Abstract
Individuals at risk of coronary heart disease (CHD) show multiple correlations across blood biomarkers. Single nucleotide polymorphisms (SNPs) indexing biomarker differences could help distinguish causal from confounded associations because of their random allocation prior to disease. We examined the association of 948 SNPs in 122 candidate genes with 12 CHD-associated phenotypes in 2775 middle aged men (a genic scan). Of these, 140 SNPs indexed differences in HDL- and LDL-cholesterol, triglycerides, C-reactive protein, fibrinogen, factor VII, apolipoproteins AI and B, lipoprotein-associated phospholipase A2, homocysteine or folate, some with large effect sizes and highly significant P-values (e.g. 2.15 standard deviations at P = 9.2 × 10−140 for F7 rs6046 and FVII levels). Top ranking SNPs were then tested for association with additional biomarkers correlated with the index phenotype (phenome scan). Several SNPs (e.g. in APOE, CETP, LPL, APOB and LDLR) influenced multiple phenotypes, while others (e.g. in F7, CRP and FBB) showed restricted association to the index marker. SNPs influencing six blood proteins were used to evaluate the nature of the associations between correlated blood proteins utilizing Mendelian randomization. Multiple SNPs were associated with CHD-related quantitative traits, with some associations restricted to a single marker and others exerting a wider genetic ‘footprint’. SNPs indexing biomarkers provide new tools for investigating biological relationships and causal links with disease. Broader and deeper integrated analyses, linking genomic with transcriptomic, proteomic and metabolomic analysis, as well as clinical events could, in principle, better delineate CHD causing pathways amenable to treatment.
INTRODUCTION
Like many common disorders, coronary heart disease (CHD) results from a complex interplay between environmental and genetic factors, complicating the identification of the causal pathways, and delaying the development of new treatments (1). By 1981, over 200 phenotypic and other differences had been shown in those with, or at higher risk of CHD (2). Alterations in circulating blood phenotypes (also referred to as biomarkers) such as lipid and lipoprotein particles, proteins involved in inflammation and coagulation, as well as metabolites and markers of oxidant stress, tend to cluster among those at higher risk, making it difficult to ascertain the nature and direction of biological relationships between biomarkers, and the independent effect of any one biomarker on CHD risk (3). These associations may be causal, but alternatively could arise because they mark subclinical disease (reverse causation), other causal factors (confounding) or some combination of the two. Thus far it has only been possible to establish unequivocally a causal role in CHD for one blood phenotype (LDL-cholesterol). This was achieved in large part by developing the HMG-CoA reductase inhibitor (statin) class of drugs that reduce blood level of LDL-cholesterol and testing their effects in randomized trials (4). However, the expense and risk associated with the development of the many selective drugs needed to target the vast array of blood phenotypes implicated in CHD, currently precludes a comprehensive and systematic approach to understanding their causal relevance and limits translation of the basic science and epidemiological findings into new treatments.
Many (if not all) of the circulating biomarkers associated with CHD behave as heritable quantitative traits, and common single nucleotide polymorphisms (SNPs) influencing their variance are now being identified both by candidate gene and genome wide studies (5–8). Unlike the associations between biomarkers, or the association of biomarkers with CHD risk, genetic associations should be protected from reverse causation because genotype is an invariant characteristic, such that there is a unidirectional flow of information from common genome variation to mRNA to protein to complex phenotype and disease. Moreover, since genotype is determined at random at conception, for SNPs affecting CHD risk, intermediate phenotypes (such as blood markers), residing off the causal pathway from SNP to disease, should be balanced evenly among the different genotypic groups, as they are in a randomized clinical trial, whereas biomarkers that mediate the effect of genomic variation on disease risks should differ by genotype. We hypothesized that SNPs, which affect the variance in biomarkers linked to CHD, could be used as unbiased tools with which to understand their causal relevance and to explore the extent to which correlation between multiple CHD-associated phenotypes are affected by confounding. This principle of Mendelian randomization has been applied to assess the causal role of individual biomarkers with CHD risk (9,10). Here we applied Mendelian randomization to help distinguish causal from confounded links between multiple biomarkers.
Formerly, association studies in the cardiovascular genetics field typed SNPs in a single gene or locus of interest. More recently, genome wide association studies (GWAS) (5–7) have extended the breadth of genetic information, but the focus of these studies has been on a single continuous trait or disease outcome, limiting the ability to address inter-relationships among traits. We utilized a genotyping strategy that exploits the recent finding that gene variants regulating expression commonly reside in the vicinity of genes (11–13), to build a panel of SNPs influencing variance in one or more CHD-related biomarkers to investigate the nature of the association between markers.
RESULTS
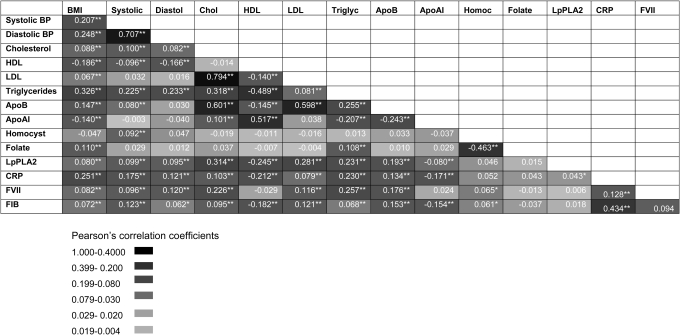
Between-phenotype correlations and associations with CHD risk
In the prospective Northwick Park Heart Study II of 3102 initially healthy men followed prospectively for a median of 13.6 years, there were 296 definite fatal or non-fatal CHD events. Measures of 14 intermediate phenotypes were available, six with annual repeat measures (on six occasions) and single measures for the remaining eight (Supplementary Material, Table S1). To delineate the potential for confounding in the associations of blood markers and other phenotypes with CHD, we assessed between-phenotype correlations. Men who developed a CHD event on follow-up exhibited multiple phenotypes which distinguished them from those who remained disease free, and these differences were highly inter-correlated (Fig. 1 and Supplementary Material, Table S1). Seventy-two of the 105 possible pair-wise correlations between blood markers and other CHD-related traits were significant with a P-value ≤ 0.05 (68 traits significant with a P-value ≤ 0.01), which significantly exceeds the five expected by chance alone (P < 1 × 10−8 for observed versus the expected). Many of the associations were highly significant and the absolute P-values are provided in Supplementary Material, Figure S1.
Figure 1.
Correlations between multiple phenotypes linked to CHD in 2775 men from the NPHSII study. Values in cells indicate Pearson's correlation coefficient R. *P < 0.01, **P < 0.001 (see colour code). Baseline and five repeat measures were available for cholesterol, triglycerides (TG), coagulation factor VII (FVIIc), fibrinogen, blood pressure (BP), smoking and body mass index (BMI) and single measures for the remaining traits.
Genic scan
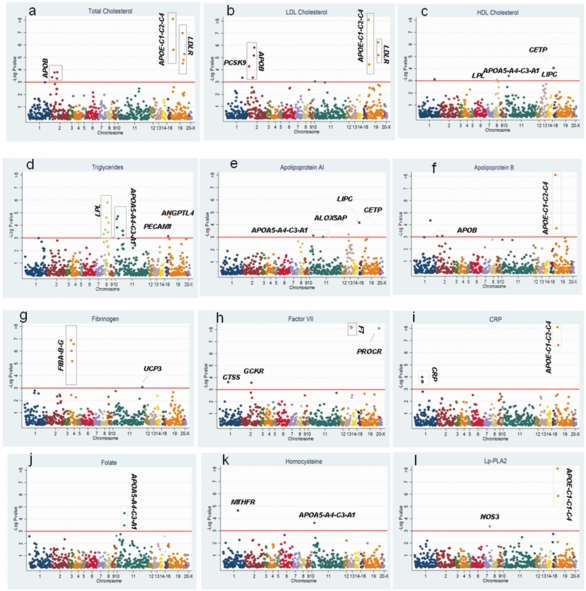
We genotyped 948 SNPs in 122 genes chosen for a high prior probability of association with CHD-associated phenotypes in NPHS II based on prior association studies and biological evidence. A total of 134 SNP-trait associations were identified distributed as follows: 12 SNPs in APOB, LDLR, PCSK9, APOE-C1-C2-C4 and LRP5 with total cholesterol; 23 SNPs in APOB, LDLR, APOE-C1-C2-C4, PLA2G7, PCSK9, CDKN, EXT2, C3 and GSTM3 with LDL-cholesterol and 6 SNPs in CETP, ALOX5AP APOA5-A4-C3-A1, LIPC and LPL with HDL cholesterol. Thirty-seven SNPs in LPL, APOA5-A4-C3-A1, TGFB1, PECAM, IL6R, C2, ILRN1, INS, LDLR, F7, ANGPTL4, APOB, GCKR, IL18RAP, PCSK9 and LRP5 were associated with triglyceride levels. Nine SNPs in LIPC, CETP, ALOX5AP, APOA5-A4-C3-A1, APOE-C1-C2-C4, IGF2, C3 and LPL were associated with apoAI levels. Twelve SNPs were associated with apoB levels; these were in APOE-C1-C2-C4, APOB, LDLR, PCSK9, LPL GSTM4 and EXT2. Three SNPs in APOE-C1-C2-C4 and NOS3 were associated with Lp-PLA2 activity. Seven SNPs in the F7 gene itself and one in PROCR exhibited very highly significant association with blood factor VII level. Seven SNPs in the FIBA-B-G cluster, UCP3, GSTM4 and the APOA5-A4-C3-A1 cluster were associated with fibrinogen. Five SNPs from two genes, APOE-C1-C2-C4 and CRP, were associated with C-reactive protein. These represented 98 unique SNPs in 36 genes. The full range of SNP-phenotype associations is illustrated in Figure 2A-l and summarized in Supplementary Material, Table S2. Because of the gene-centric strategy, the SNP-phenotype associations clustered within genes, even though SNPs had been chosen as tag SNPs. These associations remained significant even after the LD structure between them was considered (see Supplementary Material, Table S6). Thus, for some genes, several SNPs remain associated with the blood biomarker, supporting the possibility of independent associations at a given locus, as we noted previously for CRP (14).
Figure 2.
Associations of 860 SNPs by chromosome with 12 blood phenotypes. The horizontal line indicates a critical FDR threshold of 0.2, approximately equivalent to a P-value < 10−3.
Significance and validity of the genetic associations
Despite the moderate size of the data set, nearly 13% (18 of 140) of the associations achieved the level of significance conventionally applied to GWAS (P-value < 10−7). Sixteen of the 134 SNPs were also directly typed and were significant hits in recent GWASs of blood markers (Table 1) including: rs6511720 in LDLR (5,15), SNPs in LD with rs42935/rs7412 with APOE SNPs and LDL-cholesterol (5,6); as well as rs708272 in CETP with HDL-cholesterol (5,6,15).
Table 1.
SNPs from the current study showing replication with GWAS hits
| Phenotype | Previous GWAS hit (alternate phenotype previously showing association) | rs | Gene | P-value | FDR | Common versus rare HMZ | % R2trans | MAF |
|---|---|---|---|---|---|---|---|---|
| Triglycerides | (6)(LDL) | rs6589566 | APOA5-A4-C3-A1 | 1.98E−05 | 0.01 | 0.633 | 0.9 | 7.0 |
| (15) | rs328 | LPL | 2.85E−04 | 0.03 | 0.189 | 0.7 | 10.9 | |
| Cholesterol | (15) | rs6511720 | LDLR | 9.38E−08 | 0.01 | 0.432 | 1.2 | 12.3 |
| (5,7,15), (42)(AD), (43)(AD) | rs4420638 | APOE-C1-C2-C4 | 2.78E−06 | 0.01 | 0.370 | 1.0 | 19.1 | |
| LDL | (5,15) | rs6511720 | LDLR | 3.83E−06 | 0.01 | 0.776 | 1.6 | 12.3 |
| (5,7,15,42)(AD), (43)(AD) | rs4420638 | APOE-C1-C2-C4 | 8.72E−04 | 0.11 | 0.344 | 0.9 | 19.1 | |
| ApoB | (15)(LDL) | rs11591147 | PCSK9 | 6.43E−05 | 0.03 | 3.055 | 0.9 | 0.9 |
| (15)(LDL); (5)(LDL) | rs6511720 | LDLR | 1.23E−03 | 0.14 | 0.195 | 0.6 | 12.3 | |
| (15)(TG), (5)(TG), (6)(LDL), (17)(CRP) | rs780094 | GCKR | 3.21E−03 | 0.20 | 0.207 | 0.5 | 37.8 | |
| ApoAI | (15)(HDL) | rs1800588 | LIPC | 4.72E−05 | 0.03 | 0.332 | 1.0 | 20.5 |
| (15)(HDL, TG) | rs328 | LPL | 2.14E−03 | 0.19 | 0.575 | 0.5 | 10.3 | |
| CRP | (7)(LDL), (15)(LDL), (5)(LDL), (42)(AD), (43)(AD) | rs4420638 | APOE-C1-C2-C4 | 2.20E−07 | 0.01 | 0.404 | 1.4 | 19.1 |
| (17) | rs3091244 | CRP | 2.74E−04 | 0.05 | 0.182 | 0.7 | 5.5 | |
| Lp-PLA2 | (7)(LDL), (24)(LDL), (5)(LDL), (42)(AD) (43) (AD) | rs4420638 | APOE-C1-C2-C4 | 1.42E−06 | 0.01 | 0.490 | 1.48 | 19.1 |
| Homocysteine | (23)(CHD) | rs6922269 | MTHFR | 2.78E−05 | 0.03 | 0.375 | 1.5 | 31.1 |
| FVII | (15)(HDL, TG) | rs328 | LPL | 2.98E−04 | 0.04 | 0.102 | 0.0 | 10.3 |
The phenotype associated with the SNP from the GWAS, if different from the current study is given in brackets.
LDL, low density lipoprotein; TG, triglycerides; AD, Alzheimer's disease, CRP, C-reactive protein, HDL, high density lipoprotein.
Notably, for each genic scan where the outcome was a single-protein phenotype at least one cis-acting SNP in the gene encoding the cognate protein was always identified among the most significantly associated SNPs, with P-values generally between 10−4 and 10−7, but as extreme as 10−140 for rs6046 in F7 and blood factor VII levels. In line with a recent report, we refer to these genes as protein quantitative trait genes (pQTGs) (8) and the SNPs as pQTSNPs.
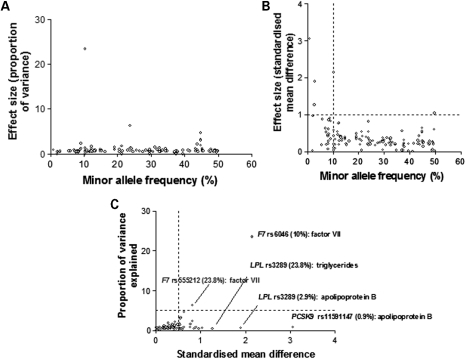
Effect size
The effect of each associating SNP was expressed both in terms of the proportion of the trait variance explained, and as the standardized mean difference in trait level between subjects homozygous for the alternative alleles (Fig. 3A and B), allowing effect sizes to be expressed on a common scale. When considered solely in terms of the variance, the effect size for individual SNPs appeared modest with only two SNPs in the F7 gene, each explaining ∼20% of the variance of factor VII (Fig. 3A), both independently significant of each other. However, for 28 SNPs (24%) crossing the FDR threshold, the difference in average trait values between homozygous subjects exceeded 0.5 standard deviations (SD) of the population distribution, with the difference exceeding 0.75 SD for 17 SNPs. For 10 SNPs, the difference in trait values between homozygous subjects exceeded 1 SD, with the most extreme differences for a common allele (>5%) being 2.15 SD for the difference in Factor VII levels between subjects homozygous for alternative alleles of rs6046 in F7 (Fig. 3C).
Figure 3.
The relationship between minor allele frequency and effect size for SNP-phenotype associations exceeding the pre-specified FDR threshold. Effect size was expressed as: (A) variance and (B) as the standardized mean difference for comparisons of homozygous subjects. (C) Examples of SNPs with extreme effects assessed in terms of standardized mean difference.
Phenome scan
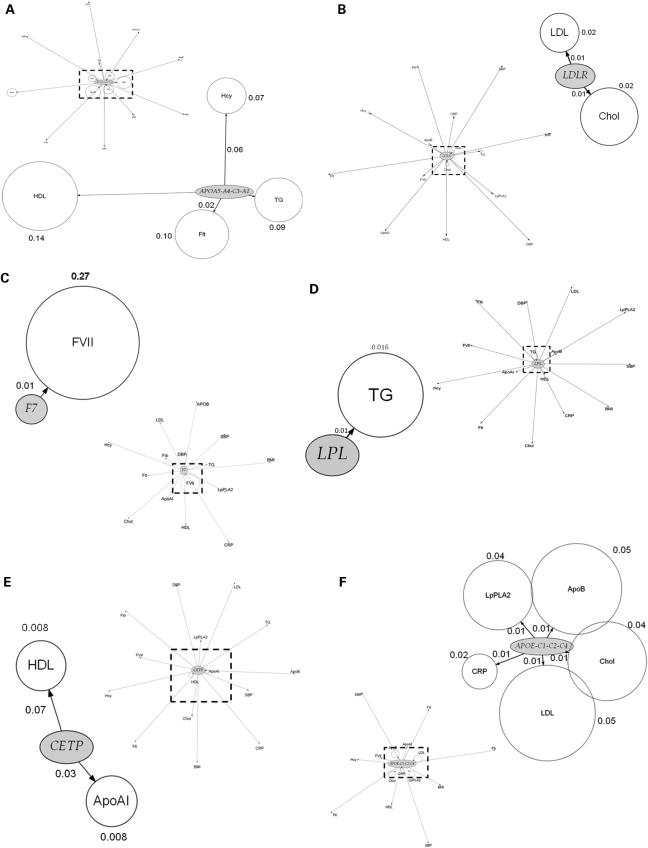
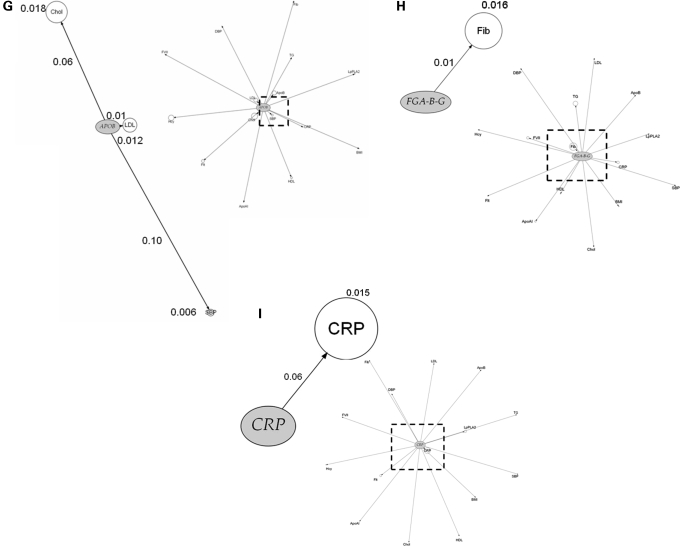
We next assessed the association of top ranking genes from the genic scan with additional phenotypes and the findings are illustrated graphically in Figure 4A–H by means of phenome plots. The phenome scan examines the relationship between variants of the gene of interest and all the intermediate traits studied. This is in contrast with the genic scan where each intermediate trait of interest is considered and all the genetic variants associated with it are examined as a Manhattan plot. In the phenome scan, the gene of interest is depicted as an ellipse and the associated phenotypic traits as circles radiating out from it. The circle diameter of each phenotype is a measure of the variance of the phenotype explained by the variation encompassing all SNPs in that gene of interest, with the value (coefficient of determination, R2) given alongside the relevant circle. In addition, for each phenotype, the distance from the gene is a measure of the significance value, adjusted for multiple testing using the false discovery rate (FDR), for the SNP with the strongest signal within the gene of interest. In the phenome scan, we applied a more stringent significance threshold, focusing on those phenotypes crossing an FDR adjusted P-value of <0.1. An inner square of dotted lines has been drawn at this threshold with the gene of interest and the phenotype(s) with FDRs <0.1 being shown in the enlarged subpanel.
Figure 4.
Phenome plots. The gene of interest is depicted as an ellipse and the associated phenotypes traits as circles. The circle diameter is a measure of the variance of the phenotype explained by the variance of the genotypes (R2) of all the SNPs in the gene of interest. The numbers given next to each phenotype are the percent values of the coefficient of determination R2 of the phenotype for the combined effect of all the SNPs of the gene. The distance from the gene to each phenotype is a measure of the significance value, adjusted for multiple testing using the FDR, for the SNP with the strongest signal within the gene of interest. The number shown next to each edge is the percent value of the FDR adjusted P-value with its length measured from ellipse (gene) centre to circle (phenotype) centre. The dashed line represents those phenotypes which fall within the <0.1 FDR with the gene of interest. Those phenotypes and the gene are then expanded alongside in the accompanying figure so that details can be seen more clearly. For the phenome scan, an even more stringent FDR adjusted P-value of <0.1 to reduce further the risk of false-positive association. (Hcy, homocysteine; Flt, folate; SBP, systolic blood pressure; DBP, diastolic blood pressure; BMI, body mass index).
SNPs in certain pQTGs (e.g. in F7, CRP and the FIBA-B-G cluster) exhibited phenotypic effects that were restricted to an alteration only in the cognate gene product. The restricted nature of the SNP effects for these genes contrasted sharply with the extensive direct phenotypic associations of the proteins they encode (Fig. 1).
SNPs in several other genes and regions, involved in lipid and lipoprotein transport or metabolism, appeared to exert more diverse phenotypic effects. For example, SNPs in the APOE-C1-C2-C4 cluster were associated with three lipid and lipoprotein traits (total- and LDL-cholesterol, apoB) consistent with previous observations (16). The phenome scan for APOE-C1-C2-C4 also revealed a strong association of SNPs in this region with the hepatocyte-derived inflammation marker C-reactive protein (CRP), but interestingly not with another hepatocyte-derived acute phase protein, fibrinogen (Fig. 4F). The APOE–CRP association was highly significant (P = 1.28 × 10−10) and the effect size was at least as large as the effect of cis-acting SNPs in the gene on CRP itself (Supplementary Material, Table S2). This association is also validated by several candidate gene studies and by two recent GWAS (17,18). In addition, APOE-C1-C2-C4 SNPs rs429358/rs7412, which together define the E2, E3 and E4 alleles, were also strongly associated with Lp-PLA2 activity (P = 2.55 × 10−14), and explained almost 4% of the variance in this trait, with a 0.21 SD difference in Lp-PLA2 activity between homozygous subjects. The association of APOE-C1-C2-C4 SNPs with apoAI was also evident but because of the more modest levels of significance, these associations are not shown in Figure 4F. The CETP SNP rs708272, which exhibited the strongest association with HDL-cholesterol in the genic scan, showed additional associations with apoAI (Fig. 4E). These associations have been corroborated by meta-analyses of genetic association studies (19).
SNPs in the APOA5-A4-C3-A1 (Fig. 4A) cluster also exhibited diverse effects on blood lipid and lipoprotein phenotypes as did variants in the LDLR gene (Fig. 4B). For the APOA5 gene cluster and LPL, the anticipated association with triglycerides was seen (20). Furthermore, for the APOA5 cluster, although the previously reported association with apoAI was evident, with an FDR of 0.14 it was not included in the enlarged phenome scan where the FDR cut-off was 0.1.
pQTSNPs as tools to distinguish causal from non-causal associations between blood protein phenotypes
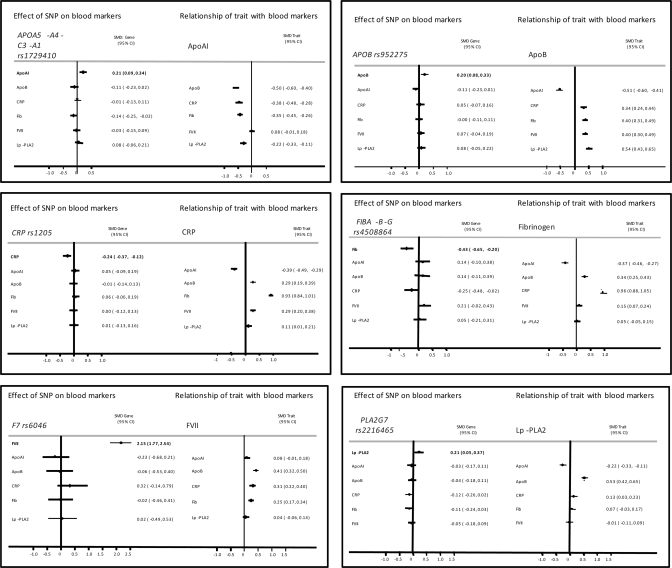
Because cis-acting variants in a gene encoding a protein trait (pQTSNPs) provide a highly specific instrument with which to investigate the causal effects of the encoded protein (utilizing the principles of Mendelian randomization), we used the most strongly associating pQTSNPs for apoAI, apoB, CRP, factor VII, fibrinogen and lipoprotein-associated phospholipase A2 (Lp-PLA2) as their unconfounded proxies to help evaluate confounding in the directly observed associations between these proteins (Supplementary Material, Fig. S2) and (Fig. 5).
Figure 5.
The magnitude of the association between protein phenotypes is illustrated as the mean difference of the standardized levels of the phenotype between top and bottom tertile of the index phenotype, approximately equal to a 2 SD difference. When the most strongly associating SNP in the pQTGs is considered, the mean difference is between the two homozygote groups. (A) apoAI, (B) apoB, (C) CRP, (D) coagulation factor VII, (E) fibrinogen, and (F) Lp-PLA2.
In Figure 5, the magnitude of the association between protein phenotypes is illustrated for a difference in the mean values of the top versus bottom tertile of the index phenotype, approximately equal to a 2 SD difference. Thus, a 2 SD difference in factor VII is associated with a standardized mean difference in apoB, CRP and fibrinogen of 0.41, 0.31 and 0.25, respectively. Individuals homozygous for the variant allele rs6046 exhibited a 2.15 SD higher blood factor VII level which is approximately equivalent to the difference in levels between the top and bottom tertile of the Factor VII distribution. However, the point estimate of the effect of rs6046 on levels of apoB and fibrinogen was null and therefore discordant from that expected from the directly observed association of factor VII with these phenotypes if this was a causal link. The point estimate of the association of rs6046 with CRP was close to that expected from a causal association, but the confidence limits spanned the null. Thus, the simplest explanation of these findings is that the factor VII is unlikely to cause an alteration in apoB or fibrinogen, but a causal effect on CRP is possible and would require a larger sample to confirm or refute reliably. For the majority of other pairwise comparisons, the point estimates of the genetic effect were either null or directionally opposite from that expected from a causal link, though because of wide confidence limits, larger studies would be required to provide definitive evidence.
The relationship between minor allele frequency and standardized effect size for 80% statistical power is plotted in Supplementary Material, Figure S3. The study can detect effects at 0.4 SD for any SNP with a MAF > 20%, but can only detect large effects (>0.8SD) from a SNP with <10% MAF.
DISCUSSION
Gene centric (genic scan) to identify variants influencing multiple blood biomarkers
We developed a genotyping strategy aimed at providing a suite of tools to help evaluate the nature of associations between multiple blood markers associated with a high risk of CHD using Mendelian randomization. We used a gene-centric approach based on genes with a high prior probability of association with the traits of interest in NPHSII. This proved to be an efficient strategy providing a high yield of SNP-biomarker associations even at conservative levels of statistical significance and a stringent FDR threshold, with134 SNP-associations out of the 948 SNPs ( for which genotype was available), distributed among ∼36 genes. These findings are in keeping with emerging data from GWAS of gene expression (11), blood proteins (8), lipid and other non-protein traits (5,6) and metabolic profiles (21). Together these provide strong evidence that regulatory SNPs are found commonly in the vicinity of genes and validate a gene-centric approach to identify genotypes influencing multiple blood traits linked to CHD and other complex disorders. The scope and breadth of gene-centric studies is likely to be extended by the emergence of comprehensive disease-specific gene-centric custom SNP arrays such as the ITMAT/BROAD/CARE (IBC) ‘cardio-metabolic chip’ that covers around 2100 genes from lipid, inflammation, coagulation, oxidants stress, matrix and other pathways linked to CHD (http://bioinf.itmat.upenn.edu/cvdsnp/index.php) (22).
Although the genotyping strategy was focused on candidate genes, some novel associations also emerged, e.g. associations of SNPs in APOE with Lp-PLA2 activity. Lp-PLA2 is physically associated with LDL particles where it could exert pro-atherogenic actions through hydrolysis of oxidized phospholipids to lysophosphatidylcholine and oxidized free fatty acids. Although APOE-C1-C2-C4 SNPs also influence the level of circulating LDL, and theoretically might therefore influence Lp-PLA2 activity simply through an effect on the concentration of this particle, other SNPs that affected LDL-cholesterol concentration (e.g. APOB, and LDLR) were not associated with Lp-PLA2 activity, suggesting a more direct link between apoE and Lp-PLA2 that is worthy of further investigation.
Though 16 of the genetic associations we identified have been corroborated by both candidate gene studies and GWAS, and can be considered robust, the relevance and utility of such associations has been debated given that the proportion of the total variance in a continuous trait explained by common alleles can be small (commonly <5%) (9). However, our study suggests that interpretations based on this metric alone may provide an incomplete picture. For many SNPs whose contribution to the variance in a phenotype was small, we found evidence of a substantial effect size when expressed in terms of the standardized mean difference in biomarker level between homozygous subjects. We noted many examples of differences over 0.5 SD, and several of over 1 SD for variants influencing triglycerides, apoB, Lp-PLA2 and factor VII. These would be considered very substantial effects for a drug designed to modify the level of one of these biomarkers. For example, standard doses of statin drugs in clinical use reduce LDL-cholesterol by about 1 SD (4). However, even SNPs with small effect could be important, because therapies that might arise from the genetic findings could be designed to have much more potent effects than the natural genetic variation. For example, variants in HMGCR that encodes the target enzyme for cholesterol-lowering statin drugs, affect LDL-cholesterol but did not emerge among the top-ranking SNPs in recent GWAS for CHD (23), although an association of these SNPs with CHD risk has been subsequently reported (24).
We also noted a trend for less common genetic variants to exert larger effects when expressed in terms of the standardized mean difference but not in terms of the variance (Fig. 3). This finding is consistent with the emerging paradigm of an inverse association between allele frequency and penetrance (25).
Phenome scans and use of SNPs as tools to distinguish causal from confounded links between biomarkers
It has been common for genome wide and candidate gene association studies to focus on SNP associations with a single trait or disease outcome, but the pathway from disease-relevant SNP to disease potentially involves multiple phenotypic perturbations both in series and in parallel. It is therefore of interest not only to determine the full spectrum of SNPs affecting a single trait or outcome, but also the range of phenotypes altered by single SNP or gene. (26). We noted several genes and SNPs with effects on multiple circulating biomarkers, suggesting that some genes may have a broad footprint of effects on the proteome and metabolome. This is often referred to as pleiotropy but it may be important to distinguish between the multiplicity of effects arising from the generation of more than one protein product as a result of alternative splicing of an mRNA transcript of a gene, from the those arising from the broad ranging downstream effects of a single protein product (such as an enzyme or transporter) involved in lipid and lipoprotein pathways (e.g. apoE or cholesteryl ester transfer protein), which is the more likely explanation for the range of SNP effects observed in the present study.
Though some SNPs exerted a broad footprint of phenotypic effects, for others (e.g. in F7 and FIB), there was a more tightly restricted association of SNPs in the encoding gene with the cognate protein but no other trait, despite the extensive directly observed correlations between the protein itself and other markers. This contrast between the range of genotypic and phenotypic associations is likely to arise from the randomized assignment of alleles, and the non-randomized (clustered) association of phenotypes (see Fig. 1) (27,28). This renders SNPs in certain pQTGs particularly well suited as proxies (instruments) with which to investigate the causal relevance for CHD of the proteins they encode because, in contrast to the proteins themselves, the genetic associations should not be prone to confounding or reverse causation bias (29–31).
For example, CRP is strongly correlated with fibrinogen (R2 = 0.43; Fig. 1). This association could arise because of a causal relationship (in either direction) or because of a common association of both these biomarkers with another factor. This makes it difficult to assess whether CRP and fibrinogen lie on the same causal pathway to CHD, and whether it is legitimate to make statistical adjustment for fibrinogen in the many observational associations of CRP with CHD (and vice versa). Statistical adjustment, which is the orthodox approach to dealing with confounding in observational epidemiology, would be legitimate only if the association of CRP with CHD was not mediated (even in part) by an elevation in the level of fibrinogen. The absence of an association of CRP SNPs with fibrinogen level (or FIBA-B-G with CRP level) (Fig. 4I), which is consistent with the findings from other studies of these variants (29,32) indicates that CRP and fibrinogen do not exhibit a direct causal association with one another and that statistical adjustment is appropriate in observational studies of these biomarkers with CHD.
Whereas cis-acting SNPs in the encoding gene offer the most specific genetic instrument with which to assess the causal role of a protein in CHD, there may not be a single best instrument for a non-protein trait such as the level of HDL- or LDL-cholesterol that are influenced by a number of different genes encoding enzymes and transporters. We noted SNPs from a range of genes influencing the same lipid trait (Supplementary Material, Table S2). Validation of the use of multiple SNPs for such causal analysis for non-protein traits is illustrated using the example of LDL-cholesterol, the only blood phenotype for which a causal role in CHD has already been firmly established.
Rare variants in LDLR, APOB and gain-of-function mutations in PCSK9 result in extreme elevations in LDL-cholesterol, as in Familial Hypercholesterolemia (33), while PCSK9 loss-of-function variants are associated with very low LDL-cholesterol levels and are protective of CHD (34). In addition, common variants of APOE (16), PSRC1/ CELSR/ SORT1, LDLR and PCSK9 (5–7,35), including some tagged by SNPs in the current study that cause more modest changes in LDL-cholesterol, have also been associated with alterations in risk of CHD (5,34,35), with the genetic effects on disease risk directionally concordant and in proportion to the size of the effect on LDL-cholesterol.
Implications of the current findings for future work
The principles applied in the current study could be extended to incorporate rare SNPs or copy number variants, or regulatory SNPs remote from genes using denser new generation GWAS platforms. The phenotypes evaluated could be extended both proximally to include mRNA expression and more distally to include more complex phenotypes linked to CHD such as carotid-intima media thickness or measures of coronary artery calcification. The breadth of the approach could also be extended laterally at the level of protein and non-protein phenotypes using proteomic and metabolomics, respectively. Our study highlights the importance of well-phenotyped population samples and emphasizes the requirement for large sample sizes. However, whether this approach will aid in developing causal networks and genotype: phenotype maps, as outlined recently by Rockman, remains to be seen (36).
In summary, we demonstrate how integrating information on genotype and blood phenotypes in humans can be used to construct association networks with a high-level of credibility, due to the particular properties of genetic variants, which are randomly allocated and unmodifiable by disease process, features which are not shared by any other natural biological exposure (e.g. mRNA levels or protein levels) Our study emphasizes the potential translational application emerging from the recent genomic advances.
MATERIAL AND METHODS
Study design and phenotypic measures
The Northwick Park Heart Study II (NPHSII) is a prospective study of 3012 healthy middle-aged men aged 50–64 years at recruitment, sampled from nine UK general practices between 1989 and 1994 (37). Men were free from disease at the time of recruitment, and information on lifestyle habits, height, weight and blood pressure were recorded at baseline and on subsequent prospective follow-up. Measures were made of at least 15 circulating blood factors associated with CHD risk that included both circulating proteins (CRP, lipid fractions and non-protein metabolites), all the measures being obtained before the development of clinical events, with repeated measurements available for some factors (Supplementary Material, Table S1). A DNA repository was established using samples from 2775 men obtained at the time of recruitment. By December 2005, after a median follow-up of 13.6 years, there had been 296 definite fatal or non-fatal CHD events (230 in 2401 of the genotyped sample). Full details of recruitment, measurements, follow-up and definitions of incident disease have been reported elsewhere (37).
Genic scan
A customized Illumina 768 SNP genotyping array was assembled to comprehensively capture common genetic variation in more than 76 genes chosen (1): for their involvement in the following pathways linked to CHD risk; lipid metabolism (10 genes), inflammation (23 genes), oxidative stress (13 genes), thrombosis and haemostasis (3 genes); (2) for a previously described association with the risk of CHD or type 2 diabetes (T2D) mellitus, including SNPs from a recent whole-genome analysis of T2D which marginally failed the P < 10−7 significance threshold for genome wide significance (Prof Philippe Froguel, personal communication); or (3) for their ability to tag copy number variation (38). For each gene or region, tagging (t) SNPs, optimized for the Illumina platform, was selected using HapMap, applying an R2 threshold of 0.8 with a minor allele frequency threshold of 0.04. Where possible, coding (c) SNPs were included in the tag set. Illumina Goldengate SNPs with pre-optimized assays were chosen where possible, however roughly 10% of the chosen SNPs failed the assay design. A complete list of genes selected and the tSNPs for each gene for which genotype was available is shown in Supplementary Material, Tables S3 and S5. Genotypes from the Illumina array were supplemented with information on 173 SNPs in a further 82 genes previously typed in this data set (Supplementary Material, Tables S4 and S6). SNPs were examined for associations with 12 blood phenotypes available in NPHSII of which six were protein phenotypes (CRP, fibrinogen, apoAI, apoB, Lp-PLA2 and factor VII) and six were non-protein metabolic phenotypes (total-cholesterol, HDL- and calculated LDL-cholesterol, triglycerides, homocysteine and folate). We refer to this as a genic scan to distinguish the focused high SNP density, gene-centric approach used here from broader, generally lower SNP density, genome-wide analyses.
Phenome scan
We examined the effect of the genes containing the highest ranking SNPs from the genic scan on additional blood biomarkers beyond the index trait and the results are illustrated by means of a phenome plot (vide infra). These findings are summarized by means of phenome plots that summarize the associations of SNPs in a gene with blood marker(s) both in terms of the FDR q-value (vide infra) and proportion of the variance of the trait explained, using a stepwise regression to find the adjusted coefficient of determination (adjR2) under the model of best fit.
Statistical analysis
Phenotypes were tested for deviation from the normal and transformed where appropriate, using the Box-Cox transformation. Pair-wise linear relationships between blood phenotypes were tested using Pearson correlation. If the phenotypic variant was measured once in the study, an ANOVA was used to test for its association with genotype. Where multiple measurements were available, the average value of the available measurements was calculated and a general linear model was fitted through the data using indicator variables. The regression coefficients obtained for the indicator variables compared to the reference category were assessed for significance using a Wald test. Based on the number of hypotheses tested, a FDR adjusted P-value was calculated using the methodology of Benjamini and Hochberg (39). Defining FDR as the proportion of falsely rejected hypotheses, i.e. for which the null was actually true, this new P-value, known as the q-value (40), is the minimum FDR when rejecting a null hypothesis from a list of tested null hypothesis, conditioned on at least one positive finding having occurred. For the construction of the phenomic plots, a stepwise regression was used to find the adjusted coefficient of determination (adjR2) under the model of best fit. The number of independent loci associated with each phenotype was calculated using the LD-based result clumping procedure in PLINK (v1.05, http://pngu.mgh.harvard.edu/purcell/plink/ (41). An extremely conservative R2 of 0.1% was used to clump SNPs, identified previously as statistically significant, in independent loci.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at HMG online.
Conflict of Interest statement. None declared.
FUNDING
This work was supported by the British Heart Foundation (grant number PG2005/014; F.D., P.J.T., J.P. and S.E.H. and a Senior Fellowship to A.D.H., FS2005/125). Funding to pay the Open Access charge was provided by the British Heart Foundation.
Supplementary Material
REFERENCES
- 1.Rader D.J., Daugherty A. Translating molecular discoveries into new therapies for atherosclerosis. Nature. 2008;451:904–913. doi: 10.1038/nature06796. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins P.N., Williams R.R. A survey of 246 suggested coronary risk factors. Atherosclerosis. 1981;40:1–52. doi: 10.1016/0021-9150(81)90122-2. [DOI] [PubMed] [Google Scholar]
- 3.Brotman D.J., Walker E., Lauer M.S., O'Brien R.G. In search of fewer independent risk factors. Arch. Intern. Med. 2005;165:138–145. doi: 10.1001/archinte.165.2.138. [DOI] [PubMed] [Google Scholar]
- 4.Baigent C., Keech A., Kearney P.M., Blackwell L., Buck G., Pollicino C., Kirby A., Sourjina T., Peto R., Collins R., et al. Efficacy and safety of cholesterol-lowering treatment: prospective meta-analysis of data from 90,056 participants in 14 randomised trials of statins. Lancet. 2005;366:1267–1278. doi: 10.1016/S0140-6736(05)67394-1. [DOI] [PubMed] [Google Scholar]
- 5.Willer C.J., Sanna S., Jackson A.U., Scuteri A., Bonnycastle L.L., Clarke R., Heath S.C., Timpson N.J., Najjar S.S., Stringham H.M., et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wallace C., Newhouse S.J., Braund P., Zhang F., Tobin M., Falchi M., Ahmadi K., Dobson R.J., Marcano A.C., Hajat C., et al. Genome-wide association study identifies genes for biomarkers of cardiovascular disease: serum urate and dyslipidemia. Am. J. Hum. Genet. 2008;82:139–149. doi: 10.1016/j.ajhg.2007.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sandhu M.S., Waterworth D.M., Debenham S.L., Wheeler E., Papadakis K., Zhao J.H., Song K., Yuan X., Johnson T., Ashford A., et al. LDL-cholesterol concentrations: a genome-wide association study. Lancet. 2008;371:483–491. doi: 10.1016/S0140-6736(08)60208-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Melzer D., Perry J.R., Hernandez D., Corsi A.M., Stevens K., Rafferty I., Lauretani F., Murray A., Gibbs J.R., Paolisso G., et al. A genome-wide association study identifies protein quantitative trait loci (pQTLs) PLoS Genet. 2008;4:e1000072. doi: 10.1371/journal.pgen.1000072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hingorani A., Humphries S. Nature's randomised trials. Lancet. 2005;366:1906–1908. doi: 10.1016/S0140-6736(05)67767-7. [DOI] [PubMed] [Google Scholar]
- 10.Davey S.G., Timpson N., Ebrahim S. Strengthening causal inference in cardiovascular epidemiology through Mendelian randomization. Ann. Med. 2008;40:524–541. doi: 10.1080/07853890802010709. [DOI] [PubMed] [Google Scholar]
- 11.Morley M., Molony C.M., Weber T.M., Devlin J.L., Ewens K.G., Spielman R.S., Cheung V.G. Genetic analysis of genome-wide variation in human gene expression. Nature. 2004;430:743–747. doi: 10.1038/nature02797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheung V.G., Spielman R.S., Ewens K.G., Weber T.M., Morley M., Burdick J.T. Mapping determinants of human gene expression by regional and genome-wide association. Nature. 2005;437:1365–1369. doi: 10.1038/nature04244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stranger B.E., Forrest M.S., Dunning M., Ingle C.E., Beazley C., Thorne N., Redon R., Bird C.P., de Grassi A., Lee C., et al. Relative impact of nucleotide and copy number variation on gene expression phenotypes. Science. 2007;315:848–853. doi: 10.1126/science.1136678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Verzilli C., Shah T., Casas J.P., Chapman J., Sandhu M.S., Debenham S.L., Boekholdt S.M., Khaw K.T., Wareham N.J., Judson R., et al. Bayesian meta-analysis of genetic association studies with different sets of markers. Am. J. Hum. Genet. 2008;82:859–872. doi: 10.1016/j.ajhg.2008.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kathiresan S., Melander O., Guiducci C., Surti A., Burtt N.P., Rieder M.J., Cooper G.M., Roos C., Voight B.F., Havulinna A.S., et al. Six new loci associated with blood low-density lipoprotein cholesterol, high-density lipoprotein cholesterol or triglycerides in humans. Nat. Genet. 2008;40:189–197. doi: 10.1038/ng.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bennet A.M., Di Angelantonio E., Ye Z., Wensley F., Dahlin A., Ahlbom A., Keavney B., Collins R., Wiman B., de Faire U., et al. Association of apolipoprotein E genotypes with lipid levels and coronary risk. JAMA. 2007;298:1300–1311. doi: 10.1001/jama.298.11.1300. [DOI] [PubMed] [Google Scholar]
- 17.Ridker P.M., Pare G., Parker A., Zee R.Y., Danik J.S., Buring J.E., Kwiatkowski D., Cook N.R., Miletich J.P., Chasman D.I. Loci related to metabolic-syndrome pathways including LEPR, HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women's Genome Health Study. Am. J. Hum. Genet. 2008;82:1185–1192. doi: 10.1016/j.ajhg.2008.03.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ruchat S.M., Despres J.P., Weisnagel S.J., Chagnon Y.C., Bouchard C., Perusse L. Genome-wide linkage analysis for circulating levels of adipokines and C-reactive protein in the Quebec family study (QFS) J. Hum. Genet. 2008;53:629–636. doi: 10.1007/s10038-008-0291-1. [DOI] [PubMed] [Google Scholar]
- 19.Thompson A., Di Angelantonio E., Sarwar N., Erqou S., Saleheen D., Dullaart R.P., Keavney B., Ye Z., Danesh J. Association of cholesteryl ester transfer protein genotypes with CETP mass and activity, lipid levels, and coronary risk. JAMA. 2008;299:2777–2788. doi: 10.1001/jama.299.23.2777. [DOI] [PubMed] [Google Scholar]
- 20.Talmud P.J., Hawe E., Martin S., Olivier M., Miller G.J., Rubin E.M., Pennacchio L.A., Humphries S.E. Relative contribution of variation within the APOC3-A4-A5 gene cluster in determining plasma triglycerides. Hum. Mol. Gen. 2002;11:3039–3046. doi: 10.1093/hmg/11.24.3039. [DOI] [PubMed] [Google Scholar]
- 21.Gieger C., Geistlinger L., Altmaier E., Hrabé de Angelis M., Kronenberg F., Meitinger T., Mewes H.W., Wichmann H.E., Weinberger K.M., Adamski J., et al. Genetics meets metabolomics: a genome-wide association study of metabolite profiles in human serum. PLoS Genet. 2008;4:e1000282. doi: 10.1371/journal.pgen.1000282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Keating B.J., Tischfield S., Murray S.S., Bhangale T., Price T.S., Glessner J.T., Galver L., Barrett J.C., Grant S.F., Farlow D.N., et al. Concept, design and implementation of a cardiovascular gene-centric 50 k SNP array for large-scale genomic association studies. PLoS ONE. 2008;3:e3583. doi: 10.1371/journal.pone.0003583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Samani N.J., Erdmann J., Hall A.S., Hengstenberg C., Mangino M., Mayer B., Dixon R.J., Meitinger T., Braund P., Wichmann H.E., et al. Genomewide association analysis of coronary artery disease. N. Engl. J. Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kathiresan S., Melander O., Anevski D., Guiducci C., Burtt N.P., Roos C., Hirschhorn J.N., Berglund G., Hedblad B., Groop L., et al. Polymorphisms associated with cholesterol and risk of cardiovascular events. N. Engl. J. Med. 2008;358:1240–1249. doi: 10.1056/NEJMoa0706728. [DOI] [PubMed] [Google Scholar]
- 25.Bodmer W., Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jones R., Pembrey M., Golding J., Herrick D. The search for genenotype/phenotype associations and the phenome scan. Paediatr. Perinat. Epidemiol. 2005;19:264–275. doi: 10.1111/j.1365-3016.2005.00664.x. [DOI] [PubMed] [Google Scholar]
- 27.Chen L., Davey S.G., Harbord R.M., Lewis S.J. Alcohol intake and blood pressure: a systematic review implementing a Mendelian randomization approach. PLoS Med. 2008;5:e52. doi: 10.1371/journal.pmed.0050052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith G.D., Lawlor D.A., Harbord R., Timpson N., Day I., Ebrahim S. Clustered environments and randomized genes: a fundamental distinction between conventional and genetic epidemiology. PLoS Med. 2007;4:e352. doi: 10.1371/journal.pmed.0040352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Casas J.P., Shah T., Cooper J., Hawe E., Mcmahon A.D., Gaffney D., Packard C.J., O'Reilly D.S., Juhan-Vague I., Yudkin J.S., et al. Insight into the nature of the CRP-coronary event association using Mendelian randomization. Int. J. Epidemiol. 2006;35:922–931. doi: 10.1093/ije/dyl041. [DOI] [PubMed] [Google Scholar]
- 30.Davey S.G., Harbord R., Milton J., Ebrahim S., Sterne J.A. Does elevated plasma fibrinogen increase the risk of coronary heart disease? Evidence from a meta-analysis of genetic association studies. Arterioscler. Thromb. Vasc. Biol. 2005;25:2228–2233. doi: 10.1161/01.ATV.0000183937.65887.9c. [DOI] [PubMed] [Google Scholar]
- 31.Keavney B., Danesh J., Parish S., Palmer A., Clark S., Youngman L., Delepine M., Lathrop M., Peto R., Collins R. Fibrinogen and coronary heart disease: test of causality by ‘Mendelian randomization. Int. J. Epidemiol. 2006;35:935–943. doi: 10.1093/ije/dyl114. [DOI] [PubMed] [Google Scholar]
- 32.Davey S.G., Lawlor D.A., Harbord R., Timpson N., Rumley A., Lowe G.D., Day I.N., Ebrahim S. Association of C-reactive protein with blood pressure and hypertension: life course confounding and mendelian randomization tests of causality. Arterioscler. Thromb. Vasc. Biol. 2005;25:1051–1056. doi: 10.1161/01.ATV.0000160351.95181.d0. [DOI] [PubMed] [Google Scholar]
- 33.Humphries S.E., Cranston T., Allen M., Middleton-Price H., Fernandez M.C., Senior V., Hawe E., Iversen A., Wray R., Crook M.A., et al. Mutational analysis in UK patients with a clinical diagnosis of familial hypercholesterolaemia: relationship with plasma lipid traits, heart disease risk and utility in relative tracing. J. Mol. Med. 2006;84:203–214. doi: 10.1007/s00109-005-0019-z. [DOI] [PubMed] [Google Scholar]
- 34.Cohen J.C., Boerwinkle E., Mosley T.H., Jr, Hobbs H.H. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 2006;354:1264–1272. doi: 10.1056/NEJMoa054013. [DOI] [PubMed] [Google Scholar]
- 35.Linsel-Nitschke P., Gotz A., Erdmann J., Braenne I., Braund P., Hengstenberg C., Stark K., Fischer M., Schreiber S., El Mokhtari N.E., et al. Lifelong reduction of LDL-cholesterol related to a common variant in the LDL-receptor gene decreases the risk of coronary artery disease—a Mendelian Randomisation study. PLoS ONE. 2008;3:e2986. doi: 10.1371/journal.pone.0002986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rockman M.V. Reverse engineering the genotype-phenotype map with natural genetic variation. Nature. 2008;456:738–744. doi: 10.1038/nature07633. [DOI] [PubMed] [Google Scholar]
- 37.Cooper J.A., Miller G.J., Bauer K.A., Morrissey J.H., Meade T.W., Howarth D.J., Barzegar S., Mitchell J.P., Rosenberg R.D. Comparison of novel hemostatic factors and conventional risk factors for prediction of coronary heart disease. Circulation. 2000;102:2816–2822. doi: 10.1161/01.cir.102.23.2816. [DOI] [PubMed] [Google Scholar]
- 38.Redon R., Ishikawa S., Fitch K.R., Feuk L., Perry G.H., Andrews T.D., Fiegler H., Shapero M.H., Carson A.R., Chen W., et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Benjamini Y., Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. Journal of the Royal Statistical Society B 57: 289-300. J. Royal Stat. Soc. B. 1995;57:289–300. [Google Scholar]
- 40.Storey J.D. A direct approach to false discovery rates. J. Royal Stat. Soc. Ser. B. 2002;64:479–498. [Google Scholar]
- 41.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., Wetten S., Li L., St Jean P.L., Upmanyu R., Surh L., Hosford D., Barnes M.R., Briley J.D., Borrie M., et al. Candidate single-nucleotide polymorphisms from a genomewide association study of Alzheimer disease. Arch. Neurol. 2008;65:45–53. doi: 10.1001/archneurol.2007.3. [DOI] [PubMed] [Google Scholar]
- 43.Webster J.A., Myers A.J., Pearson J.V., Craig D.W., Hu-Lince D., Coon K.D., Zismann V.L., Beach T., Leung D., Bryden L., et al. Sorl1 as an Alzheimer's disease predisposition gene? Neurodegener. Dis. 2008;5:60–64. doi: 10.1159/000110789. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.