Abstract
Spinal muscular atrophy (SMA) is an autosomal recessive neurodegenerative disease. Loss of the survival motor neuron (SMN1) gene, in the presence of the SMN2 gene causes SMA. SMN functions in snRNP assembly in all cell types, however, it is unclear how this function results in specifically motor neuron cell death. Lack of endogenous mouse SMN (Smn) in mice results in embryonic lethality. Introduction of two copies of human SMN2 results in a mouse with severe SMA, while one copy of SMN2 is insufficient to overcome embryonic lethality. We show that SMN(A111G), an allele capable of snRNP assembly, can rescue mice that lack Smn and contain either one or two copies of SMN2 (SMA mice). The correction of SMA in these animals was directly correlated with snRNP assembly activity in spinal cord, as was correction of snRNA levels. These data support snRNP assembly as being the critical function affected in SMA and suggests that the levels of snRNPs are critical to motor neurons. Furthermore, SMN(A111G) cannot rescue Smn−/− mice without SMN2 suggesting that both SMN(A111G) and SMN from SMN2 undergo intragenic complementation in vivo to function in heteromeric complexes that have greater function than either allele alone. The oligomer composed of limiting full-length SMN and SMN(A111G) has substantial snRNP assembly activity. Also, the SMN(A2G) and SMN(A111G) alleles in vivo did not complement each other leading to the possibility that these mutations could affect the same function.
INTRODUCTION
Spinal muscular atrophy (SMA) is a leading genetic cause of death in infants (1). This autosomal recessive disease is characterized by degeneration of motor neurons of the spinal cord and atrophy of muscles due to denervation (2,3). Two forms of the Survival Motor Neuron (SMN) gene exist in humans, SMN1 and SMN2 (4). Both genes reside in the same genomic region and produce RNA and protein (4–6). The two genes are 99.9% identical with one key difference; SMN2 has a C to T transition within an exon splice modulator of exon 7 (7–10). This single nucleotide change causes the majority of transcript from SMN2 to lack exon 7 (SMN(D7)) (4,11,12). The SMN(D7) protein does not oligomerize efficiently and is rapidly degraded, thus causing a reduction in SMN levels (5,6,13,14). SMA is caused by loss or mutation of SMN1 and retention of SMN2 such that insufficient functional SMN protein is produced for motor neurons (15). The number of copies of SMN2 retained in SMA cases modifies the phenotypic severity of the disease (15,16). However, ∼5% of SMA patients have small mutations in SMN1 and in all cases reported to date the SMN2 gene is still present (17–20), despite the fact that 10–15% of normal individuals lack SMN2 (4,19). SMN2 provides some full-length SMN that can function directly or can interact with the mutant allele to regain its function (complementation). Some of these small mutations are missense mutations that disrupt specific domains of SMN (17,19,21).
SMN is a ubiquitously expressed protein found in both the cytoplasm and nucleus where it often accumulates in structures called gems (5,6,22,23). SMN interacts with Gemins 2–8 and unrip to form the SMN complex (24–26). The SMN protein complex functions in assembling Sm proteins onto the UsnRNA to form snRNPs, which are critical for correct gene splicing (27,28). As might be expected for a housekeeping gene, complete loss of SMN is embryonic lethal (29) and loss from a tissue causes death of that tissue (30–32). Mice lack SMN2 and thus, lethality can be rescued by introduction of SMN2. Low copy number of SMN2 gives rise to SMA and a high copy number of SMN2 rescues mice that lack mouse Smn (33,34). Expression of SMN(D7) in SMA mice containing two copies of SMN2 is not detrimental, but is beneficial with mice living on average 13 days (35). The mild SMA mutation, SMN(A2G) (36), when expressed in mice with SMN2 results in mild SMA, but SMN(A2G) by itself cannot rescue Smn−/− mice (37).
SMN has a well-characterized function essential to all eukaryotic cells (snRNP assembly), yet SMA is a motor neuron-specific disorder. Why, then, does the disruption of a ubiquitous protein preferentially affect motor neurons? In SMA, there is reduction of SMN as opposed to complete loss of SMN, which has been shown to result in disruption of snRNP assembly in fibroblasts from severe, type 1, SMA patients (38). Analysis of snRNP assembly in tissue extracts shows that this activity is developmentally regulated in normal mice (39) and that the degree of its impairment in SMA mice correlates with phenotypic severity as it is strongly reduced in spinal cords of severe mice (40,41). Importantly, SMN deficiency unevenly alters the snRNP profile of tissues and appears to preferentially decrease the levels of snRNPs of the minor splicing pathway, such as the U11 splicing pathway (42), in severe SMA mice (40,41). Although array analysis has been performed and various splicing changes identified, it is still not clear which changes are due to reduced SMN levels as opposed to stress or secondary changes (41). Thus, no specific targets of SMN reduction have been identified to date.
SMN has also been found in axons and growth cones of neurons, but not associated with Sm proteins (43–45). Reduction of SMN in zebrafish results in motor axon defects (46) and motor neurons cultured from SMA mice have truncated axons and smaller growth cones (47). In addition, these growth cones have lower amounts of beta-actin mRNA, which results in an altered distribution of calcium channels (48). This has led to the suggestion that SMN functions in axonal transport of mRNA and it is this function that is disrupted in SMA (47). We have previously examined two SMN mutations, SMN(A111G) and SMN(VDQNQKE). The SMN(A111G) allele, which occurs in a type 1b/II SMA patient with two copies of SMN2 (17), has been shown in culture to perform snRNP assembly when SMN levels are knocked down by siRNA (49). SMN(VDQNQKE) is a truncated form of SMN (exons 1–6) with the added motif VDQNQKE (50). SMN(VDQNQKE) does not efficiently associate with itself, full-length SMN or Sm proteins and thus, is predicted to be inefficient in snRNP assembly (51). However, when assayed for its ability to correct axonal defects in zebrafish where endogenous SMN had been knocked down, SMN(VDQNQKE) rescued and SMN(A111G) did not. This indicated that snRNP assembly was not critical for correction of axonal defects in zebrafish. However, it has not been possible to measure snRNP assembly in zebrafish. Further, in SMA mice, no defects of axonal growth or patterning have been detected (52). To further understand these SMN alleles, we have investigated them in SMA mice. In the current paper, we show that, when expressed at high levels, the SMN(A111G) allele rescues mice that lack mouse Smn and contain one or two copies of SMN2 (33), but does not rescue the embryonic lethality of Smn−/− mice (29) that do not have SMN2. Thus, the SMN(A111G) allele is complemented by SMN2 and performs snRNP assembly. A reinvestigation of SMN(A111G) in zebrafish reveals it is capable of rescuing axonal defects when present at higher concentrations. Lastly, the SMN(A111G) allele and SMN(A2G) allele do not complement each other to rescue embryonic lethality of Smn−/− mice, indicating that these two mutations could affect the same functional domain of SMN. This data shows a direct correlation of the ability of an SMN allele to perform snRNP assembly and correct the SMA phenotype. Furthermore, we suggest that the majority of milder SMA missense alleles work by interacting with small amounts of full-length SMN from the SMN2 gene. Full-length SMN, in a hetero-oligomer with mutant SMN, compensates for the mutant to restore function; whereas the homo-oligomer composed of just mutant SMN lacks function. Indeed, mild alleles of SMA are those that can complement full-length SMN produced by SMN2.
RESULTS
Generation of transgenic mouse lines
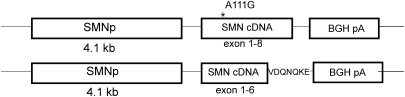
Transgenes containing SMN(A111G) or SMN(VDQNQKE) under the control of a 4.1 kb SMN promoter were microinjected into FVB/N oocytes. Ten founder lines were obtained for SMN(VDQNQKE) and 11 founder lines for SMN(A111G). The construct used for generation of the transgenic lines is diagramed in Figure 1. Of the 10 SMN(VDQNQKE) lines, 8 transmitted the transgene to offspring and 9 of 11 SMN(A111G) lines successfully transmitted the transgene. Three lines of SMN(VDQNQKE) were chosen by RT–PCR for further analysis as they had detectable RNA transcripts; lines 1946, 1947 and 1951. Line 1951 was found by real-time PCR to have 14 copies of the transgene. Three lines of SMN(A111G) were also chosen by RT–PCR for further analysis; lines 588, 591 and 2117. Line 588 was found to have 4 copies of the transgene, line 591 had 5 copies and line 2117 had 17 copies. The mice were backcrossed onto the SMA mouse background (SMN2+/+; Smn+/−) until mice that were heterozygous for the transgene, homozygous for SMN2 and homozygous for the mouse Smn knockout allele (SMN(VDQNQKE)+/−; SMN2+/+; Smn−/−) or (SMN(A111G)+/−; SMN2+/+; Smn−/−) were obtained.
Figure 1.
Diagram of transgene constructs. SMNp stands for the SMN promoter 4.1 kb fragment. BGH pA is the poly-adenylation sequence of bovine growth hormone.
Expression of transgenes
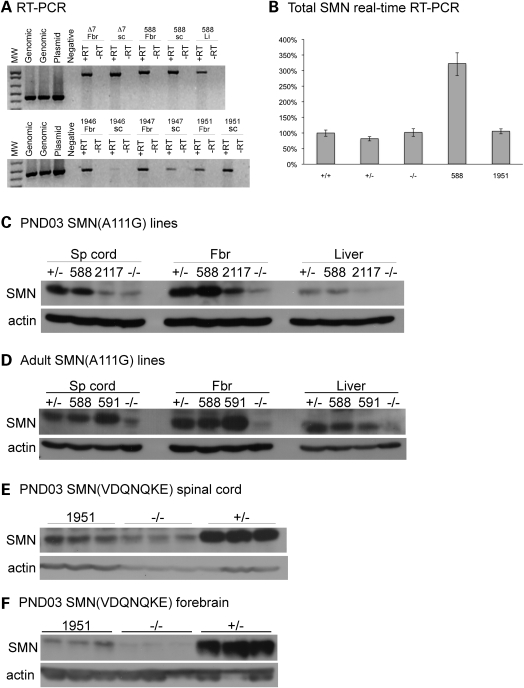
RT–PCR analysis revealed that the three SMN(VDQNQKE) lines had SMN RNA transcript expression in spinal cord, forebrain and liver (Fig. 2A bottom). Line 1951 appeared to have the highest mRNA transcript levels in spinal cord (Fig. 2A bottom). However, quantitative analysis of total human SMN RNA by real-time RT–PCR indicated that SMN(VDQNQKE) had a negligible increase in total SMN (Fig. 2B). Consistent with the real-time RT–PCR results, western blot analysis of post-natal day 3 (PND03) SMN(VDQNQKE) SMA mice revealed low SMN levels in forebrain and spinal cord (Fig. 2E and F) of all lines. Levels of SMN were quantitated for line 1951 (Fig. 2E and F). This line did not have statistically different levels of expression over SMA mice in forebrain or spinal cord. Thus, due to low SMN protein levels, this line was not expected to correct SMA.
Figure 2.
Expression analysis of SMN(A111G) and SMN(VDQNQKE) mice. (A) RT–PCR analysis of SMN expression in forebrain, spinal cord and liver of SMN(A111G) line 588 (top) using transgene specific primers. RT–PCR analysis of SMN expression in forebrain and spinal cord of SMN(VDQNQKE) lines 1946, 1947 and 1951 (bottom). Cyclophilin was used as a loading control (data not shown). (B) Quantitative real-time RT–PCR analysis of total SMN expression of SMN(VDQNQKE) line 1951 and SMN(A111G) line 588 in spinal cord using human SMN primers that detect SMN2 as well as the transgene. +/+, (SMN2 hom; mSmn +/+); +/−, (SMN2 hom; mSmn +/−); −/− (SMN2 hom; mSmn −/−). Analysis of SMN expression in brain was similar (data not shown) and limiting cycle PCR was also consistent with real-time RT–PCR (data not shown). Western blot analysis of SMN expression in spinal cord, forebrain and liver of 3-day-old SMN(A111G) mice from lines 588 and 2117 (C) and in adult SMN(A111G) mice, lines 588 and 591 (D). All mice are homozygous for SMN2 and lack mouse Smn unless otherwise noted. +/− refers to the presence of one copy of mouse Smn and −/− being absence of mouse Smn. Western blot analysis in spinal cord (E) of 3-day-old SMN(VDQNQKE) heterozygous mice and forebrain (F) of SMN(VDQNQKE) homozygous mice.
Three lines of SMN(A111G) were found by RT–PCR to express SMN mRNA. Line 588 had high RNA expression in spinal cord, forebrain and liver (Fig. 2A top) and real-time PCR revealed an ∼2-fold increase in total SMN levels (Fig. 2B). Line 591 had high levels of transcript in spinal cord, but low in forebrain and liver (data not shown). Line 2117 had expression in both spinal cord and forebrain (data not shown). Western blot analysis of PND03 tissue from SMA mice with the transgene showed that SMN(A111G) line 588 and 591 had SMN expression similar to a carrier mouse (SMN2+/+; Smn+/−) and considerably higher (6-fold) than severe SMA mice (SMN2+/+; Smn−/−) (Fig. 2C and D). Line 588 and 591 did produce SMN levels equivalent to a carrier mouse in muscle (Supplementary Material, Fig. S4). As the two lines behaved in a similar manner for both expression and survival analysis most subsequent analysis is done with SMN(A111G) line 588. The SMN(A111G) line 2117 expressed considerably less SMN protein than either 588 or 591 in all tissues analyzed (Fig. 2C and D). It does, however, express slightly more SMN than a typical SMA mouse (SMN2+/+; Smn−/−) at least in brain. Expression data is summarized in Table 1.
Table 1.
Summary of expression and survival of SMN(A111G) and SMN(VDQNQKE) lines
| Line | Copy No. | Protein expression |
Lifespan (days) | ||
|---|---|---|---|---|---|
| Brain | Spinal cord | Liver | |||
| SMN(A111G) | |||||
| 588 | 4 | +++ | +++ | + | normal (n = 11) |
| 591 | 5 | +++ | +++ | ++ | normal (n = 14) |
| 2117 | 17 | ++ | + | + | 8.2 ± 1.1 (n = 24) |
| SMN(VDQNQKE) | |||||
| 1951 | 14 | + | + | ND | 6.5 ± 0.8 (n = 18) |
| Controls | |||||
| SMA | + | + | + | 4.4 ± 0.3 (n = 90) | |
| Carrier | ++++ | ++++ | +++ | normal (n = 8) | |
Protein expression determined by western blot analysis on tissue from PND03 animals except for SMN(A111G) 591, which is adult tissue. Copy No. refers to the number of copies of the SMN(A111G) or SMN(VDQNQKE) transgenes. ND, not determined; +, low; ++, medium; +++, high expression; ++++, very high expression; n, number of mice.
Survival of SMN2; Smn−/− mice with SMN(A111G) or SMN(VDQNQKE)
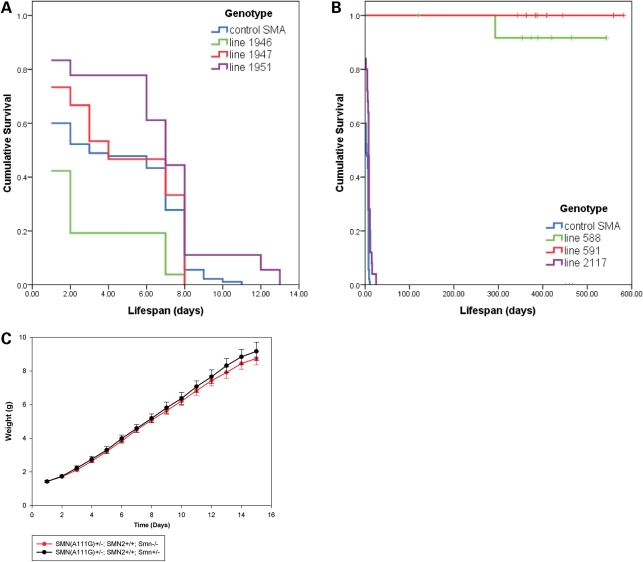
To determine the effect of SMN(A111G) and SMN(VDQNQKE) on SMA mice, we bred the mice to obtain those that lacked mouse Smn, had one or two copies of SMN2 and contained the SMN(A111G) or SMN(VDQNQKE) transgene. We had previously shown that mice lacking mouse Smn and containing two copies of SMN2 die at approximately 5 days (33), whereas mice with one copy of SMN2 show embryonic lethality at or before embryonic day 10.5 (52). Severe SMA mice (SMN2+/+; Smn−/−) have a mean life span of 4.4 ± 0.3 days (n = 90) in these experiments (Fig. 3A), consistent with previous reports (33). The presence of SMN(VDQNQKE) transgene had a minimal effect on the survival of SMA animals with line 1946, having a mean life span of 2.4 ± 0.5 days (n = 26), line 1947 having a mean life span of 4.7 ± 0.8 days (n = 15); and line 1951 had a mean life span of 6.5 ± 0.8 days (n = 18) (P = 0.03) (Fig. 3A). Making the SMN(VDQNQKE) homozygous did not result in significant enhancement of survival of SMA animals. SMA pups that contained SMN(VDQNQKE) showed a typical SMA phenotype (33) with no noticeable difference to SMA pups lacking the transgene. Thus, SMN(VDQNQKE) has a minor impact on survival, most likely due to low SMN transgene expression levels.
Figure 3.
Survival analysis and phenotype of SMN(A111G) and SMN(VDQNQKE) SMA mice. (A) Kaplan–Meier survival curve of SMN(VDQNQKE) SMA mice showing minimal or no correction of lifespan. Blue line: SMN2+/+; Smn−/− control SMA mice have a mean survival of 4.4 ± 0.3 days (n = 90). Green line: Line 1946 SMN(VDQNQKE)+/− or SMN(VDQNQKE)+/+; SMN2+/+; Smn−/− mice have a mean lifespan of 2.4 ± 0.5 days (n = 26). This is significantly fewer days (P < 0.001) than the control SMA mice. Red line: Line 1947 SMN(VDQNQKE)+/− or SMN(VDQNQKE)+/+; SMN2+/+; Smn−/− mice survive for 4.7 ± 0.8 days (n = 15). Purple line: Line 1951 SMN(VDQNQKE)+/− or SMN(VDQNQKE)+/+; SMN2+/+; Smn−/− mice survive for 6.5 ± 0.8 days (n = 18), which is a minimal improvement over control (P = 0.03). (B) Kaplan–Meier survival curves of SMN(A111G) SMA mice showing complete correction of lifespan for lines 588 and 591, while line 2117 has a minimal, but significant increase in lifespan. Blue line: SMN2+/+; Smn−/− control SMA mice have a mean survival of 4.4 ± 0.3 days (n = 90). Purple line: Line 2117 SMN(A111G)+/−; SMN2+/− or SMN2+/+; Smn−/− mice live for 8.2 ± 1.1 days (n = 24). This is a significant (P < 0.001) increase in lifespan over the control SMA mice. Red line: Line 591 SMN(A111G)+/−; SMN2+/− or SMN2+/+; Smn−/− mice survive a normal lifespan (P < 0.001). The mice can live for up to 1.5 years at which time they were removed from the study. Green line: Line 588 SMN(A111G)+/−; SMN2+/− or SMN2+/+; Smn−/− mice also survive for up to 1.5 years at which time they were removed from the study. One mouse died at 10 months of age from this cohort from an unknown cause. (C) Weight curves for SMN(A111G) line 588 mice. All mice are of the genotype SMN(A111G)+/−; SMN2+/+; Smn−/− or control carrier mice SMN(A111G)+/−; SMN2+/+; Smn+/− The SMN(A111G) mice have an identical weight to control animals. Weight data is represented in mean weight in grams each day with standard deviation.
In contrast, the SMN(A111G) transgene had a major impact on survival of SMA mice with high expressing animals (lines 588 and 591) surviving for more than a year. SMN(A111G) line 2117, with low expression of SMN, when present in SMA animals, resulted in a mean survival of 8.2 ± 1.1 days (n = 24) (P < 0.001) (Fig. 3B). In two cases in line 2117, mice have lived considerably longer than 8 days (11 months and 1 year), however, in general, this is not the case and we consider these mice to be escapers. All mice analyzed had two copies of SMN2. The presence of SMN(A111G) from lines 588 or 591 with high expression of SMN caused a marked impact on survival with the mean censored survival times being over a year and animals living for at least 1.5 years before being removed from the study (Fig. 3B). This was the same as control animals that had mouse Smn. In addition, there was no obvious phenotype in these animals and they were comparable to animals that possessed mouse Smn (Fig. 3C).
Due to the nature of the crosses used to obtain these animals, the mice could be either homozygous for SMN2 (2 copies of SMN2) or heterozygous for SMN2 (1 copy of SMN2). Again, when mice have one copy of SMN2, it is embryonic lethal on or before embryonic day 10.5 (52). However, for both SMN(A111G) lines 588 and 591 animals of the genotype SMN(A111G); SMN2+/−; Smn−/− are frequently obtained and survive for over a year with no noticeable difference to mice that have two copies of SMN2. Thus, the SMN(A111G) allele complements SMN2 to give an SMN complex that functions to rescue embryonic lethality of one copy-SMN2 (SMN2+/−; Smn−/−) mice, as well as SMA in two-copy SMN2 mice. In addition, this indicates that the SMN(A111G) transgene is expressed in all cell types in sufficient amounts to perform SMN’s essential function. This led us to ask whether SMN(A111G) by itself could rescue Smn−/− mice. Therefore, we removed SMN2 and set-up crosses to address whether SMN(A111G) could rescue Smn−/− mice in the absence of SMN2. Analysis of these crosses (Table 2) revealed that SMN(A111G) never rescued embryonic lethality. Of 55 progeny obtained, we expected to see approximately seven animals of this genotype, but this never occurred. This indicates that SMN(A111G) has an absolute requirement for full-length SMN in order to form functional heteromeric complexes.
Table 2.
Survival of SMN(A111G) animals without SMN2
| Genotype | Observed | Expected | χ2 |
|---|---|---|---|
| Cross type: SMN(A111G)+/−; Smn+/−×Smn+/−a | |||
| SMN(A111G)+/−; Smn+/+ | 11 | 7 | 2.5 |
| SMN(A111G)+/−; Smn+/− | 18 | 14 | 1.3 |
| SMN(A111G)+/−; Smn−/− | 0 | 7 | 6.9* |
| Smn+/+ | 8 | 7 | 0.2 |
| Smn+/− | 18 | 14 | 1.3 |
| Smn−/− | 0 | 7 | 6.9* |
| Total | 55 | ||
| Cross type: SMN(A111G)+/−; Smn−/−×SMN(A2G)+/−; Smn+/−b | |||
| SMN(A111G)+/− or SMN(A2G)+/−; Smn+/+ | 54 | 46 | 1.4 |
| SMN(A111G)+/− or SMN(A2G)+/−; Smn+/− | 104 | 92 | 1.6 |
| SMN(A111G)+/− and SMN(A2G)+/−; Smn−/− | 0 | 15 | 15.3* |
| Smn+/+ | 7 | 15 | 4.5* |
| Smn+/− | 34 | 31 | 0.4 |
| Total | 199 | ||
aSurvival of SMN(A111G) animals in crosses in the absence of the SMN2 transgene. All possible genotypes of offspring are noted. Mice of the genotype Smn−/− have been previously reported to be embryonic lethal (29). No mice of the genotype SMN(A111G)+/−; Smn−/− were obtained (χ2 = 6.9, P < 0.01) indicating that this transgene cannot function without SMN2.
bSurvival of SMN(A111G) and SMN(A2G) animals when crossed together. Three genotypes Smn−/−, SMN(A111G)+/−; Smn−/− and SMN(A2G)+/−; Smn−/− are omitted as they have been previously determined to be embryonic lethal and skew the χ2 results. No mice of the genotype SMN(A111G)+/−; SMN(A2G)+/−; Smn−/− were obtained (χ2 = 15.3, P < 0.001) indicating that these two transgenes do not complement each other. Fewer wild-type (Smn+/+) mice were observed than expected, but this is just at the level of significance (χ2 = 4.5, P < 0.05). All expected values are rounded to the nearest whole mouse.
*Statistically significant (P < 0.05).
Intragenic complementation occurs when a multimeric protein is formed from subunits produced by different mutant alleles of a gene. Thus, SMN produced by SMN2 and SMN(A111G) undergo intragenic complementation. We wished to test whether the two missense alleles SMN(A111G) and SMN(A2G) could also perform this type of complementation, as both are functional in the presence of SMN2. We crossed SMN(A2G)+/−; Smn+/− mice with SMN(A111G); Smn+/− mice so as to obtain SMN(A111G)+/−; SMN(A2G)+/−; Smn−/− mice. However, no animals of this genotype were observed out of 199 progeny obtained (Table 2). This indicates that the SMN(A2G) and SMN(A111G) alleles do not complement each other to form a functional SMN complex.
SnRNP assembly correlates with SMA phenotype and survival
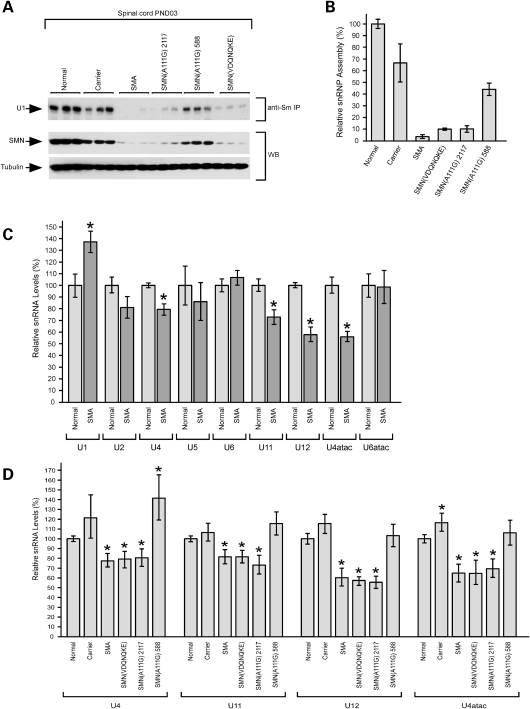
We have shown previously that SMN(A111G) self-associates efficiently and binds Sm proteins (51). In addition, assays in transiently transfected cells showed that SMN(A111G) expression is capable of increasing in vitro snRNP assembly activity of cells with reduced levels of SMN (49). We, therefore, assayed snRNP assembly activity in the spinal cord extracts of SMN(A111G) and SMN(VDQNQKE) SMA mice using in vitro transcribed radioactive U1 snRNA and immunoprecipitation with anti-Sm antibodies (Fig. 4A). Normal control animals (SMN2+/+; Smn+/+) were used to set the 100% normal snRNP assembly level in spinal cord. Animals of the carrier genotype (SMN2+/+; Smn+/−) had 65.95 ± 16.4% snRNP assembly in spinal cord and severe mice (SMN2+/+; Smn−/−) had 3.06 ± 1.49% assembly (Fig. 4B). SMN(VDQNQKE) line 1951 showed very low snRNP assembly activity at 9.40 ± 0.89%; whereas SMN(A111G) line 2117 (low expressing) had 9.57 ± 2.85% assembly and line 588 (high expressing) had 43.37 ± 5.31% assembly (Fig. 4B). The level of snRNP assembly correlated well with the survival and phenotype of each genotype. Both normal (SMN2+/+; Smn+/+) and carrier (SMN2+/+; Smn+/−) lived a normal lifespan with similar high assembly, as does the high expressing SMN(A111G) line 588. Both the SMN(VDQNQKE) line 1951 and the low SMN(A111G) line 2117 had lower snRNP capacity and correspondingly shorter lifespans of 6.2 days and 8.0 days, respectively. SMN2 +/+; Smn−/− mice lived 4.3 days and had very low snRNP capacity at ∼3%.
Figure 4.
Analysis of snRNP assembly activity and snRNA levels in the spinal cord of SMN(A111G) and SMN(VDQNQKE) mice. (A) Representative snRNP assembly reactions were carried out using in vitro transcribed radioactive U1 snRNA and 25 µg of whole spinal cord extracts from the indicated mice at post-natal day 3, followed by immunoprecipitation with anti-Sm antibodies. Immunoprecipitated U1 snRNAs were analyzed by electrophoresis on denaturing polyacrylamide gels and autoradiography. Western blot analysis of the spinal cord extracts that were probed with antibodies against SMN and tubulin is shown. Genotypes represented are: normal (SMN2+/+; Smn+/+), carrier (SMN2+/+; Smn+/−), SMA (SMN2+/+; Smn−/−), line 588 SMN(A111G)+/−; SMN2+/+; Smn−/−, line 2117 (low) SMN(A111G)+/−; SMN2+/+; Smn−/− and line 1951 SMN(VDQNQKE)+/−; SMN2+/+; Smn−/−. (B) Quantitation of snRNP assembly activity. The amount of immunoprecipitated U1 snRNA from in vitro snRNP assembly experiments as in (A) was quantified using a STORM 860 Phosphorimager (Molecular Dynamics). The normal control animals (SMN2+/+; Smn+/+) were set as the 100% normal snRNP assembly level and values are presented as mean±SEM. Animals of the carrier genotype (SMN2+/+; Smn+/−) had 65.95 ± 16.4% snRNP assembly and severe mice (SMN2+/+; Smn−/−) had 3.06 ± 1.49% assembly. SMN(VDQNQKE) line 1951 showed very low snRNP assembly activity at 9.40 ± 0.89%; whereas SMN(A111G) line 2117 (low expressing) had 9.57 ± 2.85% assembly and line 588 (high expressing) had 43.37 ± 5.31% assembly. (C) Quantitation of major and minor snRNAs in normal (SMN2+/+; Smn+/+) and severe SMA (SMN2+/+; Smn−/−) mice. Total RNA from the spinal cord of post-natal day 3 normal and SMA mice were analyzed by real-time RT–PCR for the levels of specific snRNAs. Data are from three independent biological replicates. The levels of U4, U11, U12 and U4atac were significantly reduced in SMA mice (P < 0.05). (D) Analysis of snRNA levels in transgenic mice. Total RNA from the same spinal cord extracts as in (A) were used for real-time RT–PCR quantitation of snRNA levels. Compared with normal mice, the levels of U11, U12, U4atac and U4 were significantly reduced in SMN(VDQNQKE) and SMN(A111G) low expressing lines, but were restored in the SMN(A111G) high expressing line (P < 0.05).
We then measured the levels of major and minor snRNAs in the spinal cord of normal and SMA animals to determine which snRNAs were reduced (Fig. 4C). The levels of U4, U11, U12 and U4atac were significantly reduced in SMA mice. The levels of the other snRNAs were not decreased in SMA animals and this is consistent with previously reported real-time PCR results (41). The four reduced snRNAs were then assayed in SMN(VDQNQKE), SMN(A111G) line 2117 and SMN(A111G) line 588 mice and compared with normal, carrier and SMA mice (Fig. 4D). The levels of these snRNAs are restored in the high expressing SMN(A111G) line 588, but not in SMN(VDQNQKE) line 1951 and SMN(A111G) line 2117 mice. Thus, restoration of a normal snRNA profile in the spinal cord correlates with correction of the SMA phenotype in mice.
Rescue of axonal defects in zebrafish by SMN(A111G)
We have previously reported that in transient assays, SMN(A111G) did not rescue motor neuron axonal defects caused by low Smn in zebrafish (51). In contrast, SMN(VDQNQKE) rescued these axonal defects (51). Since SMN(A111G) expression (at high levels) rescues transgenic SMA mice, we re-examined the rescue of axonal defects by SMN(A111G) and SMN(VDQNQKE) in zebrafish using varied concentrations of RNA for rescue. As shown in Supplementary Material, Figures S1 and S2, we found that at high concentration the SMN(A111G) construct was capable of rescue. In the case of SMN(VDQNQKE), rescue did still occur, but at reduced concentration of RNA this form of SMN was less effective at rescue than full-length SMN (Supplementary material).
Muscle analysis of SMN(A111G) SMA animals
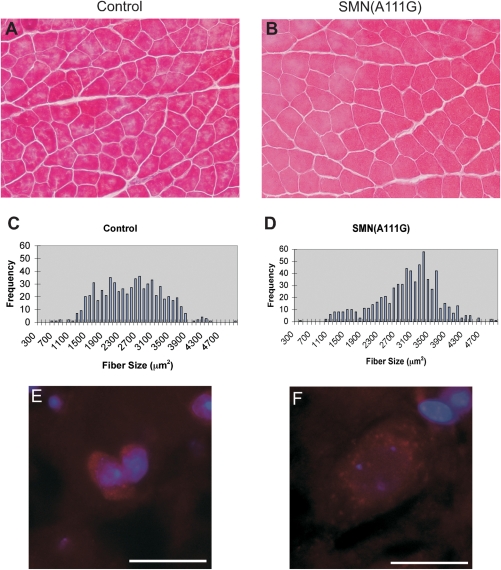
Gastrocnemius muscle was examined from 10-month-old adult SMN(A111G) transgenic mice (line 588) (Fig. 5A and B). The SMN(A111G) mice show hypertrophy of gastrocnemius fibers. They also have patches of smaller atrophic fibers, but the majority of fibers at this stage are larger with an average size of 2870.2 ± 28.5 µm2 versus the control with a mean size of 2456.1 ± 28.8 µm2. The distribution of fiber sizes was shifted in the SMN(A111G) mice to a larger range of 2800–3900 µm2 (Fig. 5C and D). While the survival and phenotype of SMN(A111G) mice is rescued, there is still a change in muscle morphology suggesting that the muscle has experienced denervation followed by re-innervation. This could be due to the fact that these mice express less SMN than normal and carrier mice and have an overall reduction in snRNP assembly, which could contribute to this change. Furthermore, calf hypertrophy is occasionally seen in mild SMA patients (53,54).
Figure 5.
Muscle morphology and immunohistochemical localization of SMN in the spinal cord of SMN(A111G) line 588. (A and B) Hematoxylin and eosin staining of gastrocnemius muscle from 10-month-old adult mice reveal larger muscle fibers in SMN(A111G) animals. (A) Control is of the genotype SMN2+/+; Smn+/− and (B) is of the genotype SMN(A111G)+/−; SMN2+/+; Smn−/−. (C and D) represent the distribution of fiber types in control and SMN(A111G) animals. The mean fiber size for the control was 2456.1 ± 28.8 µm2 (SE) and the mean fiber size for SMN(A111G) was 2870.2 ± 28.5 µm2. This is a mean difference of 414.0 µm2. The median fiber size for the control was 2480.5 µm2 and 2962.0 µm2 for SMN(A111G). The median fiber sizes are statistically different by Mann–Whitney and two-sample Kolmogorov–Smirnov tests (P < 0.001). (E and F) Immunostaining of 8 µm sections of spinal cord from 10-month-old control (E) and SMN(A111G) (F) mice. Nuclear and cytoplasmic staining of SMN (red) is clearly present in SMN(A111G) animals. DAPI (blue) was used as a nuclear stain. Scalebar is 20 µm.
SMN expression and rescue in motor neurons of SMN(A111G) animals
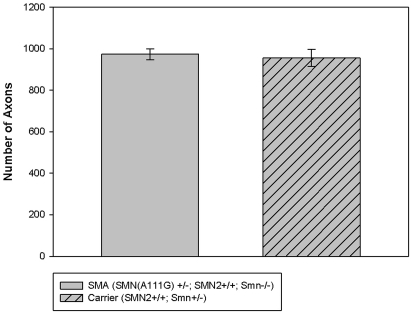
The expression of SMN in the spinal cords of SMN(A111G) line 588 mice was examined (Fig. 5E and F). Cross-sections of adult spinal cord were immunostained for SMN. Both nuclear (Gems) and cytoplasmic staining of SMN was visible in spinal cord motor neurons. Further, axons of the L4 ventral root of 6-month-old SMN(A111G)+/−; SMN2+/+; Smn−/− (line 588) spinal cords were counted (Fig. 6). The SMN(A111G) spinal cords had normal root counts when compared with age-matched carrier controls (SMN2+/+; Smn+/−). Normal root counts are consistent with a model of denervation, followed by re-innervation of muscle resulting in hypertrophy.
Figure 6.
L4 ventral root count in 6-month-old SMN(A111G) mice. The SMN(A111G) mice do not differ from carrier mice. Group means are represented with standard errors.
DISCUSSION
Mutations of SMN
A number of studies have examined in vitro how missense mutations found in SMA patients affect SMN’s ability to bind either to itself or members of the SMN complex (13,37,51,55–58). A large number of mutants have been described in exon 6 (20,59) that disrupt the ability of SMN to oligomerize and this reduces the capacity of the SMN complex to bind Sm proteins (13,56). The loss of SMN exon 7 (SMN(D7)) has a similar effect on these SMN interactions (13,56). SMN protein levels have been examined in lymphoblasts from a type 1 patient with a missense mutation that disrupts oligomerization (5). In this case, it was found that SMN protein levels were reduced to similar levels that occur in classic type 1 SMA patients with loss of SMN1. Furthermore, it is known that loss of SMN exon 7 impairs its ability to associate with itself or full-length SMN (13,51,56) and results in rapid degradation of SMN (60).
Furthermore, it was shown that severe SMA missense mutations that disrupt SMN oligomerization such as SMN(Y272C) and SMN(G279V) undergo rapid degradation in a similar manner to SMN(D7) (60). In contrast to SMN(D7) and SMN(G279V), the addition of amino acids to the C-terminus of SMN(exons 1–6) stabilizes SMN and gives some functionality at least in transient assays in cultured cells (61,62,63). One suggested strategy for therapeutics in SMA is to induce read-through of the translational stop codon in exon 8 of SMN transcripts lacking exon 7 (62). It will be important to determine whether transgenes with these read-through constructs can correct SMA. Certainly one type of severe SMA allele, as indicated in Figure 7, are those that disrupt SMN’s ability to oligomerize and result in rapid degradation. We have previously shown that SMN(VDQNQKE) could rescue axonal defects in zebrafish caused by knockdown of endogenous Smn (51). In this case, SMN protein was produced at sufficient levels to mediate this correction and was comparable to full-length SMN and SMN(A111G). In the current paper, SMN(VDQNQKE) did not rescue SMA mice. However, the SMN levels obtained from this transgene were low. The difference in protein levels accounts for the different results between mice and zebrafish. Further studies with stronger expression will be required to resolve this issue.
Figure 7.
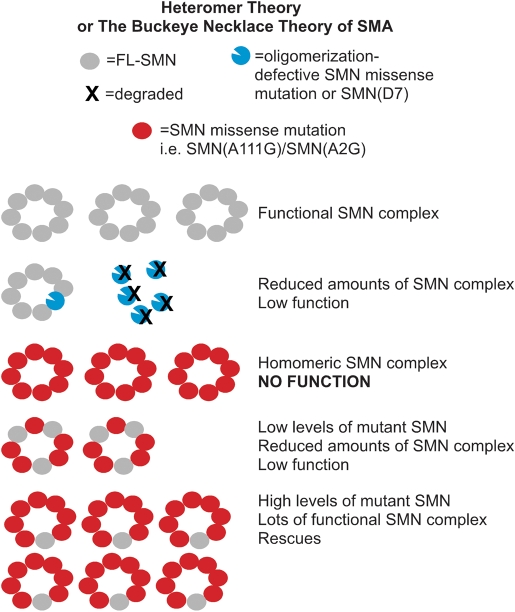
The heteromer theory of SMA to explain why retention of SMN2 is critical for phenotypic rescue by mild missense alleles. Mutations of SMN defective in oligomerization or SMN(D7) are rapidly degraded and result in a severe phenotype as the level of competent SMN complexes is insufficient for motor neurons (Row 2). Mild missense mutations of SMN are those that can complement full-length SMN from SMN2. Expression of mild mutant SMN transgenes in the absence of full-length SMN results in lethality due to the inability of the mutant to function on its own (Row 3). The addition of SMN2 results in functional SMN complexes wherein full-length SMN provides a missing property necessary for the activity of the complex. Thus, any SMN complex that contains a molecule of full-length SMN will be functional and any complex that does not contain full-length SMN will not be functional. In other words, there is no dominant negative effect of SMN(A111G) and the SMN from SMN2 is limiting. The maximum number of functional SMN complexes will be obtained when SMN(A111G) levels are high and each complex contains one full-length SMN (Row 4), while lower levels of SMN(A111G) will result in complexes containing more than one full-length SMN, thus limiting the number of functional complexes (Row 5). The SMN ring is shown as an octamer based on (74) and our data suggest that the complex is more than a dimer because much greater amounts of SMN(A111G) than SMN from SMN2 are necessary to obtain full rescue (in other words the ratio is not 1:1).
The missense mutation, SMN(A111G), can associate with full-length SMN to form an oligomer in vitro (17,51). In addition, in vitro binding studies demonstrate that SMN(A111G) does bind Sm proteins, but with reduced affinity (51). Furthermore, when endogenous SMN is knocked down by siRNA and SMN(A111G) introduced, this construct does produce SMN that can perform assembly of Sm proteins onto snRNA much like other milder SMA missense mutations (49). This contrasts with other Tudor domain mutations, which severely disrupt the binding of Sm proteins and do not perform snRNP assembly (49). In this study, we show that SMN(A111G) can rescue SMA when expressed at high levels in the presence of one copy of SMN2. This results in mice that have no obvious phenotype and live for 1 year or more. Furthermore, SMN(A111G) SMA mice with two copies of SMN2 have substantial snRNP assembly and the levels of snRNAs are restored to normal when compared with SMA mice.
The heteromer theory of mild SMA mutations
In the current study, we show that SMN(A111G) does not rescue the SMN knockout allele on its own and we suggest that on its own, this allele has no activity in snRNP assembly. This is similar to the SMN(A2G) allele that does not correct lethality in the absence of Smn (37). In our experience, a single copy of SMN2 in mice lacking mouse Smn results in an embryonic lethal phenotype, presumably due to insufficient SMN. However, when one or two copies of SMN2 and SMN(A111G) are on a Smn knockout background, then there is rescue of lethality with animals living for more than a year with no signs of SMA. Thus, SMN2 and SMN(A111G) transgenes undergo allelic complementation so as to give substantially greater function than either allele on its own. Intragenic or allelic complementation occurs when proteins oligomerize to form a functional unit from two different alleles. The resulting heteromer has greater activity than either homomeric allele (64,65). Allelic complementation has been reported in a number of recessive conditions, particularly certain enzyme deficiencies such as argininosuccinate lyase (64) and propionyl-CoA carboxylase (66). In these cases, the interaction of two mutant proteins creates an active site in the heteromer, which does not exist in either of the homomeric mutants. The SMN complex can be viewed as an enzyme that acts to place Sm proteins onto snRNA (67). The demonstration that the SMN(A111G) allele can undergo allelic complementation with limited SMN from a single copy of SMN2 indicates that the critical functional unit of SMN in SMA and snRNP assembly is a heteromer between full-length SMN from SMN2 and mutant SMN (Fig. 7). While an SMN complex comprised solely of SMN(A111G) must lack function, the addition of at least a single full-length SMN to SMN(A111G) yields a heteromeric SMN complex, where activity is created. Moreover, that high levels of SMN(A111G) by themselves have no effect and that high levels of SMN(A111G) rescue phenotype more than does additional SMN from a second copy of SMN2 suggests that the SMN complex is a heteromer of multiple subunits. High levels of SMN(A111G) in the presence of low levels of full-length SMN should result in complexes that contain, on average, one molecule of full-length SMN. As full-length SMN from low copy numbers of SMN2 is limiting, this would maximize the number of SMN complexes capable of functioning. Low levels of SMN(A111G) in the presence of low levels of full-length SMN results in complexes that, on average, contain more than one molecule of full-length SMN and thus, there are fewer active complexes in total. This leads to low function and limited rescue with a slight extension of lifespan (as in line 2117). Further, we hypothesize that in the heteromer the mutant SMN(A111G) functions to bind the Sm heptamer, while the full-length SMN functions to allow a single snRNA per complex to be loaded with Sm proteins.
Severe missense mutations in SMA are those that disrupt oligomerization resulting in low SMN levels or they disrupt a critical function and cannot complement with full-length SMN produced by SMN2; whereas, mild mutations are those that can complement with SMN produced by SMN2. Alleles that cannot interact efficiently with full-length SMN are severe, as are those that have severe disruption of Sm binding (i.e. E134K). In the case of mild alleles, we have found that SMN(A2G) and SMN(A111G) together cannot rescue the loss of Smn and thus, these alleles do not complement each other. One interpretation of this result is that the SMN(A2G) and SMN(A111G) mutations affect the same function. It would be interesting to determine whether any point mutations, such as SMN(A2G) or SMN(A111G), can complement mild C-terminal mutations. C-terminal mutations have been shown to reduce, but not to eliminate the ability to bind SMN. If self-association is the primary function affected by C-terminal mutations, then at high expression levels, these alleles are predicted to still retain function in the absence of full-length SMN. We suspect that all mild SMN missense mutations will require some full-length SMN from SMN2 to act as mild alleles and have some function.
The connection of SMA to snRNP assembly
Even though SMN is a ubiquitous protein, reduction of SMN primarily affects motor neurons. Two theories exist to explain why this specificity occurs: the axonal theory and the snRNP theory. Evidence for a role of SMN in axons has come from studies in zebrafish (46) and cultured motor neurons (47) where SMA motor axons are truncated in development. Also, reduced amounts of β-actin mRNA in the growth cones of motor neurons (47) leads to altered distribution of Ca2+ channels (48). However, no alteration of axon growth is detected in vivo in SMA mice (52). Winkler et al. (68) reported that knockdown of SMN or Gemin 2 resulted in motor axon defects in zebrafish. In contrast, we have reported that knockdown of SMN, but not Gemin 2 results in axon defects (69). One notable difference between these studies is whether or not the fish scored had altered morphology. In the case of Winkler et al. (68), as indicated in Supplementary material, Tables, over 50% of the fish scored were morphologically altered; whereas, in the study by McWhorter et al. (69), no morphologically abnormal fish were scored (46,68). In addition, Plastin 3 has been identified as a potential modifier of SMA (70); however, whether overexpression of Plastin 3 acts to alter splicing or simply to encourage axon growth in general is currently unclear.
To further investigate SMN’s function and the correction of axon defects in zebrafish, we have examined various SMN mutants for their ability to correct (51). It was found that co-injection of morpholino with SMN(A111G) or the synthetic mutant, SMN(Q282A), did not result in rescue even though the mutant SMN (A111G) is capable of snRNP assembly in the presence of low amounts of full-length SMN (49). The mutant SMN(Q282A) has not been assayed for its ability to perform snRNP assembly, although it does retain its ability to self-associate and bind Sm proteins in vitro (51). In contrast, SMN(VDQNQKE) did rescue in this assay, even though it is predicted not to be capable of snRNP assembly. These experiments lent support to an axonal function of SMN separate from that of snRNP assembly; however, it has proved difficult to measure snRNP assembly in zebrafish. Given our finding that SMN(A111G) rescues SMA mice, we reinvestigated the ability of this allele to correct axonal defects in zebrafish. We found that at higher concentrations SMN(A111G) did indeed correct the axonal defects indicating that this mutant cannot be used to distinguish an axonal function from an snRNP function.
A disruption of snRNP function, however, could be the likely cause of SMA as it could also produce an axonal phenotype if abnormal splicing of axonal-specific transcripts occurs. Previously, it has been shown that reduction in snRNP assembly results in altered levels of snRNPs. In particular, there is a reduction in the levels of minor snRNPs (40). This leads to the prediction that the minor splicing pathway could be specifically affected in SMA, perhaps affecting a gene(s) of importance for motor neurons. Microarray studies revealed a large number of expression and splicing changes in SMA animals (41). However, the splicing changes cannot be explained by the observation that only snRNAs of the minor splicing pathway were found to be decreased in the SMA tissues analyzed. Possible secondary alterations due to the state of the animals could occur and a series of changes that are detected, such as the increase in cytochrome P450 (41), suggest non-specific stress responses. It is currently not clear what constitutes the primary targets and secondary effects of reduced snRNPs. The availability of mice with various levels of snRNP assembly activity, as well as different levels of snRNPs, would allow changes observed in the arrays to be correlated with these factors enhancing the ability to identify true targets of reduced snRNPs.
In the current paper, we show that SMN(A111G) rescues SMA mice and provides substantial snRNP assembly activity. The levels of snRNP assembly activity in spinal cord correlate with the level of rescue obtained with each line of mice. The high expressing line 588 lives the longest with the highest snRNP assembly; whereas, the low expressing line (2117), which lives 8 days, has minimal snRNP assembly levels. Investigation of the levels of major and minor snRNAs in the spinal cord confirm that SMA animals experience a preferential reduction of minor snRNAs (U11, U12 and U4atac) compared with major snRNAs (U4). Importantly, analysis of snRNA levels in transgenic SMA mice expressing mutant SMN alleles showed correlation of the rescue of SMA to the restoration of normal snRNA levels in SMN(A111G) mice. Thus, both snRNP assembly activity and snRNA levels correlate with severity of SMA animals and correction of SMA.
In summary, SMN alleles that are capable of snRNP assembly correct SMA mice if expressed at sufficient levels and this also results in correction of snRNP profiles. Thus, snRNP assembly activity is highly correlated with the ability to correct SMA and no alleles analyzed to date separate SMA correction from snRNP assembly. However, whether SMN is critical for assembly of protein complexes onto mRNA for transport in axons remains unknown. Currently, it is possible that reduced snRNP levels results in altered splicing of a gene that functions in axons, thus, uniting the axon and snRNP hypothesis. Alternatively, SMN-dependent RNP assembly reactions critical for axons could be disrupted in SMA. It, thus, becomes important to determine what the targets of reduced snRNPs are and what RNP assembly reactions are affected by reduced SMN. The SMN missense alleles examined to date do not rescue Smn−/− mice, presumably because they lack snRNP assembly activity by themselves. The missense alleles examined, in particular SMN(A111G), can complement one copy of SMN2, indicating that small amounts of full-length SMN in a heteromeric SMN complex with mutant SMN restore function. In addition, this indicates that an oligomer is the functional unit in vivo, perhaps with the active site being formed by the interaction of subunits, as in other cases of intragenic complementation.
MATERIALS AND METHODS
Generation of transgenic mice and breeding
Both the SMN(A111G) and SMN(VDQNQKE) transgenes were made similarly. A 4.1 kb SMN promoter (SMNp) fragment in the pcDNA3 vector was isolated using BglII and BamHI. The SMN cDNA was isolated with BglII and BamHI. The SMN cDNA was ligated to the pcDNA3 vector containing SMNp using T4 DNA ligase (USB). The SMNp was then screened by PCR for directionality using the forward primer SMNPF tggagttcgagacgaggcctaagc and the reverse primer 3.2R agtagatcggacagattttgct. Also, EcoRI and SacII digests to confirm direction of promoter. Subsequently, mutant SMN cDNAs were then inserted by BamHI and EcoRI restriction digest and ligation. The construct was confirmed by sequencing. Seventy-five micrograms of SMNp: SMN(A111G) pcDNA3 or SMNp:SMN(VDQNQKE) pcDNA3 plasmid was then linearized using PvuI and DraIII. The constructs were then given to the Ohio State Transgenic Animal Facility (TAF) and were microinjected into FVB/N oocytes. Animals born at TAF were screened for potential founders by PCR using the same SMNPF and 3.2R primers. Founders were then backcrossed to SMA carrier mice (SMN2+/+; Smn+/−) to add a low level of human SMN while eliminating endogenous mouse Smn. Mice resulting from these crosses were genotyped for the mutant cDNA with SMNPF and 3.2 R primers. The presence of SMN2 and mouse Smn was screened as previously described (35). The SMN(A111G) high expressing lines have the following Jackson Laboratories stock numbers: JR8782 FVB-Tg(SMN2*A111G)588Ahmb/J and JR9134 FVB-Tg(SMN2*A111G)591Ahmb/J.
Determination of transgene copy number
A TaqMan Real-time qPCR protocol was used for transgene copy number determination (71). For the SMN transgene, the following primers and probe were used: forward primer 5′TGCTGGCTGCCTCCATTT3′, reverse primer 5′ GCATCATCAAGAGAATCTGGACAT 3′, and probe 5′ FAM-CTTCTGGACCACCAATAATTCCCCCACC-TAMRA 3′. The internal control used was apolipoprotein B and the following primer and probe sequences were used: forward primer 5′CACGTGGGCTCCAGCATT 3′, reverse primer 5′ TCACCAGTCATTTCTGCCTTTG 3′, probe 5′ VIC-CCAATGGTCGGGCACTGCTCAA-TAMRA 3′. A multiplex PCR reaction was set up as follows: 12.5 ml of 2× TaqMan Universal PCR master mix (ABI, CA), 0.25 ml of each primer (40 mm), 0.75 ml of each probe (5 mm) and 10 ml of diluted template DNA (2.5 ng/ml). The reaction was carried out at 50°C for 2 min, 95°C for 10 min, followed by 40 cycles of 95°C for 15 s and 60°C for 1 min on an ABI prism 7000 sequence detection system with 96-well format. For each sample, triplicate PCR reactions were carried out.
RT–PCR and western blot analysis
Total RNA was isolated using a combined TRIzol (Invitrogen) and RNeasy Mini Kit protocol (QIAGEN). Thirty micrograms of tissue was used and 1 ml of TRIzol reagent was added. Tissue was homogenized, incubated at room temperature for 5 mins. Two hundred microliters of chloroform was added, shaken vigorously and incubated for 5 min at room temperature. The samples were then centrifuged for 15 min at 4°C at 12 000g. The top layer was removed to a new tube and then 1 volume 70% ethanol was added. The QIAGEN manufacturer’s protocol was then resumed at this step. Total RNA was treated for DNA contamination using the DNA-free Kit (Ambion) as per manufacturer’s instructions. First strand cDNA synthesis was carried out using 6 µg of total DNA-free RNA. This was added to 250 ng random hexamer in nuclease-free water. This mixture was heated at 70°C for 10 min and then cooled on ice for 10 min to allow primers to anneal. A master mix was added to the tube that contained 1 mm dNTPs, 4 units of RNase-OUT (Invitrogen), 1× Reverse Transcriptase enzyme buffer and 7 units of RT (Life Sciences, Inc.). The samples were then incubated at 42°C for 1 h. PCR was carried out on the cDNA using either forward primer 4F gtgagaactccaggtctcctgg and reverse primer BGH-R tagaaggcacagtcgagg or forward primer 1F gcggcggcagtggtggcggc and reverse primer 4R tggagcagatttgggcttga. PCR conditions were: 95°C for 3 min for denaturation, followed by 35 cycles of 95°C for 1 min, 59°C for 1 min, 72°C for 1 min, then extension. Control primers used were cyclophilin forward agacgccgctgtctcttttcg and cyclophilin reverse primer ccacagtcggagatggtgatc.
Western blot analysis was carried out by homogenizing tissue in 200 µl of blending buffer [62.5 mm Tris, pH 6.8, 5 mm EDTA, 10% SDS, Mini Complete Protease Inhibitor Cocktail (Roche)]. The samples were sonicated briefly, mixed with 6× SDS Loading Buffer (350 mm Tris, pH 6.8, 10.28% SDS, 36% glycerol, 600 mm DTT, 0.012% bromophenol blue) and loaded onto a 12% SDS–polyacrylamide gel. The gel was transferred onto Hybond P PVDF membrane (GE Healthcare). The membrane was blocked for 1 h at room temperature in 5% milk, 1% BSA, 0.1% Tween-20 in 1× PBS. The blot was then probed with primary antibody 8F7 against SMN at 1: 1000 for 1 h. Secondary antibody conjugated to horseradish peroxidase (Rockland) was added at 1: 5000 and incubated for 1 h. The blot was then exposed to ECL substrate (GE Healthcare) and developed. The blot was stripped and re-probed with a loading control β-actin primary antibody (Sigma-Aldrich) at 1:10 000 for 1 h followed by the same Rockland secondary. For the western blot experiment in Figure 4, anti-SMN clone 8 (BD Transduction Laboratories) and anti-tubulin DM 1A (Sigma-Aldrich) primary monoclonal antibodies were used. Quantitation of western blots was carried out using NIH Image software and comparison of protein expression from western blots was carried out using two-sample t-tests.
Analysis of snRNP assembly activity and snRNA levels
In vitro snRNP assembly reactions with radioactive U1 snRNA and mouse tissue extracts were carried out essentially as described previously (40). Following addition of heparin and urea to a final concentration of 5 mg/ml and 2 M, respectively, reactions were incubated for 15 min at room temperature and then analyzed by immunoprecipitation with anti-SmB antibodies (72). Immunoprecipitations were carried out in RSB-500 buffer (500 mm NaCl, 10 mm Tris-HCl pH 7.4, 2.5 mm MgCl2) containing 0.1% NP40, EDTA-free protease inhibitor cocktail (Roche) and phosphatase inhibitors (50 mm NaF, 0.2 mm Na3VO4) for 2 h at 4°C. Immunoprecipitated U1 snRNA was analyzed by electrophoresis on denaturing polyacrylamide gels and autoradiography. Quantitation was performed using a STORM 860 Phosphorimager (Molecular Dynamics) and the ImageQuant version 4.2 software.
Quantitation of snRNAs was carried out using a real-time RT–PCR method. Total RNA was purified with Trizol reagent (Invitrogen). DNase I - RNase-free (Roche) was then applied to eliminate genomic DNA contamination from total RNA samples. 5.8S rRNA- and snRNA-specific primers were used to generate cDNA using Advantage RT-for-PCR kit (Clontech): 5.8S rRNA forward GCGCTAGCTGCGAGAATTAATGTG, 5.8S rRNA reverse CAAGTGCGTTCGAAGTGTCGATGA, U1 forward ATTGCACTTTGGGGTGTGCTGA, U1 reverse TGCAGTCGAGTTTCCCGCATTT, U2 forward CGGCCTTTTGGCTAAGATCAAGTG, U2 reverse TCCTCGGATAGAGGACGTATCAGA, U4 forward GAGGTTTATCCGAGGCGCGATTAT, U4 reverse CATGGCGGGGTATTGGGAAAAGTT, U5 forward TTTCCGTGGAGAGGAACAACTCTG, U5 reverse CTTGCCAAGACAAGGCCTCAAAA, U6 forward TCGCTTCGGCAGCACATATACT, U6 reverse CGCTTCACGAATTTGCGTGTCA, U4atac forward TTTCTTGGGGTTGCGCTACTGT, U4atac reverse AAAGCAGAGCTCTAACCGATGCAG, U6atac forward TAAGCACTCCCCTTGACAGGGAT, U6atac reverse ACACAGTGGCTAGATGCCATGA, U11 forward CGTGCGGAATCGACATCAAGAGA, U11 reverse CAACGATCACCAGCTGCCCAATTA, U12 forward GCCCGAGTCCTCACTGCTTATGT, U12 reverse AAAGTAGGCGGGTCGCTCAGAT.
One microgram of total RNA was utilized as template in a 20 µl reverse transcriptase reaction. 2.5% of the cDNA was used for each real-time PCR reaction. Real-time PCR reactions were carried out on an Eppendorf Mastercycler ep Realplex4 real-time PCR system using Power SYBR® Green PCR Master Mix (ABI). The same reverse primers were used in both RT and real-time PCR. Each measurement was carried out in triplicates in a standard 3-step PCR reaction. Absolute quantification was performed using Realplex 2.0 software. Data were calculated according to the ΔCt method. The same procedure was used for quantitative real-time RT–PCR analyisis of total SMN from transgenic mouse tissue. Primers used were GAPDH forward AATGTGCCGTCGTGGATCTGA and reverse GATGCCTGCTTCACCACCTTCT as a control. SMN human-specific primers SMN hsa forward AAGCCCAAATCTGCTCCATGGAAC and reverse TGGCTTTCCTGGTCCCAGTCTT were used as well as mouse SMN-specific primers SMN mmu forward TGCTCCGTGGACCTCATTTCTT and reverse TGGCTTTCCTGGTCCTAATCCTGA.
Zebrafish injections
Injections were performed as previously described (51).
Histology and immunofluoresence
Gastrocnemius muscles from 6-month-old animals were sectioned at 12 µm and stained with hematoxylin and eosin as previously described (35). Root counts were done on spinal cords of 6-month-old mice as previously described (73). Immunofluorescence was performed on 8 µm spinal cord sections of 6-month-old adult mice as described (73).
Statistical analysis
Survival curves (Kaplan–Meier) and statistical analysis was carried out with SPSS v16 (SPSS, Inc.). Kaplan–Meier analysis was done using the log rank method. Muscle fiber size medians were tested using the Wilcoxon Mann–Whitney and two-sample Kolmogorov–Smirnov test. Survival statistics of mouse crosses were obtained with χ2 distributions.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by NIH grant NS038650, the SMA Angels and the Madison Fund to A.H.M.B. Transgenic creation was supplemented by grant P30NS04578. Zebrafish work was supported by NIH grant NS050414 to C.E.B. L.P. is supported by grants from the SMA Foundation and the Motor Neuron Center of Columbia University. C.L. is supported by the SMA Foundation.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Dr Matthew E.R. Butchbach for assistance with tissue preparation, Dr Jill Rafael-Fortney for the use of the SPOT software for analysis of microscope images and Dr Vicki McGovern for providing technical advice.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Roberts D.F., Chavez J., Court S.D. The genetic component in child mortality. Arch. Dis. Child. 1970;45:33–38. doi: 10.1136/adc.45.239.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Crawford T.O., Pardo C.A. The neurobiology of childhood spinal muscular atrophy. Neurobiol. Dis. 1996;3:97–110. doi: 10.1006/nbdi.1996.0010. [DOI] [PubMed] [Google Scholar]
- 3.Dubowitz V. Muscle Disorders in Childhood. London: W.B. Saunders; 1995. [Google Scholar]
- 4.Lefebvre S., Burglen L., Reboullet S., Clermont O., Burlet P., Viollet L., Benichou B., Cruaud C., Millasseau P., Zeviani M., et al. Identification and characterization of a spinal muscular atrophy-determining gene. Cell. 1995;80:155–165. doi: 10.1016/0092-8674(95)90460-3. [DOI] [PubMed] [Google Scholar]
- 5.Lefebvre S., Burlet P., Liu Q., Bertrandy S., Clermont O., Munnich A., Dreyfuss G., Melki J. Correlation between severity and SMN protein level in spinal muscular atrophy. Nat. Genet. 1997;16:265–269. doi: 10.1038/ng0797-265. [DOI] [PubMed] [Google Scholar]
- 6.Coovert D.D., Le T.T., McAndrew P.E., Strasswimmer J., Crawford T.O., Mendell J.R., Coulson S.E., Androphy E.J., Prior T.W., Burghes A.H. The survival motor neuron protein in spinal muscular atrophy. Hum. Mol. Genet. 1997;6:1205–1214. doi: 10.1093/hmg/6.8.1205. [DOI] [PubMed] [Google Scholar]
- 7.Lorson C.L., Hahnen E., Androphy E.J., Wirth B. A single nucleotide in the SMN gene regulates splicing and is responsible for spinal muscular atrophy. Proc. Natl Acad. Sci. USA. 1999;96:6307–6311. doi: 10.1073/pnas.96.11.6307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Monani U.R., Lorson C.L., Parsons D.W., Prior T.W., Androphy E.J., Burghes A.H., McPherson J.D. A single nucleotide difference that alters splicing patterns distinguishes the SMA gene SMN1 from the copy gene SMN2. Hum. Mol. Genet. 1999;8:1177–1183. doi: 10.1093/hmg/8.7.1177. [DOI] [PubMed] [Google Scholar]
- 9.Cartegni L., Krainer A.R. Disruption of an SF2/ASF-dependent exonic splicing enhancer in SMN2 causes spinal muscular atrophy in the absence of SMN1. Nat. Genet. 2002;30:377–384. doi: 10.1038/ng854. [DOI] [PubMed] [Google Scholar]
- 10.Kashima T., Manley J.L. A negative element in SMN2 exon 7 inhibits splicing in spinal muscular atrophy. Nat. Genet. 2003;34:460–463. doi: 10.1038/ng1207. [DOI] [PubMed] [Google Scholar]
- 11.Gennarelli M., Lucarelli M., Capon F., Pizzuti A., Merlini L., Angelini C., Novelli G., Dallapiccola B. Survival motor neuron gene transcript analysis in muscles from spinal muscular atrophy patients. Biochem. Biophys. Res. Commun. 1995;213:342–348. doi: 10.1006/bbrc.1995.2135. [DOI] [PubMed] [Google Scholar]
- 12.Parsons D.W., McAndrew P.E., Monani U.R., Mendell J.R., Burghes A.H., Prior T.W. An 11 base pair duplication in exon 6 of the SMN gene produces a type I spinal muscular atrophy (SMA) phenotype: further evidence for SMN as the primary SMA-determining gene. Hum. Mol. Genet. 1996;5:1727–1732. doi: 10.1093/hmg/5.11.1727. [DOI] [PubMed] [Google Scholar]
- 13.Lorson C.L., Strasswimmer J., Yao J.M., Baleja J.D., Hahnen E., Wirth B., Le T., Burghes A.H., Androphy E.J. SMN oligomerization defect correlates with spinal muscular atrophy severity. Nat. Genet. 1998;19:63–66. doi: 10.1038/ng0598-63. [DOI] [PubMed] [Google Scholar]
- 14.Lorson C.L., Androphy E.J. An exonic enhancer is required for inclusion of an essential exon in the SMA-determining gene SMN. Hum. Mol. Genet. 2000;9:259–265. doi: 10.1093/hmg/9.2.259. [DOI] [PubMed] [Google Scholar]
- 15.Burghes A.H. When is a deletion not a deletion? When it is converted. Am. J. Hum. Genet. 1997;61:9–15. doi: 10.1086/513913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.McAndrew P.E., Parsons D.W., Simard L.R., Rochette C., Ray P.N., Mendell J.R., Prior T.W., Burghes A.H. Identification of proximal spinal muscular atrophy carriers and patients by analysis of SMNT and SMNC gene copy number. Am. J. Hum. Genet. 1997;60:1411–1422. doi: 10.1086/515465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sun Y., Grimmler M., Schwarzer V., Schoenen F., Fischer U., Wirth B. Molecular and functional analysis of intragenic SMN1 mutations in patients with spinal muscular atrophy. Hum. Mutat. 2005;25:64–71. doi: 10.1002/humu.20111. [DOI] [PubMed] [Google Scholar]
- 18.Clermont O., Burlet P., Benit P., Chanterau D., Saugier-Veber P., Munnich A., Cusin V. Molecular analysis of SMA patients without homozygous SMN1 deletions using a new strategy for identification of SMN1 subtle mutations. Hum. Mutat. 2004;24:417–427. doi: 10.1002/humu.20092. [DOI] [PubMed] [Google Scholar]
- 19.Prior T.W. Spinal muscular atrophy diagnostics. J. Child Neurol. 2007;22:952–956. doi: 10.1177/0883073807305668. [DOI] [PubMed] [Google Scholar]
- 20.Alias L., Bernal S., Fuentes-Prior P., Barcelo M.J., Also E., Martinez-Hernandez R., Rodriguez-Alvarez F.J., Martin Y., Aller E., Grau E., et al. Mutation update of spinal muscular atrophy in Spain: molecular characterization of 745 unrelated patients and identification of four novel mutations in the SMN1 gene. Hum. Genet. 2009;125:29–39. doi: 10.1007/s00439-008-0598-1. [DOI] [PubMed] [Google Scholar]
- 21.Cusco I., Barcelo M.J., del Rio E., Baiget M., Tizzano E.F. Detection of novel mutations in the SMN Tudor domain in type I SMA patients. Neurology. 2004;63:146–149. doi: 10.1212/01.wnl.0000132634.48815.13. [DOI] [PubMed] [Google Scholar]
- 22.Liu Q., Dreyfuss G. A novel nuclear structure containing the survival of motor neurons protein. EMBO J. 1996;15:3555–3565. [PMC free article] [PubMed] [Google Scholar]
- 23.Young P.J., Le T.T., thi Man N., Burghes A.H., Morris G.E. The relationship between SMN, the spinal muscular atrophy protein, and nuclear coiled bodies in differentiated tissues and cultured cells. Exp. Cell Res. 2000;256:365–374. doi: 10.1006/excr.2000.4858. [DOI] [PubMed] [Google Scholar]
- 24.Gubitz A.K., Feng W., Dreyfuss G. The SMN complex. Exp. Cell Res. 2004;296:51–56. doi: 10.1016/j.yexcr.2004.03.022. [DOI] [PubMed] [Google Scholar]
- 25.Eggert C., Chari A., Laggerbauer B., Fischer U. Spinal muscular atrophy: the RNP connection. Trends Mol. Med. 2006;12:113–121. doi: 10.1016/j.molmed.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 26.Pellizzoni L. Chaperoning ribonucleoprotein biogenesis in health and disease. EMBO Rep. 2007;8:340–345. doi: 10.1038/sj.embor.7400941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Pellizzoni L., Yong J., Dreyfuss G. Essential role for the SMN complex in the specificity of snRNP assembly. Science. 2002;298:1775–1779. doi: 10.1126/science.1074962. [DOI] [PubMed] [Google Scholar]
- 28.Meister G., Buhler D., Pillai R., Lottspeich F., Fischer U. A multiprotein complex mediates the ATP-dependent assembly of spliceosomal U snRNPs. Nat. Cell Biol. 2001;3:945–949. doi: 10.1038/ncb1101-945. [DOI] [PubMed] [Google Scholar]
- 29.Schrank B., Gotz R., Gunnersen J.M., Ure J.M., Toyka K.V., Smith A.G., Sendtner M. Inactivation of the survival motor neuron gene, a candidate gene for human spinal muscular atrophy, leads to massive cell death in early mouse embryos. Proc. Natl Acad. Sci. USA. 1997;94:9920–9925. doi: 10.1073/pnas.94.18.9920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Frugier T., Tiziano F.D., Cifuentes-Diaz C., Miniou P., Roblot N., Dierich A., Le Meur M., Melki J. Nuclear targeting defect of SMN lacking the C-terminus in a mouse model of spinal muscular atrophy. Hum. Mol. Genet. 2000;9:849–858. doi: 10.1093/hmg/9.5.849. [DOI] [PubMed] [Google Scholar]
- 31.Cifuentes-Diaz C., Frugier T., Tiziano F.D., Lacène E., Roblot N., Joshi V., Moreau M.H., Melki J. Deletion of murine SMN exon 7 directed to skeletal muscle leads to severe muscular dystrophy. J. Cell Biol. 2001;152:1107–1114. doi: 10.1083/jcb.152.5.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Vitte J.M., Davoult B., Roblot N., Mayer M., Joshi V., Courageot S., Tronche F., Vadrot J., Moreau M.H., Kemeny F., et al. Deletion of murine Smn exon 7 directed to liver leads to severe defect of liver development associated with iron overload. Am. J. Pathol. 2004;165:1731–1741. doi: 10.1016/S0002-9440(10)63428-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Monani U.R., Sendtner M., Coovert D.D., Parsons D.W., Andreassi C., Le T.T., Jablonka S., Schrank B., Rossol W., Prior T.W., et al. The human centromeric survival motor neuron gene (SMN2) rescues embryonic lethality in Smn(−/−) mice and results in a mouse with spinal muscular atrophy. Hum. Mol. Genet. 2000;9:333–339. doi: 10.1093/hmg/9.3.333. [DOI] [PubMed] [Google Scholar]
- 34.Hsieh-Li H.M., Chang J.G., Jong Y.J., Wu M.H., Wang N.M., Tsai C.H., Li H. A mouse model for spinal muscular atrophy. Nat. Genet. 2000;24:66–70. doi: 10.1038/71709. [DOI] [PubMed] [Google Scholar]
- 35.Le T.T., Pham L.T., Butchbach M.E., Zhang H.L., Monani U.R., Coovert D.D., Gavrilina T.O., Xing L., Bassell G.J., Burghes A.H. SMNDelta7, the major product of the centromeric survival motor neuron (SMN2) gene, extends survival in mice with spinal muscular atrophy and associates with full-length SMN. Hum. Mol. Genet. 2005;14:845–857. doi: 10.1093/hmg/ddi078. [DOI] [PubMed] [Google Scholar]
- 36.Parsons D.W., McAndrew P.E., Iannaccone S.T., Mendell J.R., Burghes A.H., Prior T.W. Intragenic telSMN mutations: frequency, distribution, evidence of a founder effect, and modification of the spinal muscular atrophy phenotype by cenSMN copy number. Am. J. Hum. Genet. 1998;63:1712–1723. doi: 10.1086/302160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Monani U.R., Pastore M.T., Gavrilina T.O., Jablonka S., Le T.T., Andreassi C., DiCocco J.M., Lorson C., Androphy E.J., Sendtner M., et al. A transgene carrying an A2G missense mutation in the SMN gene modulates phenotypic severity in mice with severe (type I) spinal muscular atrophy. J. Cell Biol. 2003;160:41–52. doi: 10.1083/jcb.200208079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wan L., Battle D.J., Yong J., Gubitz A.K., Kolb S.J., Wang J., Dreyfuss G. The survival of motor neurons protein determines the capacity for snRNP assembly: biochemical deficiency in spinal muscular atrophy. Mol. Cell Biol. 2005;25:5543–5551. doi: 10.1128/MCB.25.13.5543-5551.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gabanella F., Carissimi C., Usiello A., Pellizzoni L. The activity of the spinal muscular atrophy protein is regulated during development and cellular differentiation. Hum. Mol. Genet. 2005;14:3629–3642. doi: 10.1093/hmg/ddi390. [DOI] [PubMed] [Google Scholar]
- 40.Gabanella F., Butchbach M.E., Saieva L., Carissimi C., Burghes A.H., Pellizzoni L. Ribonucleoprotein assembly defects correlate with spinal muscular atrophy severity and preferentially affect a subset of spliceosomal snRNPs. PLoS ONE. 2007;2:e921. doi: 10.1371/journal.pone.0000921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Lotti F., Dittmar K., Younis I., Wan L., Kasim M., Dreyfuss G. SMN deficiency causes tissue-specific perturbations in the repertoire of snRNAs and widespread defects in splicing. Cell. 2008;133:585–600. doi: 10.1016/j.cell.2008.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kolossova I., Padgett R.A. U11 snRNA interacts in vivo with the 5′ splice site of U12-dependent (AU-AC) pre-mRNA introns. RNA. 1997;3:227–233. [PMC free article] [PubMed] [Google Scholar]
- 43.Rossoll W., Kronig A.-K., Ohndorf U.-M., Steegborn C., Jablonka S., Sendtner M. Specific interaction of Smn, the spinal muscular atrophy determining gene product, with hnRNP-R and gry-rbp/hnRNP-Q: a role for Smn in RNA processing in motor neuron axons? Hum. Mol. Genet. 2002;11:93–105. doi: 10.1093/hmg/11.1.93. [DOI] [PubMed] [Google Scholar]
- 44.Sharma A., Lambrechts A., Hao le T., Le T.T., Sewry C.A., Ampe C., Burghes A.H., Morris G.E. A role for complexes of survival of motor neurons (SMN) protein with gemins and profilin in neurite-like cytoplasmic extensions of cultured nerve cells. Exp. Cell Res. 2005;309:185–197. doi: 10.1016/j.yexcr.2005.05.014. [DOI] [PubMed] [Google Scholar]
- 45.Zhang H., Xing L., Rossoll W., Wichterle H., Singer R.H., Bassell G.J. Multiprotein complexes of the survival of motor neuron protein SMN with Gemins traffic to neuronal processes and growth cones of motor neurons. J. Neurosci. 2006;26:8622–8632. doi: 10.1523/JNEUROSCI.3967-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.McWhorter M.L., Monani U.R., Burghes A.H., Beattie C.E. Knockdown of the survival motor neuron (Smn) protein in zebrafish causes defects in motor axon outgrowth and pathfinding. J. Cell Biol. 2003;162:919–931. doi: 10.1083/jcb.200303168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Rossoll W., Jablonka S., Andreassi C., Kroning A.K., Karle K., Monani U.R., Sendtner M. Smn, the spinal muscular atrophy-determining gene product, modulates axon growth and localization of beta-actin mRNA in growth cones of motoneurons. J. Cell Biol. 2003;163:801–812. doi: 10.1083/jcb.200304128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jablonka S., Beck M., Lechner B.D., Mayer C., Sendtner M. Defective Ca2+ channel clustering in axon terminals disturbs excitability in motoneurons in spinal muscular atrophy. J. Cell Biol. 2007;179:139–149. doi: 10.1083/jcb.200703187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Shpargel K.B., Matera A.G. Gemin proteins are required for efficient assembly of Sm-class ribonucleoproteins. Proc. Natl Acad. Sci. USA. 2005;102:17372–17377. doi: 10.1073/pnas.0508947102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhang H., Xing L., Singer R.H., Bassell G.J. QNQKE targeting motif for the SMN-Gemin multiprotein complex in neurons. J. Neurosci. Res. 2007;85:2657–2667. doi: 10.1002/jnr.21308. [DOI] [PubMed] [Google Scholar]
- 51.Carrel T.L., McWhorter M.L., Workman E., Zhang H., Wolstencroft E.C., Lorson C., Bassell G.J., Burghes A.H., Beattie C.E. Survival motor neuron function in motor axons is independent of functions required for small nuclear ribonucleoprotein biogenesis. J. Neurosci. 2006;26:11014–11022. doi: 10.1523/JNEUROSCI.1637-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.McGovern V.L., Gavrilina T.O., Beattie C.E., Burghes A.H. Embryonic motor axon development in the severe SMA mouse. Hum. Mol. Genet. 2008;17:2900–2909. doi: 10.1093/hmg/ddn189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bouwsma G., Van Wijngaarden G.K. Spinal muscular atrophy and hypertrophy of the calves. J. Neurol. Sci. 1980;44:275–279. doi: 10.1016/0022-510x(80)90136-7. [DOI] [PubMed] [Google Scholar]
- 54.Pearn J., Hudgson P. Anterior-horn cell degeneration and gross calf hypertrophy with adolescent onset. A new spinal muscular atrophy syndrome. Lancet. 1978;1:1059–1061. doi: 10.1016/s0140-6736(78)90910-8. [DOI] [PubMed] [Google Scholar]
- 55.Buhler D., Raker V., Luhrmann R., Fischer U. Essential role for the tudor domain of SMN in spliceosomal U snRNP assembly: implications for spinal muscular atrophy. Hum. Mol. Genet. 1999;8:2351–2357. doi: 10.1093/hmg/8.13.2351. [DOI] [PubMed] [Google Scholar]
- 56.Pellizzoni L., Charroux B., Dreyfuss G. SMN mutants of spinal muscular atrophy patients are defective in binding to snRNP proteins. Proc. Natl Acad. Sci. USA. 1999;96:11167–11172. doi: 10.1073/pnas.96.20.11167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Young P.J., Man N.T., Lorson C.L., Le T.T., Androphy E.J., Burghes A.H., Morris G.E. The exon 2b region of the spinal muscular atrophy protein, SMN, is involved in self-association and SIP1 binding. Hum. Mol. Genet. 2000;9:2869–2877. doi: 10.1093/hmg/9.19.2869. [DOI] [PubMed] [Google Scholar]
- 58.Otter S., Grimmler M., Neuenkirchen N., Chari A., Sickmann A., Fischer U. A comprehensive interaction map of the human survival of motor neuron (SMN) complex. J. Biol. Chem. 2007;282:5825–5833. doi: 10.1074/jbc.M608528200. [DOI] [PubMed] [Google Scholar]
- 59.Hahnen E., Schönling J., Rudnik-Schöneborn S., Raschke H., Zerres K., Wirth B. Missense mutations in exon 6 of the survival motor neuron gene in patients with spinal muscular atrophy (SMA) Hum. Mol. Genet. 1997;6:821–825. doi: 10.1093/hmg/6.5.821. [DOI] [PubMed] [Google Scholar]
- 60.Burnett B.G., Munoz E., Tandon A., Kwon D.Y., Sumner C.J., Fischbeck K.H. Regulation of SMN protein stability. Mol. Cell Biol. 2009;29:1107–1115. doi: 10.1128/MCB.01262-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Zhang H.L., Pan F., Hong D., Shenoy S.M., Singer R.H., Bassell G.J. Active transport of the survival motor neuron protein and the role of exon-7 in cytoplasmic localization. J. Neurosci. 2003;23:6627–6637. doi: 10.1523/JNEUROSCI.23-16-06627.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mattis V.B., Bowerman M., Kothary R., Lorson C.L. A SMNDelta7 read-through product confers functionality to the SMNDelta7 protein. Neurosci. Lett. 2008;442:54–58. doi: 10.1016/j.neulet.2008.06.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Heier C.R., DiDonato C.J. Translational readthrough by the aminoglycoside geneticin (G418) modulates SMN stability in vitro and improves motor function in SMA mice in vivo. Hum. Mol. Genet. 2009;18:1310–1322. doi: 10.1093/hmg/ddp030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Yu B., Howell P.L. Intragenic complementation and the structure and function of argininosuccinate lyase. Cell. Mol. Life Sci. 2000;57:1637–1651. doi: 10.1007/PL00000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Howell P.L., Turner M.A., Christodoulou J., Walker D.C., Craig H.J., Simard L.R., Ploder L., McInnes R.R. Intragenic complementation at the argininosuccinate lyase locus: reconstruction of the active site. J. Inherit. Metab. Dis. 1998;21(Suppl. 1):72–85. doi: 10.1023/a:1005361724967. [DOI] [PubMed] [Google Scholar]
- 66.Rodriguez-Pombo P., Perez-Cerda C., Perez B., Desviat L.R., Sanchez-Pulido L., Ugarte M. Towards a model to explain the intragenic complementation in the heteromultimeric protein propionyl-CoA carboxylase. Biochim. Biophys. Acta. 2005;1740:489–498. doi: 10.1016/j.bbadis.2004.10.009. [DOI] [PubMed] [Google Scholar]
- 67.Fischer U., Liu Q., Dreyfuss G. The SMN–SIP1 complex has an essential role in spliceosomal snRNP biogenesis. Cell. 1997;90:1023–1029. doi: 10.1016/s0092-8674(00)80368-2. [DOI] [PubMed] [Google Scholar]
- 68.Winkler C., Eggert C., Gradl D., Meister G., Giegerich M., Wedlich D., Laggerbauer B., Fischer U. Reduced U snRNP assembly causes motor axon degeneration in an animal model for spinal muscular atrophy. Genes Dev. 2005;19:2320–2330. doi: 10.1101/gad.342005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.McWhorter M.L., Boon K.L., Horan E.S., Burghes A.H., Beattie C.E. The SMN binding protein Gemin2 is not involved in motor axon outgrowth. Dev. Neurobiol. 2008;68:182–194. doi: 10.1002/dneu.20582. [DOI] [PubMed] [Google Scholar]
- 70.Oprea G.E., Krober S., McWhorter M.L., Rossoll W., Muller S., Krawczak M., Bassell G.J., Beattie C.E., Wirth B. Plastin 3 is a protective modifier of autosomal recessive spinal muscular atrophy. Science. 2008;320:524–527. doi: 10.1126/science.1155085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Liu D.P., Schmidt C., Billings T., Davisson M.T. Quantitative PCR genotyping assay for the Ts65Dn mouse model of Down syndrome. BioTechniques. 2003;35:1170–1174, 1176, 1178. doi: 10.2144/03356st02. passim. [DOI] [PubMed] [Google Scholar]
- 72.Carissimi C., Saieva L., Baccon J., Chiarella P., Maiolica A., Sawyer A., Rappsilber J., Pellizzoni L. Gemin8 is a novel component of the survival motor neuron complex and functions in snRNP assembly. J. Biol. Chem. 2006;281:37009–37016. doi: 10.1074/jbc.M512243200. [DOI] [PubMed] [Google Scholar]
- 73.Gavrilina T.O., McGovern V.L., Workman E., Crawford T.O., Gogliotti R.G., DiDonato C.J., Monani U.R., Morris G.E., Burghes A.H. Neuronal SMN expression corrects spinal muscular atrophy in severe SMA mice while muscle-specific SMN expression has no phenotypic effect. Hum. Mol. Genet. 2008;17:1063–1075. doi: 10.1093/hmg/ddm379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Man N.t., Humphrey E., Lam L.T., Fuller H.R., Lynch T.A., Sewry C.A., Goodwin P.R., Mackenzie A.E., Morris G.E. A two-site ELISA can quantify upregulation of SMN protein by drugs for spinal muscular atrophy. Neurology. 2008;71:1757–1763. doi: 10.1212/01.wnl.0000313038.34337.b1. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.