hybridize than typical exon array probes, we used a set of criteria to identify and remove individual probes with abnormal probe intensities, for example probes that cross-hybridize to off-target transcripts. These criteria were described in detail before (23). After probe filtering, we collected a final list of 328 exon array probesets, with at least three reliable probes in each probeset to infer the splicing profiles of MIR-derived exons.
For each exon, we calculated the Pearson correlation co-efficient of individual probes' intensities with the overall gene expression levels in 11 tissues [estimated from all exons of a gene, see (54,56)]. As in (23), we defined a probe to be ‘correlated’ with gene expression levels if the Pearson correlation co-efficient was above 0.6. We defined an exon to be ‘correlated’ if it had at least three probes correlated with gene expression levels.
RT–PCR and sequencing analysis of MIR-derived exons in 10 human tissues
Total RNA samples from 10 human tissues were purchased from Clontech (Mountain View, CA, USA). RNAs of three tissues (liver, pancreas and spleen) were from single donors. RNAs of all other seven tissues were pooled from multiple donors (cerebellum, 24 donors; heart, 3 donors; kidney, 14 donors; skeletal muscle, 7 donors; prostate, 32 donors; testis, 39 donors; thyroid, 64 donors). Single-pass cDNA was synthesized using High-Capacity cDNA Reverse Transcription Kit (Applied Biosystems, Foster City, CA, USA) according to manufacturer's instructions. For each tested MIR-derived exon, we designed a pair of forward and reverse PCR primers at flanking constitutive exons using PRIMER3 (57). Primer sequences are described in Supplementary Material, Table S1. Two micrograms of total RNA were used for each 20 µl cDNA synthesis reaction. For each candidate MIR exonization event, 1 µl of cDNA were used for the amplification in a 25 µl PCR reaction. PCR reactions were run for 40 cycles in a Bio-Rad thermocycler with an annealing temperature of 62°C. The reaction products were resolved on 2% TAE/agarose gels. Candidate DNA fragments corresponding to exon inclusion and exon skipping forms were cloned for sequencing using Zero Blunt TOPO PCR Cloning Kit (Invitrogen, Carlsbad, CA, USA).
RT–PCR analysis of MIR-derived exons in RNAs of non-human primates and mice
Primary fibroblast cell cultures of chimpanzee, rhesus macaque and marmoset were purchased from Coriell Institute for Medical Research (Camden, NJ, USA). Frozen chimpanzee, rhesus macaque and marmoset kidneys were generously provided by Southwest National Primate Research Center (San Antonio, TX, USA). Human fibroblast cell culture was provided by Steven Moore (University of Iowa, IA, USA). Mouse kidney was collected from one wild-type male C57BL/6.
RNA was prepared using TRIzol (Invitrogen) according to the manufacturer's instructions. Single-pass cDNA was synthesized using the High Capacity cDNA Reverse Transcription Kit (Applied Biosystems). RT–PCR analysis was performed using primers designed from flanking exons of the target exon in chimpanzee, rhesus macaque, marmoset and mouse genomes. In any non-human primate, if the sequences of flanking exons were unavailable in the low-coverage genome assembly, we used RT–PCR primers designed for flanking exons in the human genome. Primer sequences are described in Supplementary Material, Table S2.
SUPPLEMENTARY MATERIAL
FUNDING
We thank David Eichmann, Ben Rogers and the University of Iowa Institute for Clinical and Translational Science (NIH grant UL1 RR024979) for computer support. This study used biological materials obtained from the Southwest National Primate Research Center, which is supported by NIH-NCRR grant P51 RR013986. This work was supported by the National Institutes of Health (1R01HG004634), Roy J. Carver Trust, and a research startup fund from the University of Iowa.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Peter Stoilov, Song Liu, Russ Carstens for discussions and comments on this manuscript. We thank Mary Jo Aivaliotis, Jerilyn Pecotte and Anne Tye for assistance.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Long M., Betran E., Thornton K., Wang W. The origin of new genes: glimpses from the young and old. Nat. Rev. Genet. 2003;4:865–875. doi: 10.1038/nrg1204. [DOI] [PubMed] [Google Scholar]
- 2.Prince V.E., Pickett F.B. Splitting pairs: the diverging fates of duplicated genes. Nat. Rev. Genet. 2002;3:827–837. doi: 10.1038/nrg928. [DOI] [PubMed] [Google Scholar]
- 3.Lynch M., Katju V. The altered evolutionary trajectories of gene duplicates. Trends Genet. 2004;20:544–549. doi: 10.1016/j.tig.2004.09.001. [DOI] [PubMed] [Google Scholar]
- 4.Li W.H., Yang J., Gu X. Expression divergence between duplicate genes. Trends Genet. 2005;21:602–607. doi: 10.1016/j.tig.2005.08.006. [DOI] [PubMed] [Google Scholar]
- 5.Pal C., Papp B., Lercher M.J. An integrated view of protein evolution. Nat. Rev. Genet. 2006;7:337–348. doi: 10.1038/nrg1838. [DOI] [PubMed] [Google Scholar]
- 6.Tautz D. Evolution of transcriptional regulation. Curr. Opin. Genet. Dev. 2000;10:575–579. doi: 10.1016/s0959-437x(00)00130-1. [DOI] [PubMed] [Google Scholar]
- 7.Chen K., Rajewsky N. The evolution of gene regulation by transcription factors and microRNAs. Nat. Rev. Genet. 2007;8:93–103. doi: 10.1038/nrg1990. [DOI] [PubMed] [Google Scholar]
- 8.Xing Y., Lee C. Alternative splicing and RNA selection pressure–evolutionary consequences for eukaryotic genomes. Nat. Rev. Genet. 2006;7:499–509. doi: 10.1038/nrg1896. [DOI] [PubMed] [Google Scholar]
- 9.Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13:1603–1608. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhang X.H., Chasin L.A. Comparison of multiple vertebrate genomes reveals the birth and evolution of human exons. Proc. Natl Acad. Sci. USA. 2006;103:13427–13432. doi: 10.1073/pnas.0603042103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sela N., Mersch B., Gal-Mark N., Lev-Maor G., Hotz-Wagenblatt A., Ast G. Comparative analysis of transposed element insertion within human and mouse genomes reveals Alu's unique role in shaping the human transcriptome. Genome Biol. 2007;8:R127. doi: 10.1186/gb-2007-8-6-r127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kondrashov F.A., Koonin E.V. Origin of alternative splicing by tandem exon duplication. Hum. Mol. Genet. 2001;10:2661–2669. doi: 10.1093/hmg/10.23.2661. [DOI] [PubMed] [Google Scholar]
- 13.Letunic I., Copley R.R., Bork P. Common exon duplication in animals and its role in alternative splicing. Hum. Mol. Genet. 2002;11:1561–1567. doi: 10.1093/hmg/11.13.1561. [DOI] [PubMed] [Google Scholar]
- 14.Alekseyenko A.V., Kim N., Lee C.J. Global analysis of exon creation versus loss and the role of alternative splicing in 17 vertebrate genomes. RNA. 2007;13:661–670. doi: 10.1261/rna.325107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wang W., Zheng H., Yang S., Yu H., Li J., Jiang H., Su J., Yang L., Zhang J., McDermott J., et al. Origin and evolution of new exons in rodents. Genome Res. 2005;15:1258–1264. doi: 10.1101/gr.3929705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gerber A., O'Connell M.A., Keller W. Two forms of human double-stranded RNA-specific editase 1 (hRED1) generated by the insertion of an Alu cassette. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 17.Mersch B., Sela N., Ast G., Suhai S., Hotz-Wagenblatt A. SERpredict: detection of tissue- or tumor-specific isoforms generated through exonization of transposable elements. BMC Genet. 2007;8:78. doi: 10.1186/1471-2156-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lander E.S., Linton L.M., Birren B., Nusbaum C., Zody M.C., Baldwin J., Devon K., Dewar K., Doyle M., FitzHugh W., et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 19.Makalowski W., Mitchell G.A., Labuda D. Alu sequences in the coding regions of mRNA: a source of protein variability. Trends Genet. 1994;10:188–193. doi: 10.1016/0168-9525(94)90254-2. [DOI] [PubMed] [Google Scholar]
- 20.Sorek R., Ast G., Graur D. Alu-containing exons are alternatively spliced. Genome Res. 2002;12:1060–1067. doi: 10.1101/gr.229302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lev-Maor G., Sorek R., Shomron N., Ast G. The birth of an alternatively spliced exon: 3′ splice-site selection in Alu exons. Science. 2003;300:1288–1291. doi: 10.1126/science.1082588. [DOI] [PubMed] [Google Scholar]
- 22.Muotri A.R., Marchetto M.C., Coufal N.G., Gage F.H. The necessary junk: new functions for transposable elements. Hum. Mol. Genet. 2007;16(Spec No. 2):R159–R167. doi: 10.1093/hmg/ddm196. [DOI] [PubMed] [Google Scholar]
- 23.Lin L., Shen S., Tye A., Cai J.J., Jiang P., Davidson B.L., Xing Y. Diverse splicing patterns of exonized Alu elements in human tissues. PLoS Genet. 2008;4:e1000225. doi: 10.1371/journal.pgen.1000225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Singer S.S., Mannel D.N., Hehlgans T., Brosius J., Schmitz J. From ‘junk’ to gene: curriculum vitae of a primate receptor isoform gene. J. Mol. Biol. 2004;341:883–886. doi: 10.1016/j.jmb.2004.06.070. [DOI] [PubMed] [Google Scholar]
- 25.Smit A.F., Riggs A.D. MIRs are classic, tRNA-derived SINEs that amplified before the mammalian radiation. Nucleic Acids Res. 1995;23:98–102. doi: 10.1093/nar/23.1.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Krull M., Petrusma M., Makalowski W., Brosius J., Schmitz J. Functional persistence of exonized mammalian-wide interspersed repeat elements (MIRs) Genome Res. 2007;17:1139–1145. doi: 10.1101/gr.6320607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Krull M., Brosius J., Schmitz J. Alu-SINE exonization: en route to protein-coding function. Mol. Biol. Evol. 2005;22:1702–1711. doi: 10.1093/molbev/msi164. [DOI] [PubMed] [Google Scholar]
- 28.Modrek B., Lee C. A genomic view of alternative splicing. Nat. Genet. 2002;30:13–19. doi: 10.1038/ng0102-13. [DOI] [PubMed] [Google Scholar]
- 29.Sorek R., Basechess O., Safer H.M. Expressed sequence tags: clean before using. Correspondence re: Z. Wang et al., computational analysis and experimental validation of tumor-associated alternative RNA splicing in human cancer. (Cancer Res., 63: 655–657, 2003) Cancer Res. 2003;63:6996. author reply 6996–6997. [PubMed] [Google Scholar]
- 30.Kuhn R.M., Karolchik D., Zweig A.S., Trumbower H., Thomas D.J., Thakkapallayil A., Sugnet C.W., Stanke M., Smith K.E., Siepel A., et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eng L., Coutinho G., Nahas S., Yeo G., Tanouye R., Babaei M., Dork T., Burge C., Gatti R.A. Nonclassical splicing mutations in the coding and noncoding regions of the ATM Gene: maximum entropy estimates of splice junction strengths. Hum. Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 32.Wang Z., Burge C.B. Splicing regulation: from a parts list of regulatory elements to an integrated splicing code. RNA. 2008;14:802–813. doi: 10.1261/rna.876308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fairbrother W.G., Yeh R.F., Sharp P.A., Burge C.B. Predictive identification of exonic splicing enhancers in human genes. Science. 2002;297:1007–1013. doi: 10.1126/science.1073774. [DOI] [PubMed] [Google Scholar]
- 34.Affymetrix. Affymetrix Tissue Panel Exon Array Data. 2005 http://www.affymetrix.com/support/technical/sample_data/exon_array_data.affx. (last accessed February 4, 2009) [Google Scholar]
- 35.Gardina P.J., Clark T.A., Shimada B., Staples M.K., Yang Q., Veitch J., Schweitzer A., Awad T., Sugnet C., Dee S., et al. Alternative splicing and differential gene expression in colon cancer detected by a whole genome exon array. BMC Genomics. 2006;7:325. doi: 10.1186/1471-2164-7-325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Clark T.A., Schweitzer A.C., Chen T.X., Staples M.K., Lu G., Wang H., Williams A., Blume J.E. Discovery of tissue-specific exons using comprehensive human exon microarrays. Genome Biol. 2007;8:R64. doi: 10.1186/gb-2007-8-4-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L., Zheng S. Studying alternative splicing regulatory networks through partial correlation analysis. Genome Biol. 2009;10:R3. doi: 10.1186/gb-2009-10-1-r3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li C., Wong W.H. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Irizarry R.A., Warren D., Spencer F., Kim I.F., Biswal S., Frank B.C., Gabrielson E., Garcia J.G., Geoghegan J., Germino G., et al. Multiple-laboratory comparison of microarray platforms. Nat. Methods. 2005;2:345–350. doi: 10.1038/nmeth756. [DOI] [PubMed] [Google Scholar]
- 40.Chen X.H., Shi H., Liu X.L., Su B. The testis-specific apoptosis related gene TTL.6 underwent adaptive evolution in the lineage leading to humans. Gene. 2006;370:58–63. doi: 10.1016/j.gene.2005.11.014. [DOI] [PubMed] [Google Scholar]
- 41.Terman J.R., Mao T., Pasterkamp R.J., Yu H.H., Kolodkin A.L. MICALs, a family of conserved flavoprotein oxidoreductases, function in plexin-mediated axonal repulsion. Cell. 2002;109:887–900. doi: 10.1016/s0092-8674(02)00794-8. [DOI] [PubMed] [Google Scholar]
- 42.Newland C.F., Beeson D., Vincent A., Newsom-Davis J. Functional and non-functional isoforms of the human muscle acetylcholine receptor. J. Physiol. 1995;489(Pt 3):767–778. doi: 10.1113/jphysiol.1995.sp021090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Masuda A., Shen X.M., Ito M., Matsuura T., Engel A.G., Ohno K. hnRNP H enhances skipping of a nonfunctional exon P3A in CHRNA1 and a mutation disrupting its binding causes congenital myasthenic syndrome. Hum. Mol. Genet. 2008;17:4022–4035. doi: 10.1093/hmg/ddn305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xing Y., Lee C. Protein modularity of alternatively spliced exons is associated with tissue-specific regulation of alternative splicing. PLoS Genet. 2005;1:e34. doi: 10.1371/journal.pgen.0010034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gotea V., Makalowski W. Do transposable elements really contribute to proteomes? Trends Genet. 2006;22:260–267. doi: 10.1016/j.tig.2006.03.006. [DOI] [PubMed] [Google Scholar]
- 46.Piriyapongsa J., Rutledge M.T., Patel S., Borodovsky M., Jordan I.K. Evaluating the protein coding potential of exonized transposable element sequences. Biol. Direct. 2007;2:31. doi: 10.1186/1745-6150-2-31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Lee J.Y., Ji Z., Tian B. Phylogenetic analysis of mRNA polyadenylation sites reveals a role of transposable elements in evolution of the 3′-end of genes. Nucleic Acids Res. 2008;36:5581–5590. doi: 10.1093/nar/gkn540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jurka J., Kapitonov V.V., Pavlicek A., Klonowski P., Kohany O., Walichiewicz J. Repbase Update, a database of eukaryotic repetitive elements. Cytogenet. Genome Res. 2005;110:462–467. doi: 10.1159/000084979. [DOI] [PubMed] [Google Scholar]
- 49.Boue S., Letunic I., Bork P. Alternative splicing and evolution. Bioessays. 2003;25:1031–1034. doi: 10.1002/bies.10371. [DOI] [PubMed] [Google Scholar]
- 50.Pan Q., Shai O., Misquitta C., Zhang W., Saltzman A.L., Mohammad N., Babak T., Siu H., Hughes T.R., Morris Q.D., et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- 51.Modrek B., Lee C. Alternative splicing in the human, mouse and rat genomes is associated with an increased rate of exon creation/loss. Nat. Genet. 2003;34:177–180. doi: 10.1038/ng1159. [DOI] [PubMed] [Google Scholar]
- 52.Lee J.A., Xing Y., Nguyen D., Xie J., Lee C.J., Black D.L. Depolarization and CaM kinase IV modulate NMDA receptor splicing through two essential RNA elements. PLoS Biol. 2007;5:e40. doi: 10.1371/journal.pbio.0050040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Johnson W.E., Li W., Meyer C.A., Gottardo R., Carroll J.S., Brown M., Liu X.S. Model-based analysis of tiling-arrays for ChIP-chip. Proc. Natl Acad. Sci. USA. 2006;103:12457–12462. doi: 10.1073/pnas.0601180103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kapur K., Xing Y., Ouyang Z., Wong W.H. Exon array assessment of gene expression. Genome Biol. 2007;8:R82. doi: 10.1186/gb-2007-8-5-r82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Affymetrix. Affymetrix Exon Array Design Datasheet. 2005 http://www.affymetrix.com/support/technical/datasheets/exon_arraydesign_datasheet.pdf. (last accessed February 4, 2009) [Google Scholar]
- 56.Xing Y., Kapur K., Wong W.H. Probe selection and expression index computation of affymetrix exon arrays. PLoS ONE. 2006;1:e88. doi: 10.1371/journal.pone.0000088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.