Abstract
G protein-coupled receptor kinase interacting protein 1 (GIT1) belongs to the family of Arf GAP proteins, and has been implicated in the regulation of G protein coupled receptor (GPCR) sequestration, cell migration, synapse formation and dendritic spine morphogenesis in neurons. To extend these cellular studies on GIT1 to an in vivo system, we generated mice with globally inactivated Git1 gene by breeding mice carrying a conditional Git1flox allele with mice expressing the CMV-Cre transgene. Although many GIT1 knockout (GIT1-KO) animals died shortly after birth, homozygous mutants that survived the early post-partum period developed normally into adulthood and were fertile. Behavioral analyses of adult GIT1-KO mice revealed normal exploratory, anxiety- and depressive-like behaviors. However, GIT1-KO mice show impaired responses to fear conditioning and fear-potentiated startle. Overall, these findings suggest that GIT1 is involved in the regulation of amygdala-mediated experience-based emotional behaviors.
Keywords: GIT1, GIT2, knockout mice, behavior, fear conditioning
GIT1 and GIT2 are structurally and functionally highly homologous, and comprise one subfamily of Arf GAP proteins [9]. GIT proteins have been shown to regulate internalization of activated GPCRs, and to promote focal adhesion turnover and cell migration [9]. GIT proteins homo- and heterodimerize and associate with PIX proteins to form high affinity oligomeric complexes [18]. PIX proteins are guanine nucleotide exchange factors (GEFs) for the Rho family members, Rac1 and Cdc42, and also bind to the Rac1/Cdc42 p21-activated kinases (PAKs) [15]. This GIT-PIX-PAK complex has been shown to regulate spino- and synaptogenesis in chicken retinal and mouse hippocampal neurons [8, 9, 12, 20]. In humans, the α-PIX (ARHGEF6) [13] and PAK3 [1] genes cause X-linked mental retardation, and the β-PIX gene (ARHGEF7) has been mapped to a genomic region associated with autosomal mental retardation [25]. The implication of GIT binding partners PIX and PAK in human mental retardation suggests a role for the GIT proteins in neurological disorders. Indeed, we recently demonstrated that mice lacking GIT2 display anxiety-like behaviors [23]. To identify in vivo functions of GIT1 in brain-mediated processes, we generated mouse lines bearing conditional or deleted alleles (knockout, GIT1-KO) of the Git1 gene using the Cre-loxP system and assessed emotional behaviors of GIT1-KO mice.
Mice harboring a conditional Git1 allele (Git1flox) were engineered using a triple-loxP targeting vector and Cre-mediated deletion (Figure 1A), as described [7]. Mice were genotyped using the following oligonucleotide primers: wt allele sense (F1) primer 5′-GATCTGCCGTGTAGCCTTTG-3′, del allele sense (F2) primer 5′-CCCAGAATGTCAAGCTGGTT-3′, and common antisense (R1) primer 5′-CTGCATAGGCTGTGATGGAA-3′. For Western Blot analysis, 20 μg protein lysates from brain tissues were resolved on a 10% PAGE gel and GIT1 and GIT2 detected with the crossreacting p95-PKL monoclonal antibody (1:1000, BD Biosciences). As a loading control, β-actin was detected with a mouse monoclonal antibody (1:1000, Santa Cruz Biotechnologies, Santa Cruz, CA). For immunostaining, NeuN was detected using a mouse monoclonal antibody (1:1000, Chemicon) as described previously [23].
Fig. 1. Generation of GIT1-deficient mice.
(A) Murine Git1 targeting vector. Protein-encoding exons are indicated by solid boxes. The lox-P sites are depicted by filled shaded triangles, and the thymidine kinase (HSV-TK), neomycin (PKG-Neo), and diphtheria toxin (PKG-DT) selection markers are indicated. Rearrangements following Cre recombinase treatment are shown as Git1flox for the conditional allele and Git1del for the knockout allele. F1, F2 and R1 indicate the location and orientation of genotyping primers. (B) Results from genotyping of GIT1 mice by genomic PCR. (C) Confirmation of GIT1 protein deficiency in cerebellar lysates from GIT1-KO animals by Western blot. (D) Morphology of WT and GIT1-KO brains assessed by neuronal staining using NeuN antibody.
Animals were housed and treated as described previously [23]. A total of 10 WT (8 males, 2 females) and 8 GIT1 KO (7 males, 1 female) were used for all behavioral experiments. Behavioral testing was conducted during the light phase in the following order: zero-maze, light-dark emergence test, tail suspension, open field testing, fear conditioning, and fear potentiated startle. All testing was completed within 11 weeks, with the average age at testing as follows; elevated zero maze, 22.5 ±1.4wks; light dark emergence test, 23.4±1.4wks; tail suspension, 24.8±1.4wks; open field 28.0±1.4wks; fear conditioning, 31±1.4wks, and fear potentiated startle 33.4±1.4wks. For assessment of locomotor activity, mice were given 60 min to freely explore the open field (VersaMax activity monitoring system, AccuScan Instruments Inc.) illuminated at ~340 lux. For the assessment of anxiety-like behaviors, mice were tested for 5 min in both the elevated zero maze and light-dark emergence test. Open areas of the zero maze were illuminated at ~50 lux, whereas the light-chamber during light-dark emergence testing was illuminated at ~870 lux. Immobility in the tail suspension test was assessed over a 6 min period. All procedures have recently been described in detail [23].
Fear potentiated startle testing was conducted in a Med-Associates (St. Albans, VT) apparatus as outlined [19]. Testing occurred over 4 days, with baseline startle responses obtained on day one, pre-conditioning potentiation to a 30-sec 12 kHz 70 dB conditioning tone (CS) on day 2, the pairing of 0.4 mA foot-shock (UCS) with the CS on day 3, and an assessment of fear-potentiated startle to the CS on day 4.
For fear conditioning, training and testing were conducted in a mouse fear conditioning apparatus (Med-Associates, St. Albans, VT) as noted [19]. For conditioning, mice were allowed to freely explore the apparatus for 2 min, after which the CS (72 dB 29 kHz tone) was presented for 30 sec and it was terminated with a 2 sec 0.4 mA scrambled foot-shock. Each mouse remained in the chamber for an additional 30 sec before being placed into its home cage. Twenty-four hours later, animals were returned to the conditioning chamber and examined in context testing for 5 min, where no CS or UCS was presented. Cued testing was conducted 24 hrs later, with the mouse placed into a novel chamber that was visually and tactilely distinct from that used for conditioning on day 1. After 2 min, the CS was presented continuously for 3 min but no UCS was given. Behavior was videotaped across all conditioning and testing; time spent freezing was scored with the Noldus Observer (Noldus Information Technologies, Leesburg, VA) by a trained observer. Freezing was defined as the absence of movement other than respiration.
Results are presented as means and standard errors of the mean. For tests where behaviors were serially scored for the same animal, repeated measures ANOVA (RMANOVA) was used. Time spent in immobility in tail suspension or in freezing for fear conditioning was used as the within subjects effect. For all RMANOVA, genotype was the between subjects effect. Independent measures t-test was used for WT and GIT1-KO comparisons for zero maze behaviors and total immobility time in tail suspension. Bonferroni corrected pair-wise comparisons were used for post-hoc assessments. In all cases a p<0.05 was considered significant.
Intercross breeding between mice heterozygous for the Git1del allele produced mice homozygous for the Git1del allele, GIT1-KO, as assessed by PCR genotyping (Figure 1B). Western blot analysis revealed that the GIT1-KO animals lacked GIT1 protein expression (Figure 1C), and neural staining with the NeuN antibody showed that GIT1-KO had normal overall brain morphology (Figure 1D). Although viable GIT1-KO mice were obtained from heterozygote crosses, the number of GIT1-KO animals surviving to adulthood was low (data not shown). GIT1-KO mice had similar weight compared to WT littermates at birth but were of slightly lower weight at weaning and as adults (data not shown). In comparison, GIT2-KO mice had a normal genotype distribution at birth and in adulthood (data not shown). Surviving GIT1-KO animals did not show any gross anatomical abnormalities and were fertile (data not shown).
In the initial assessment of spontaneous exploration in the open field, behaviors were not distinguished by genotype either during the first 5 min (activity; WT: 195.0 ±15.6, GIT1-KO: 223.1 ±32.1 cm; thigmotaxis WT: 106.6 ±13.7, GIT1-KO: 115.7 ±26.6 cm; center time WT: 153.1 ±15.7, GIT1-KO: 144.7 ±10.8 sec) or over the first 30 min of testing (activity; WT: 774.2 ±70.2, GIT1-KO: 856.7 ±141.1 cm; thigmotaxis WT: 418.6 ±47.8, GIT1-KO: 435.0 ±62.1; center time WT: 292.5 ±46.9, GIT1-KO: 228.8 ±56.1 sec; center activity WT: 355.6 ±35.8, GIT1-KO: 421.7 ±70.7). These results suggested normal exploratory and anxiety-like behaviors of GIT1-KO mice in a novel environment during the critical first 5 min, when anxiety is highest.
Mice were next subjected to the zero maze and light-dark emergence tests (Figure 2). In the zero-maze, GIT1-KO animals did not differ from WT controls according to time spent in open areas (Figure 2A), numbers of open area transitions (Figure 2B), average time spent for each visit to the open areas (Figure 2C), numbers of head-dips (Figure 2D, inset) or stretch-attend postures (Figure 2D), or percent time freezing (WT = 0.1 ±0.1, GIT1-KO = 0.5 ±0.3). By contrast, GIT1-KO animals showed reduced latencies to enter the open areas (Figure 2A, inset) and reduced latencies for transitions to any part of the maze (Figure 2B, inset).
Fig 2. Anxiety-like behaviors of WT and GIT1-KO mice are similar in the zero maze and light-dark emergence tests.
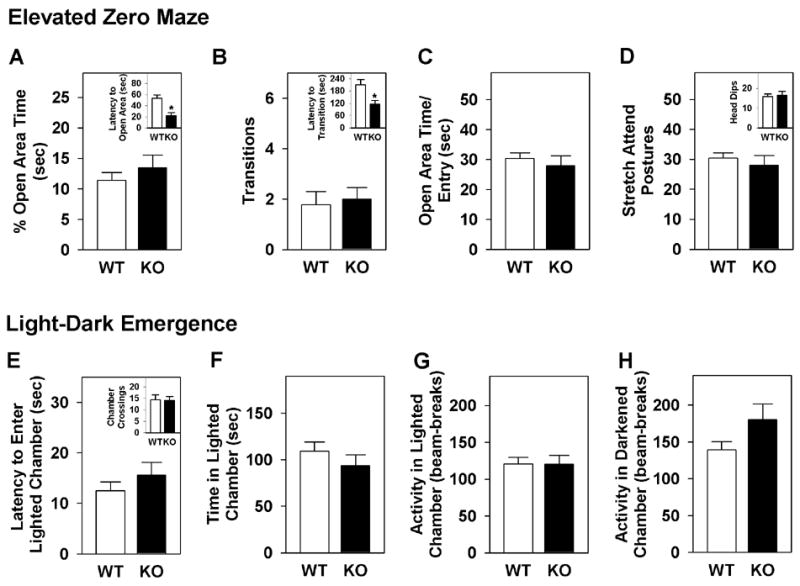
(A) Percent time in the open areas of the maze, and latency to enter an open arm (inset). (B) Numbers of closed-to-open-to-closed area transitions, and latency to transition (inset). (C) Average visit time to the open areas. (D) Numbers of stretch-attend postures, and numbers of head-dips over the edge of the maze (inset). (E) Latency to first enter the lighted chamber, and numbers of chamber crossings (inset). (F) Time spent in the lighted chamber. (G) Activity (beam-breaks) in the lighted chamber. (H) Activity (beam-breaks) in the darkened chamber. n=10 (WT), n=8 (KO). *p<0.05, WT versus KO mice.
In the light-dark emergence test, no genotype differences were noted between GIT1-KO mice and WT controls in the latency to enter the lighted chamber (Figure 2E), time spent (Figure 2F) or activity (Figure 2G) there, or in the darkened chamber (Figure 2H), or in the number of crossings between the lighted and darkened chambers (Fig. 2E, inset). Taken together, these findings show that GIT1-KO animals do not display anxiety-like behaviors in the open field, zero maze, or light-dark box.
Since GIT2-KO mice exhibited anti-depressive-like behaviors [23], the GIT1-KO mice were evaluated in the tail suspension test [6]. As in tests for anxiety-like responses, no genotype differences were observed between GIT1-KO and WT mice (Figure 3). This result suggests that GIT1-KO mice do not present with depressive-like behavior.
Fig 3. Absence of depressive-like responses of GIT1-KO mice in the tail suspension test.

(A) Immobility times for each minute of the 6 min test. (B) Cumulative immobility times. n=10 (WT), n=8 (KO).
Extending our investigations to other emotional responses, the mice were assessed in tests of fear-associated learning. The mice were first tested in fear conditioning. During fear conditioning on day 1, freezing behaviors of GIT1-KO mice did not differ from WT controls (Figure 4A). When conditioned animals were tested for contextual fear on day 2, freezing behavior was low in GIT1-KO mice over the first 4 min, but reached levels of the WT animals by the end of testing (Figure 4B). When tested for cued fear on day 3, freezing was not distinguished by genotype prior to presentation of the CS (Figure 4C). Upon CS presentation, freezing was greatly augmented in WT mice, whereas only a weak and transient response was seen in the GIT1-KO mice.
Fig 4. GIT1-KO mice are deficient in contextual and cued fear conditioning.
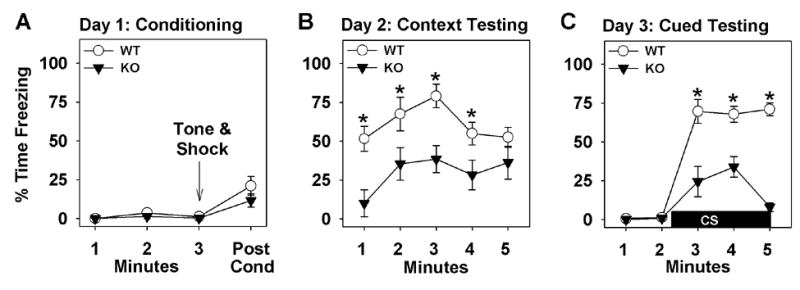
(A) Day 1, fear conditioning. (B) Day 2, context testing. (C) Day 3, cued testing. n=10 (WT), n=8 (KO); *p<0.05, WT versus KO mice.
The deficiency in both contextual and cued fear conditioning suggests that amygdala function may be abnormal in the homozygous mutants. To examine this idea further, mice were tested for fear-potentiated startle. Baseline startle responses of GIT1 mice on day 1 were not distinguished by genotype (data not shown). On day 2, prior to conditioning, potentiation to the 70 dB tone did not differ between WT and GIT1-KO animals (Figure 5A). Subsequently, on day 4, WT mice exhibited strong fear potentiation in response to the startle stimulus, whereas no change in potentiation was observed in GIT1-KO animals (Figure 5B).
Fig 5. Impaired responses of GIT1-KO mice in fear-potentiated startle.

(A) Day 2, pre-conditioning to the 70 dB CS. (B) Day 4, testing for fear-potentiated startle. n=10 (WT), n=8 (KO); *p<0.05, WT versus KO mice.
The data presented herein describe the first observations of in vivo neurological functions for GIT1. While GIT1-KO mice are susceptible to early postnatal death, as also noted recently in a distinct GIT1-KO mouse line [16], survivors develop normally and show normal exploratory, anxiety- and depressive-like behaviors. However, when tested for associative learning in two distinct paradigms for conditioned fear, GIT1-KO mice exhibit impaired responses. The deficiency in contextual and cued fear conditioning and in fear-potentiated startle suggest that extra hippocampal and/or amygdala functions are aberrant in GIT1-KO animals [4, 10]. Associative learning of CS and UCS and the formation of fear memory during fear conditioning may be mediated by long-term potentiation (LTP) [24]. In turn, LTP is facilitated by an increase in synaptic strength triggered by activation and recruitment of NMDA and AMPA receptors [14] and changes in synapse morphology [3]. Previous reports have identified β-PIX as part of a signaling cascade facilitating NMDA receptor stimulated spine morphogenesis [20] and GIT1 in AMPA receptor translocation to synapses [11]. Since GIT and PIX proteins form a tight complex [18], the GIT1/PIX complex may play a central role in orchestrating NMDA/AMPA activation and LTP formation important for proper acquisition and expression of fear memories. Considering the broad expression of GIT and PIX proteins in the brain and the linkage of the PIX (ARHGEF6, ARHGEF7) and PAK3 genes to mental retardation, deficits in learning and memory formation may be expected in GIT1-deficient mice.
Our results here with GIT1-KO mice and previously with GIT2-KO mice [23] indicate that these two proteins play different functional roles in vivo. While GIT1-KO mice did not display anxiety-like behaviors, GIT2-KO mice showed prominent anxiety-like behaviors [23]. Biochemical assessments of GIT1 and GIT2 functions have generally found these two proteins to function in nearly identical manners. Both act as PIP3-stimulated GTPase-activating proteins for all Arf GTP-binding proteins, can homodimerize and heterodimerize, and can bind tightly to PIX proteins to form oligomeric GIT/PIX complexes [9]. Several binding partners appear to be shared [9, 17], although many GIT1 partners remain untested for interaction with GIT2. Given the very similar patterns of GIT1 and GIT2 expression throughout the brain [22], functional redundancy of the two GIT proteins might be expected. To date, the only hints for functional differences between the two GIT proteins come from studies of focal adhesion dynamics [2, 5, 9, 21]. Whether these behavioral differences result from distinct functions for GIT1 and GIT2 in the same neurons, or from relative expression levels or expression in distinct neuron populations, remains to be clarified.
In conclusion, the behavioral deficits in conditioned fear observed in GIT1-KO mice suggest an important and unique role for GIT1 function in aversive emotional learning and memory.
Acknowledgments
The authors would like to thank Liping Du and Jiechun Zhou for assistance in behavioral testing, Dr. Jonathan C. Poe (Department of Immunology, Duke University Medical Center) for providing the CMV-Cre transgenic mice, Cheryl Bock and the Duke Comprehensive Cancer Center Transgenic Core Facility for helping with the creation of the GIT1-KO mice. Supported by NIH grants GM59989 and DA016347, and the American Heart Association Grant-in-Aid 0655464U to RTP, and an American Psychological Association Diversity Fellowship to RMR.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Allen KM, Gleeson JG, Bagrodia S, Partington MW, MacMillan JC, Cerione RA, Mulley JC, Walsh CA. PAK3 mutation in nonsyndromic X-linked mental retardation. Nat Genet. 1998;20:25–30. doi: 10.1038/1675. [DOI] [PubMed] [Google Scholar]
- 2.Brown MC, Cary LA, Jamieson JS, Cooper JA, Turner CE. Src and FAK Kinases Cooperate to Phosphorylate Paxillin Kinase Linker, Stimulate Its Focal Adhesion Localization, and Regulate Cell Spreading and Protrusiveness. Mol Biol Cell. 2005;16:4316–28. doi: 10.1091/mbc.E05-02-0131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen LY, Rex CS, Casale MS, Gall CM, Lynch G. Changes in synaptic morphology accompany actin signaling during LTP. J Neurosci. 2007;27:5363–72. doi: 10.1523/JNEUROSCI.0164-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Fendt M, Fanselow MS. The neuroanatomical and neurochemical basis of conditioned fear. Neurosci Biobehav Rev. 1999;23:743–60. doi: 10.1016/s0149-7634(99)00016-0. [DOI] [PubMed] [Google Scholar]
- 5.Frank SR, Adelstein MR, Hansen SH. GIT2 represses Crk- and Rac1-regulated cell spreading and Cdc42-mediated focal adhesion turnover. Embo J. 2006;25:1848–59. doi: 10.1038/sj.emboj.7601092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fukui M, Rodriguiz RM, Zhou J, Jiang SX, Phillips LE, Caron MG, Wetsel WC. Vmat2 heterozygous mutant mice display a depressive-like phenotype. J Neurosci. 2007;27:10520–9. doi: 10.1523/JNEUROSCI.4388-06.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gainetdinov RR, Bohn LM, Walker JK, Laporte SA, Macrae AD, Caron MG, Lefkowitz RJ, Premont RT. Muscarinic supersensitivity and impaired receptor desensitization in G protein-coupled receptor kinase 5-deficient mice. Neuron. 1999;24:1029–36. doi: 10.1016/s0896-6273(00)81048-x. [DOI] [PubMed] [Google Scholar]
- 8.Hayashi K, Ohshima T, Hashimoto M, Mikoshiba K. Pak1 regulates dendritic branching and spine formation. Dev Neurobiol. 2007;67:655–69. doi: 10.1002/dneu.20363. [DOI] [PubMed] [Google Scholar]
- 9.Hoefen RJ, Berk BC. The multifunctional GIT family of proteins. J Cell Sci. 2006;119:1469–75. doi: 10.1242/jcs.02925. [DOI] [PubMed] [Google Scholar]
- 10.Kim M, Davis M. Electrolytic lesions of the amygdala block acquisition and expression of fear-potentiated startle even with extensive training but do not prevent reacquisition. Behav Neurosci. 1993;107:580–95. doi: 10.1037//0735-7044.107.4.580. [DOI] [PubMed] [Google Scholar]
- 11.Ko J, Kim S, Valtschanoff JG, Shin H, Lee JR, Sheng M, Premont RT, Weinberg RJ, Kim E. Interaction between liprin-alpha and GIT1 is required for AMPA receptor targeting. J Neurosci. 2003;23:1667–77. doi: 10.1523/JNEUROSCI.23-05-01667.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kreis P, Thevenot E, Rousseau V, Boda B, Muller D, Barnier JV. The p21-activated kinase 3 implicated in mental retardation regulates spine morphogenesis through a Cdc42-dependent pathway. J Biol Chem. 2007;282:21497–506. doi: 10.1074/jbc.M703298200. [DOI] [PubMed] [Google Scholar]
- 13.Kutsche K, Yntema H, Brandt A, Jantke I, Nothwang HG, Orth U, Boavida MG, David D, Chelly J, Fryns JP, Moraine C, Ropers HH, Hamel BC, van Bokhoven H, Gal A. Mutations in ARHGEF6, encoding a guanine nucleotide exchange factor for Rho GTPases, in patients with X-linked mental retardation. Nat Genet. 2000;26:247–50. doi: 10.1038/80002. [DOI] [PubMed] [Google Scholar]
- 14.Malenka RC, Bear MF. LTP and LTD: an embarrassment of riches. Neuron. 2004;44:5–21. doi: 10.1016/j.neuron.2004.09.012. [DOI] [PubMed] [Google Scholar]
- 15.Manser E, Loo TH, Koh CG, Zhao ZS, Chen XQ, Tan L, Tan I, Leung T, Lim L. PAK kinases are directly coupled to the PIX family of nucleotide exchange factors. Mol Cell. 1998;1:183–92. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 16.Pang J, Hoefen R, Pryhuber GS, Wang J, Yin G, White RJ, Xu X, O’Dell MR, Mohan A, Michaloski H, Massett MP, Yan C, Berk BC. G-protein-coupled receptor kinase interacting protein-1 is required for pulmonary vascular development. Circulation. 2009;119:1524–32. doi: 10.1161/CIRCULATIONAHA.108.823997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Premont RT, Claing A, Vitale N, Perry SJ, Lefkowitz RJ. The GIT family of ADP-ribosylation factor GTPase-activating proteins. Functional diversity of GIT2 through alternative splicing. J Biol Chem. 2000;275:22373–80. doi: 10.1074/jbc.275.29.22373. [DOI] [PubMed] [Google Scholar]
- 18.Premont RT, Perry SJ, Schmalzigaug R, Roseman JT, Xing Y, Claing A. The GIT/PIX complex: an oligomeric assembly of GIT family ARF GTPase-activating proteins and PIX family Rac1/Cdc42 guanine nucleotide exchange factors. Cell Signal. 2004;16:1001–11. doi: 10.1016/j.cellsig.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 19.Rodriguiz RM, Wetsel WC. Assessments of cognitive deficits in mutant mice. In: Levin ED, Buccafusco JJ, editors. Animal Models of Cognitive Impairment. CRC Press; Boca Raton, FL: 2006. pp. 223–282. [PubMed] [Google Scholar]
- 20.Saneyoshi T, Wayman G, Fortin D, Davare M, Hoshi N, Nozaki N, Natsume T, Soderling TR. Activity-Dependent Synaptogenesis: Regulation by a CaM-Kinase Kinase/CaM-Kinase I/betaPIX Signaling Complex. Neuron. 2008;57:94–107. doi: 10.1016/j.neuron.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmalzigaug R, Garron ML, Roseman JT, Xing Y, Davidson CE, Arold ST, Premont RT. GIT1 utilizes a focal adhesion targeting-homology domain to bind paxillin. Cell Signal. 2007;19:1733–44. doi: 10.1016/j.cellsig.2007.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmalzigaug R, Phee H, Davidson CE, Weiss A, Premont RT. Differential expression of the ARF GAP genes GIT1 and GIT2 in mouse tissues. J Histochem Cytochem. 2007;55:1039–48. doi: 10.1369/jhc.7A7207.2007. [DOI] [PubMed] [Google Scholar]
- 23.Schmalzigaug R, Rodriguiz RM, Phillips LE, Davidson CE, Wetsel WC, Premont RT. Anxiety-like behaviors in mice lacking GIT2. Neurosci Lett. 2009;451:156–61. doi: 10.1016/j.neulet.2008.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sigurdsson T, Doyere V, Cain CK, LeDoux JE. Long-term potentiation in the amygdala: a cellular mechanism of fear learning and memory. Neuropharmacology. 2007;52:215–27. doi: 10.1016/j.neuropharm.2006.06.022. [DOI] [PubMed] [Google Scholar]
- 25.Walczak-Sztulpa J, Wisniewska M, Latos-Bielenska A, Linne M, Kelbova C, Belitz B, Pfeiffer L, Kalscheuer V, Erdogan F, Kuss AW, Ropers HH, Ullmann R, Tzschach A. Chromosome deletions in 13q33–34: Report of four patients and review of the literature. Am J Med Genet A. 2008;146:337–42. doi: 10.1002/ajmg.a.32127. [DOI] [PubMed] [Google Scholar]



