SUMMARY
Endothelial cells play essential roles in maintenance of vascular integrity, angiogenesis and wound repair. We show that an endothelial cell-restricted microRNA (miR-126) mediates developmental angiogenesis in vivo. Targeted deletion of miR-126 in mice causes leaky vessels, hemorrhaging, and partial embryonic lethality, due to a loss of vascular integrity and defects in endothelial cell proliferation, migration and angiogenesis. The subset of mutant animals that survives displays defective cardiac neovascularization following myocardial infarction. The vascular abnormalities of miR-126 mutant mice resemble the consequences of diminished signaling by angiogenic growth factors, such as VEGF and FGF. Accordingly, miR-126 enhances the pro-angiogenic actions of VEGF and FGF and promotes blood vessel formation by repressing the expression of Spred-1, an intracellular inhibitor of angiogenic signaling. These findings have important therapeutic implications for a variety of disorders involving abnormal angiogenesis and vascular leakage.
INTRODUCTION
Endothelial cells (ECs) line the internal surfaces of vascular structures and play essential roles in vascular development, function and disease (Carmeliet, 2003). During blood vessel formation, known as vasculogenesis, ECs proliferate, migrate, and associate to form a primitive vascular labyrinth that serves as a scaffold for recruitment of smooth muscle cells. Subsequent sprouting of vessels, through angiogenesis, allows for further expansion of the vascular system and tissue vascularization.
Numerous peptide growth factors promote angiogenesis by enhancing EC migration, proliferation, survival and cell-cell interactions. VEGF and FGF, the most potent angiogenic growth factors, are required for neoangiogenesis during embryogenesis and adulthood (Cross and Claesson-Welsh, 2001). Binding of these factors to their cell surface receptors activates the MAP kinase pathway, which promotes angiogenic growth and maturation. Conversely, inhibition of MAP kinase signaling diminishes angiogenesis (Eliceiri et al., 1998; Giroux et al., 1999; Hood et al., 2002), and has been advanced as an anti-angiogenic therapy (Hood et al., 2002; Panka et al., 2006).
Recent studies have revealed important roles for microRNAs in the response of the cardiovascular system to injury and stress (Latronico et al., 2007; Van Rooij and Olson, 2007). miRNAs represent a class of ~22 nucleotide noncoding RNAs that regulate gene expression by targeting mRNAs for cleavage or translational repression (Bartel, 2004). More than 500 miRNAs have been identified in humans and other eukaryotic species, with about a third encoded by introns of protein coding genes. miRNAs are initially transcribed as large pri-miRNAs that are processed through sequential steps to give rise to a heteroduplex RNA. The miRNA strand of the heteroduplex becomes incorporated into the RNA-induced silencing complex (RISC), where it is enabled to target specific mRNAs through complementary sequences in 3’ untranslated regions (He and Hannon, 2004). The opposite strand of the heteroduplex, known as the star (*) strand, is generally degraded.
Here we show that an endothelial cell-specific miRNA, miR-126, modulates angiogenesis in vivo. Targeted deletion of miR-126 in mice results in vascular leakage, hemorrhaging, and embryonic lethality in a subset of mutant mice. These vascular abnormalities can be attributed to diminished angiogenic growth factor signaling, resulting in reduced EC growth, sprouting and adhesion. The subset of mutant animals that survives is prone to cardiac rupture and lethality following myocardial infarction with defective vascularization of the infarct. The pro-angiogenic actions of miR-126 correlate with its repression of Spred-1, a negative regulator of MAP kinase signaling. Thus, in the absence of miR-126, increased expression of Spred-1 diminishes the transmission of intracellular angiogenic signals by VEGF and FGF. We conclude that miR-126 functions as an endothelial cell-specific regulator of angiogenic signaling.
RESULTS
Endothelial-specific Expression of miR-126
In light of recent studies implicating miRNAs in cardiovascular development and disease, we searched publicly available databases for miRNAs that appeared to be restricted to cardiovascular tissues. Among several such miRNAs, miR-126 appeared enriched in tissues with a high vascular component, such as heart and lung (Lagos-Quintana et al., 2002). A survey of miRNA expression patterns in zebrafish also showed miR-126 to be specific for the vascular system. (Wienholds et al., 2005).
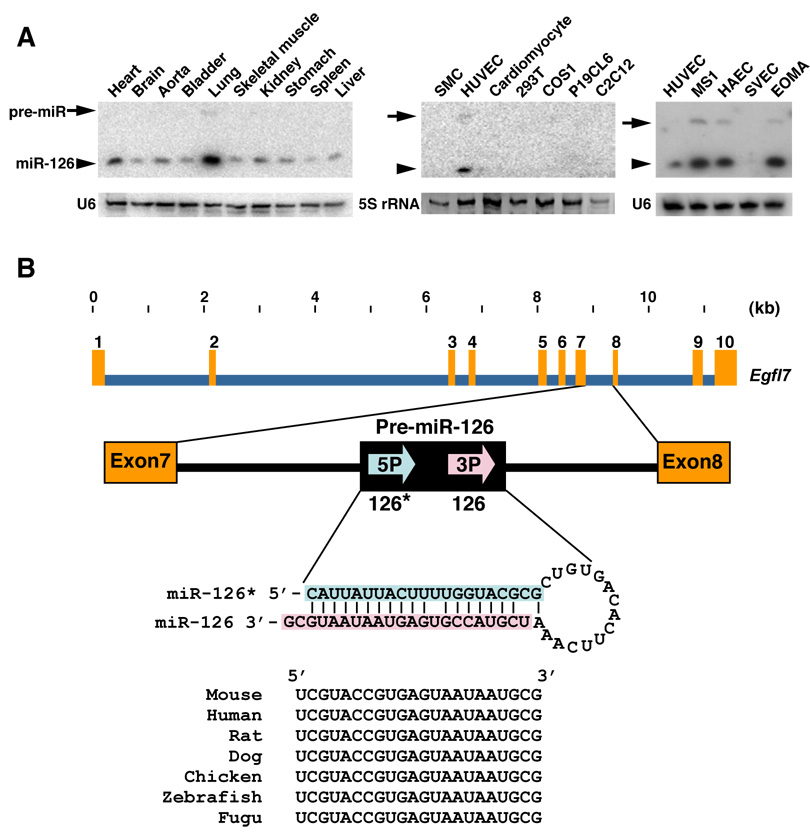
Northern blot analysis showed miR-126 to be expressed in a broad range of tissues, with highest expression in lung and heart (Fig. 1A), consistent with prior studies (Harris et al., 2008; Lagos-Quintana et al., 2002; Musiyenko et al., 2008). miR-126* was detectable at only trace levels of expression (data not shown). A survey of cell lines revealed miR-126 to be expressed in primary human umbilical vein ECs (HUVECs) and in numerous EC cell lines, including the MS1, HAEC and EOMA cell lines, but not in SV40 transformed ECs (SVECs) or non-endothelial cell types (Fig. 1A).
Figure 1. Endothelial cell-specific expression and gene structure of miR-126.
(A) Expression of miR-126 in different tissues and cell lines, as detected by Northern blot. U6 or 5S rRNA serve as a loading control. Arrows indicate the position of pre-miR-126, and arrowheads indicate the mature miR-126. SMC: mouse smooth muscle cell line; HUVEC: human umbilical vein endothelial cell (EC) line; P19CL6: derivative of P19 embryonic carcinoma cells; C2C12: mouse myoblast cell line. MS1: mouse primary islet ECs transformed with SV40 large T antigen; HAEC: human aortic ECs; SVEC: SV40 transformed mouse EC line; and EOMA: mouse hemangioendothelioma-derived line.
(B) Structure of the mouse Egfl7 gene. miR-126 (miR-126-3p) and miR-126* (miR-126-5p) are generated as a stem loop encoded by intron 7. Evolutionary conservation of miR-126 is shown.
miR-126 (also referred to as miR-126-3p) and miR-126* (miR-126-5p) are conserved from Fugu to homo sapiens (http://microrna.sanger.ac.uk/sequences/index.shtml). In mammals and birds, miR-126 and -126* are encoded by intron 7 of the EGF-like domain 7 (Egfl7) gene (Fig. 1B), which encodes an EC-specific secreted peptide that has been reported to act as a chemoattractant and inhibitor of smooth muscle cell migration (Campagnolo et al., 2005; Fitch et al., 2004; Parker et al., 2004; Soncin et al., 2003). The expression pattern of miR-126 in tissues and cell lines parallels that of Egfl7 (Fitch et al., 2004; Soncin et al., 2003), consistent with the conclusion that the miRNA is processed from intronic RNA sequence of the pre-Egfl7 mRNA. RT-PCR using cDNA from human placenta with primers upstream and downstream of intron 7 of Egfl7 showed that miR-126 was generated from a subset of Egfl7 transcripts in which intron 7 was retained (unpublished results). In situ hybridization with mouse embryo sections using a portion of intron 7 encompassing pri-miR-126 as a probe revealed EC-specific expression of miR-126 from E7.5 to adulthood (Supplemental Fig. 1), similar to Egfl7.
EC-specific Transcription of Egfl7/miR-126
To further visualize the expression pattern of miR-126 in vivo, we cloned 5.4 kb of genomic DNA immediately 5’ of the Egfl7/miR-126 gene into a lacZ reporter gene and generated transgenic mice. This DNA fragment was sufficient to direct expression specifically in ECs throughout embryogenesis and in adult tissues (Fig. 2A and data not shown).
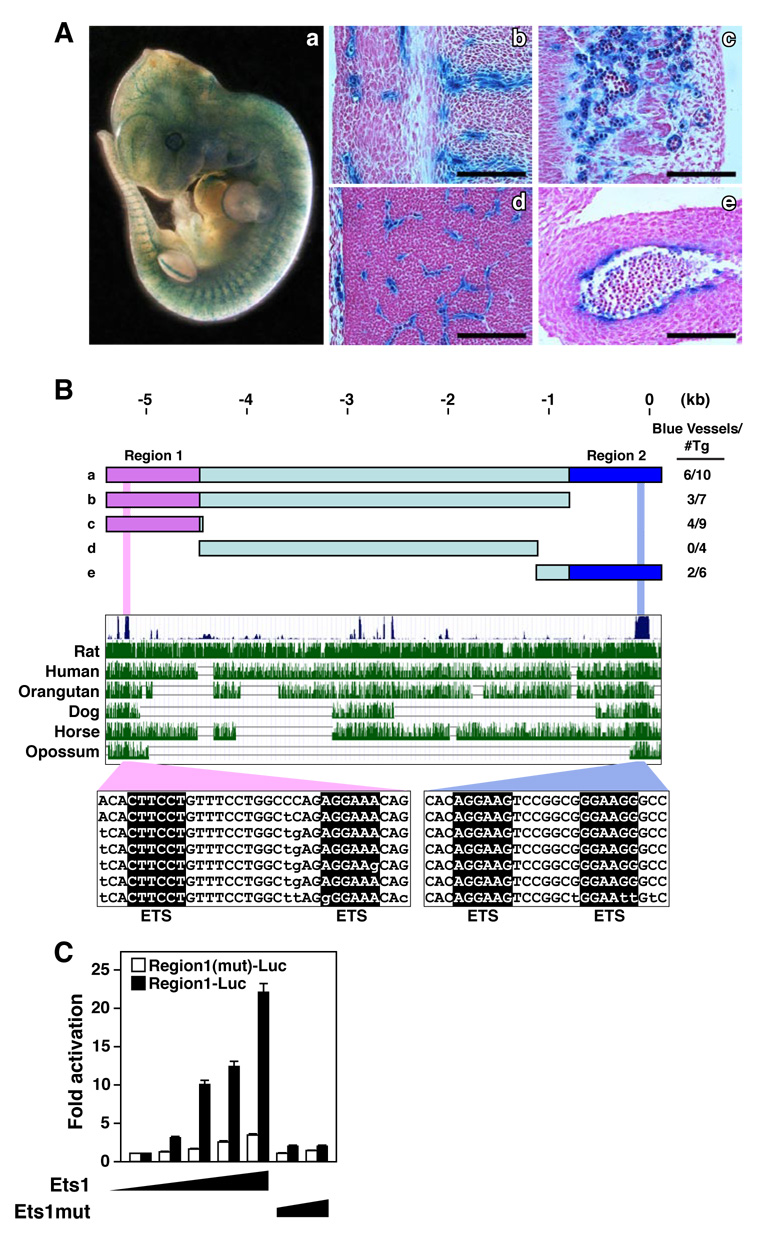
Figure 2. Cis-regulatory sequences that direct endothelial-specific expression of Egfl7/miR-126.
(A) E12.5 transgenic mouse embryo harboring a lacZ transgene controlled by 5.4kb 5’ flanking DNA upstream of the Egfl7/miR-126 gene. EC specific lacZ expression showing a) whole mount embryo, and sagittal section in b) perichondral region, c) dermis, d) brain, and e) outflow tract. Scale bar equals 50um.
(B) Schematic diagrams of genomic regions upstream of the Egfl7/miR-126 gene tested for regulation of lacZ in transgenic mice at E12.5. The fraction of transgenic embryos showing endothelial-specific expression of lacZ is shown. Evolutionary conservation of the Egfl7/miR-126 5’ flanking region is shown below. The conserved ETS binding sites are highlighted in pink and light blue.
(C) Genomic fragment for regulatory region 1(Region1-Luc) was tested for activation by increasing amounts of an expression plasmid encoding Ets1 or Ets1 mutant lacking the DNA binding domain (Ets1mut) in COS-7 cells. A deletion mutation was introduced into the ETS binding site (Region1(mut)-Luc). Ets1 activated Region1-Luc but not Region1(mut)-Luc, while Ets1mut failed to activate either construct.
The 5.4 kb DNA fragment contained two regions (Region 1 and Region 2) of high evolutionary conservation, each of which was sufficient to direct endothelial-specific expression in vivo (Fig. 2B). Both regions contained conserved consensus sequences for binding of Ets transcription factors, which have been implicated in endothelial-specific transcription (Lelievre et al., 2001). Ets1 potently transactivated these regulatory regions in transfected COS-7 cells, whereas an Ets1 mutant lacking the DNA binding domain was devoid of activity. Moreover, mutations in the Ets sites blunted transcriptional activation by Ets1 (Fig. 2C) and abolished expression of the lacZ transgene in ECs in vivo (data not shown). These findings suggest that Ets transcription factors are sufficient and necessary for endothelial-specific transcription of Egfl7/miR-126.
Creation of miR-126 Null Mice
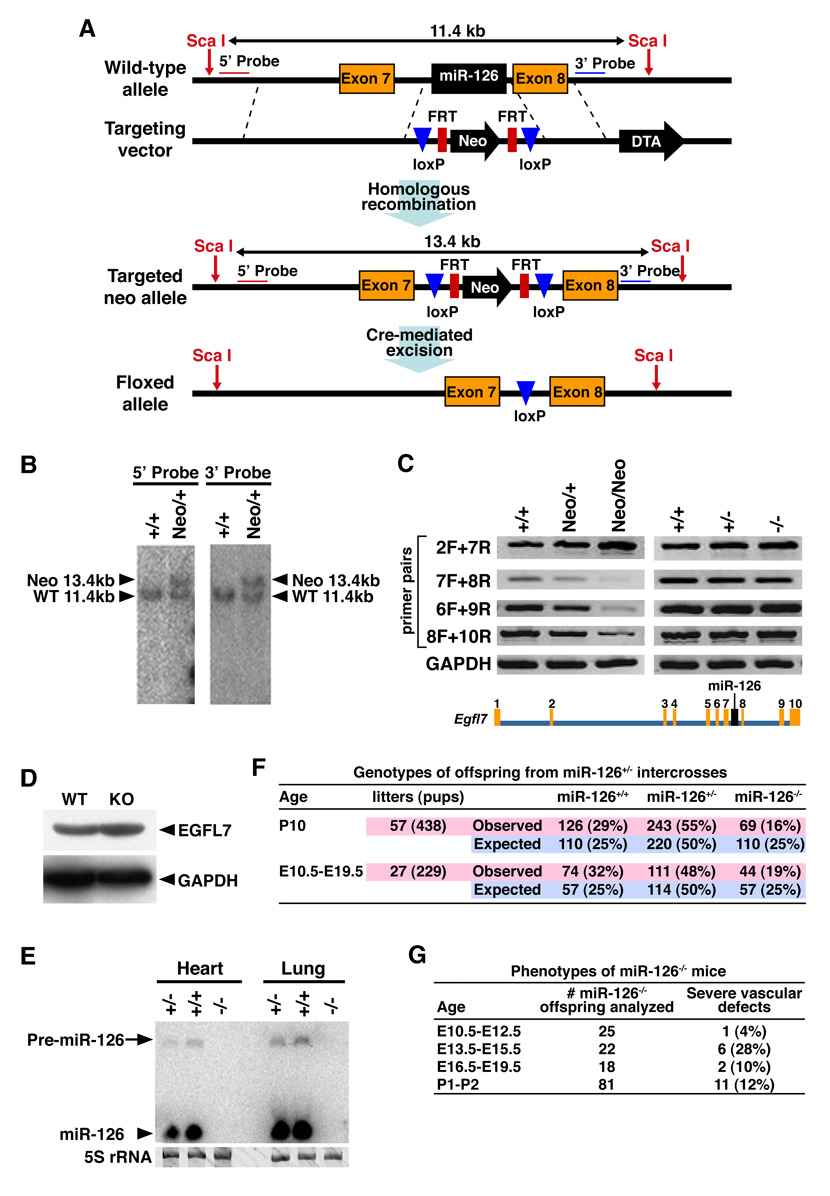
To explore the functions of miR-126 in vivo, we deleted the region of intron 7 of the Egfl7 gene encoding miR-126 and inserted a neomycin-resistance cassette flanked by loxP sites (Fig. 3A and B). Mice heterozygous for the mutant miR-126 allele were intercrossed to obtain miR-126neo/neo mutants. The presence of the neomycin cassette in the Egfl7 intron altered the splicing of surrounding exons, as detected by RT-PCR (Fig. 3C, left panel). Removal of the neomycin cassette from the Egfl7 intron by breeding miR-126neo/+ mice to mice expressing Cre recombinase under control of the CAG promoter normalized the transcription and splicing of Egfl7 (Fig. 3C, right panel).
Figure 3. Targeting of the miR-126 gene.
(A) Strategy to generate miR-126 mutant mice by homologous recombination. The 96bp Egfl7 intron 7 sequence, which contains miR-126, was replaced with a neomycin resistance cassette (Neo) flanked by loxP sites. Neo was removed in the mouse germ line by crossing heterozygous mice to CAG-Cre transgenic mice. DTA, diphtheria toxin A.
(B) Southern blot analysis of genomic DNA from ES cells. DNA was digested with Sca I. Using either the 5’ probe or 3’ probe, the sizes of the wild-type and mutant (miR-126neo allele) are 11.4kb and 13.4kb, respectively. Genotypes are shown on the top.
(C) Analysis of Egfl7 transcripts in heart (left panel) or lung (right panel) of miR-126neo/neo or miR126−/− mice, as detected by RT-PCR. The Egfl7 gene structure and the exon numbers are shown on the bottom. Primers used for RT-PCR were named based on the exon number in the forward (F) and reverse (R) direction. Genotypes are shown on the top. GAPDH was used as a control. Note that Egfl7 expression is disrupted in the miR-126neo/neo mutants, as shown by RT-PCR with primers 7F and 8R, 6F and 9R, 8F and 10R; and normalized upon deletion of the neo cassette, as shown by RT-PCR with the primers indicated.
(D) Detection of EGFL7 and GAPDH protein by Western blot of heart extracts from WT and miR-126 KO mice.
(E) Detection of miR-126 transcripts by Northern analysis of hearts and lungs. 5S rRNA serves as a loading control.
(F) Genotypes of offspring from miR-126+/− intercrosses. The actual and expected number of mice for each genotype at the indicated stages is shown.
(G) Genotypes of embryos from miR-126+/− intercrosses. The number of miR-126−/− offspring analyzed at each age is shown. Severe vascular defects were defined as edema, hemorrhage, severe growth retardation and lethality. Less than 1% of wild-type or miR-126+/− embryos or neonates showed vascular abnormalities.
miR-126+/− mice were intercrossed to obtain miR-126−/− mice. Neither the mature miR-126 nor the stem-loop was expressed in these animals (Fig. 3E). The targeted mutation did not alter the expression of Egfl7 mRNA (Fig. 3C, right panel) or EGFL7 protein (Fig. 3D) in tissues from homozygous mutant mice.
Vascular Abnormalities in miR-126 Mutant Mice
miR-126−/− mice were obtained at a lower than predicted frequency from miR-126+/− intercrosses (Fig. 3F). At postnatal day 10 (P10), 16% of offspring obtained from heterozygous intercrosses were homozygous mutants, versus the expected 25%. Thus, about 40% of the miR-126−/− mice died embryonically or perinatally. Analysis of embryos obtained from timed matings revealed miR-126−/− embryos that were dead or dying with severe systemic edema, multifocal hemorrhages, and ruptured blood vessels throughout embryogenesis (Fig. 4A and B). The highest percentage of embryos with vascular abnormalities was observed from E13.5 to E15.5 (Fig. 3G). However, a failure in growth of the cranial vessels was observed as early as E10.5 (Fig. 4B), prior to systemic edema, hemorrhage or overall embryo demise, indicating that the vascular defects represent a primary effect of miR-126 deletion. Similarly, vascularization of the retina, which begins at P0, and involves the outward migration of ECs from the central retinal artery, was severely impaired in miR-126−/− mice (Fig. 4C) in the absence of other morphological abnormalities.
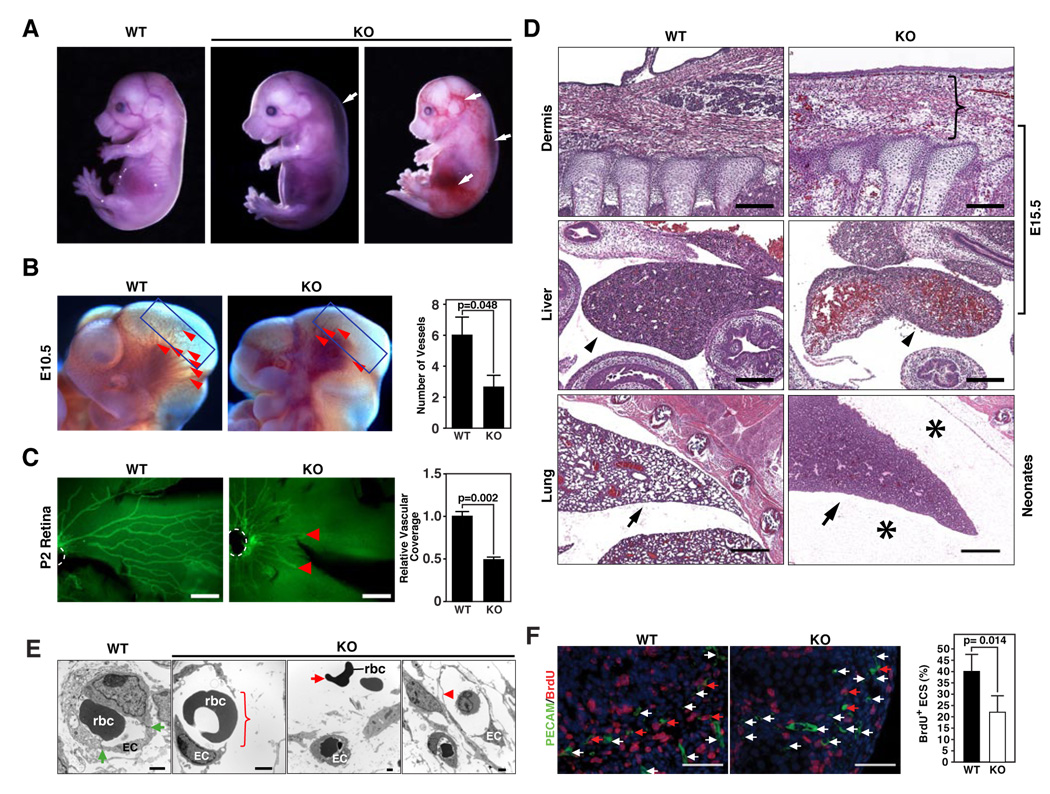
Figure 4. Vascular abnormalities in miR-126 null mice.
(A) Wild-type (WT) and miR-126−/− (KO) embryos at E15.5. A subset of KO embryos shows systemic edema and hemorrhages as indicated by the arrows.
(B) Lateral views of cranial regions of WT and miR-126 KO embryos at E10.5. Superficial cranial vessels, shown by arrowheads, are apparent in WT embryos, but are severely deficient in the mutant. The number of vessels in the cranial region indicated by boxes, is shown on the right (n=6).
(C) Vascularization of the retina at P2, as visualized by PECAM staining. The position of the central retinal artery is demarcated by dashed white lines and the termini of retinal vessels in the mutant by red arrows. Bar = 200 um. Relative vascular coverage is shown on the right (n=3).
(D) H&E staining of sagittal sections of the dermis and liver of E15.5 embryos and lung of neonates of the indicated genotypes. Scale bar in the upper and middle panel equals 200um, and scale bar in the bottom panel equals 500um. The bracket indicates the thickening of dermis with erythrocytes and inflammatory cells in the tissue space in the KO embryo. The arrowheads indicate congestion of red blood cells in KO liver compared to the WT liver. The arrows point to the lungs, in which the alveoli fail to inflate in the KO mice. Asterisks show edema in the thoracic cavity in KO neonates.
(E) Electron microscopy of capillaries in WT and KO embryos at E15.5. The bracket shows the breakdown of vessels in KO embryos. The green arrows point to tight junctions in WT endothelial cells. The red arrow points to the red blood cells floating outside of the vessels in KO embryos, while the arrow head indicates the thinning of the endothelial layer in the vessel of KO embryo. EC: endothelial cell; rbc: red blood cell.
(F) Endothelial cell proliferation in E15.5 KO embryos. Significantly less BrdU (red) and PECAM1 (green) double positive cells were observed in KO compared to WT embryos. The red arrow points to the PECAM/BrdU double positive cells, while the white arrow points to the PECAM single positive cells. Nuclei were stained with DAPI (blue). The statistics are shown on the bar graph (p=0.014).
Histological analysis of mutant embryos and neonates showed abnormal thickening of the epidermis, a hallmark of edema, with erythrocytes and inflammatory cells in the tissue spaces, as well as congestion of red blood cells in the liver, which may reflect compensatory erythropoiesis caused by hypoxia (Fig. 4D). Of the miR-126−/− mice that survived to birth, approximately 12% died by P1 and contained excessive protein-rich fluid in the pleural spaces of the thoracic cavity, an indication of severe edema. The lungs were also not inflated, possibly secondary to the severe edema (Fig. 4D). Edema and hemorrhage were also observed in the thoracic cavity outside of the pericardial space in some miR-126−/− newborn mice. These abnormalities suggested a role for miR-126 in maintenance of endothelial integrity. Indeed, electron microscopy of miR-126−/− embryos confirmed the lack of endothelial integrity and revealed extensive rupture of blood vessels and lack of tight cell-cell interactions (Fig. 4E).
PECAM-positive ECs from vascularized tissues of miR-126−/− embryos at E15.5 displayed diminished proliferation compared to wild-type ECs, as detected by BrdU staining (Fig 4F), whereas proliferation of non-ECs was not significantly different in wild-type and mutant embryos (data not shown). We detected no difference in apoptosis between wild-type and mutant ECs at E15.5 by TUNEL staining (data not shown).
The surviving miR-126−/− mice appeared normal to adulthood and displayed no obvious abnormalities based on histological analysis of tissues. Male mutant mice were fertile. However, females were sub-fertile with reduced litter size (data not shown). We conclude that miR-126 plays an important role in maintenance of vascular integrity during embryogenesis, but is not essential for vascular homeostasis after birth.
Defective Angiogenesis of miR-126−/− ECs
To explore the angiogenic functions of miR-126−/−, we analyzed sprouting angiogenesis using an ex vivo aortic ring assay. Aortic rings from 4-week old miR-126−/− mice and wild-type littermates were isolated and cultured on matrigel with endothelial growth medium containing FGF-2 and VEGF and supplemented with 3% mouse serum. ECs from wild-type mice showed extensive outgrowth between days 4 and 6 of culture, whereas endothelial outgrowth was dramatically impaired in aortic rings obtained from miR-126−/− mice (Fig. 5A). Staining for PECAM confirmed the identity of ECs in aortic ring cultures (data not shown).
Figure 5. Impaired angiogenesis of miR-126 KO ECs.
(A) Representative images of cultured aortic rings isolated from wild-type (WT) and miR-126−/− (KO) mice at days 4–6 are shown. Extensive endothelial outgrowth can be seen in WT explants, but not in mutants. Relative migratory activity under each condition was quantified as shown in the bar graphs with statistics.
(B) Representative images of PECAM1 (Green) staining of matrigel plugs implanted into mice of the indicated genotypes are shown. Significant less angiogenesis was observed in the matrigel with FGF-2 in KO mice compared to WT mice. No significant angiogenesis was observed in the matrigel plugs with lacking FGF-2 in WT or KO mice. Scale bar equals 60um.
(C) The extent of angiogenesis in the matrigel plug assay was quantified by determining PECAM staining area using Image J software. P = 0.0008 for KO compared to WT matrigel plugs.
(D) Survival of WT and KO mice following MI. P value equals 0.05 and 0.014 for the survival of KO mice compared to WT for 1 week and 3 week post MI.
(E) Histological analysis of hearts from WT and KO mice following MI. Panels a–d show longitudinal sections through the right ventricle (RV) and left ventricle (LV). Note thrombi in the atria of the mutant heart, indicative of heart failure. Panels e and f show transverse sections stained with Masson’s trichrome to reveal scar formation. Note the extensive loss of myocardium in the KO mice. Panels g and h show PECAM1 staining in the boxed infarct region from e and f. Note the deficiency of vasculature in KO mice following MI. The scale bar in a–f equals 1cm, and the scale bar in g and h equals 40um.
We further analyzed the angiogenic response of ECs in miR-126−/− mice in vivo using a matrigel plug EC invasion assay in which mice were injected subcutaneously with a matrigel plug containing the pro-angiogenic factor FGF-2, or PBS as control. In response to angiogenic growth factor signaling, ECs typically migrate into the matrigel plug and assemble into a primitive vascular network, which can be detected by PECAM staining one week later. EC invasion requires angiogenic growth factors and is not observed in PBS control matrigel plugs. ECs from miR-126−/− mice showed a dramatically diminished angiogenic response to FGF-2 compared to controls (Fig. 5B, C).
Reduced Survival of miR-126−/− Mice Following Myocardial Infarction
The diminished angiogenic response of miR-126−/− ECs revealed in the matrigel EC invasion assay suggested that miR-126 might play an important role in neoangiogenesis of adult tissues, as occurs in response to injury. Neoangiogenesis is essential for cardiac repair following myocardial infarction (MI), when collateral vessels form at the site of the infarct to maintain blood flow to ischemic cardiac tissue (Kutryk and Stewart, 2003). Myocardial vascularization following MI requires signaling by VEGF and FGF (Scheinowitz et al., 1997; Syed et al., 2004).
We therefore compared the response of wild-type and miR-126 null mice to MI following surgical ligation of the left coronary artery. MI in wild-type mice typically results in an infarct, followed by the formation of a scar. Under the surgical conditions for these experiments, 70% of wild-type mice survived for at least three weeks following MI (Fig. 5D). In contrast, half of miR-126−/− mice died by one-week post-MI, and nearly all died by three weeks (Fig. 5D).
Unoperated hearts from wild-type and miR-126 mutant mice were indistinguishable histologically (Fig. 5E, a and b). One week after MI, mutant mice showed ventricular dilatation compared to wild-type hearts and commonly developed atria thrombi (Fig. 5E, c and d), indicative of heart failure. By 3 weeks post-MI, histological analysis showed more extensive fibrosis and loss of functional myocardium in miR-126 mutants compared to wild-type controls (Fig. 5E, e and f). Many miR-126 mutant animals that died during this period also displayed ventricular rupture, a known consequence of inadequate myocardium that can result from deficient blood flow (data not shown), whereas myocardial rupture was never observed in wild-type mice following MI.
PECAM staining revealed extensive vascularization of the injured myocardium in wild-type mice 3 weeks following MI. In contrast, there was a relative paucity of new vessels in the mutants, and those vessels that were observed appeared truncated and fragmentary (Fig. 5E). Thus, miR-126 appears to be important for normal neovascularization following MI.
Modulation of Angiogenic Signaling by miR-126
The vascular defects in miR-126−/− embryos, combined with the impaired angiogenic activity of mutant ECs, suggested that miR-126 was essential for normal responsiveness of ECs to angiogenic growth factors. To further test this possibility, we infected HUVECs with a miR-126 expressing adenovirus (Ad-miR-126) and examined MAP kinase activation by FGF-2, as detected by phosphorylation of ERK1/2. As shown in Figure 6A, activation of ERK1/2 phosphorylation by FGF-2 was enhanced approximately 2-fold by Ad-miR-126 compared to an Ad-lacZ control. Conversely, knockdown of miR-126 expression with a 2’-0-methyl-miR-126 antisense oligonucleotide diminished ERK phosphorylation in response to VEGF, compared to a control oligonucleotide (Fig. 6B). These findings suggested that miR-126 augments MAP kinase pathway activation by FGF and VEGF.
Figure 6. Modulation of angiogenic growth factor signaling by miR-126.
(A) miR-126 enhances FGF-dependent phosphorylation of ERK1/2. HUVEC cells were infected with adenovirus expressing lacZ or miR-126, and treated with FGF-2 (10ng/ml) for the indicated periods of time. Cell lysates were immunoblotted with the indicated antibodies to determine the level of phosphorylated and total ERK1/2. Ad-miR-126 enhanced FGF-2 dependent phosphorylation of ERK1/2.
(B) Knockdown of miR-126 diminishes VEGF-dependent phosphorylation of ERK1/2. HAEC cells were transfected with 2’-O-methyl-miR-126 antisense oligonucleotide or control oligonucleotide, and treated with VEGF (10ng/ml) for 10 min. Cell lysates were immunoblotted with the indicated antibodies to determine the level of phosphorylated and total ERK1/2. GAPDH was used as a loading control.
(C) Sequence alignment of miR-126 with Spred-1 3’ untranslated regions (UTRs) from different species.
(D) Detection of Spred-1, CRK and GAPDH protein by Western Blot of yolk sac extracts of E15.5 WT and miR-126 KO embryos.
(E) miR-126 targets the Spred-1 3’ UTR. The 3’ UTR of Spred-1 mRNA, and the Spred-1m 3’ UTR with mutations engineered in the region complementary to the miR-126 seed region (GGTACGA to TTGGAAG), was inserted into the pMIR-REPORT vector (Ambion). The miR-126 mutant (miR-126m) construct consists of the miR-126-3p sequence CGTACC mutated to GCATGG, and the corresponding miR-126-5p sequence GGTACG mutated to CCATGC. Transfection of COS-7 cells was performed using the indicated combination of plasmids. CMV-βGAL was used as an internal control for transfection efficiency. Error bars indicate standard deviation. The P values are shown. ns, not significant.
(F) Relative Spred-1 mRNA expression level upon miR-126 over-expression or knockdown. HUVEC or HAEC cells were subjected to the indicated treatments, and the level of Spred-1 mRNA was determined by Real-time RT-PCR. GAPDH served as a control. Error bars indicate standard deviation. The P values are shown.
(G) Up-regulation of Spred-1 mRNA in miR-126−/− endothelial cells. The level of Spred-1 mRNA was determined by Real-time RT-PCR with L7 as control. Error bars indicate standard deviation. P=0.01 for KO compared to WT.
(H) Representative images of cultured aortic rings isolated from wild-type (WT) and miR-126 KO mice at days 5 and 6 are shown. Adenoviral over-expression of Spred-1 in WT explants impairs endothelial outgrowth, whereas siRNA-mediated knockdown of Spred-1 in explants from miR-126 KO mice enhances endothelial outgrowth. Relative migratory activity under each condition was quantified as shown in the bar graphs with statistics.
(I) Scratch-wound assay of HUVEC cells response to VEGF. Knockdown of miR-126 expression with anti-sense RNA impairs EC migration, whereas knockdown of Spred-1 with siRNA restores migration in the presence of miR-126 antisense RNA. The edges of the scratch-wound are shown by red dashed lines.
Inhibition of Spred-1 Expression by miR-126
To identify potential mRNA targets of miR-126 that might contribute to the endothelial abnormalities of miR-126 mutant mice, we compared the gene expression profiles by microarray analysis of ECs isolated from adult kidneys of wild-type and miR-126 null mice. Since most miRNAs promote the degradation of their target mRNAs (Jackson and Standart, 2007), we focused on mRNAs that were up-regulated in miR-126−/− ECs (Supplemental Table 1). Numerous mRNAs involved in angiogenesis, cell adhesion, inflammatory/cytokine signaling and cell cycle control were up-regulated in miR-126−/− EC cells. Among this group of transcripts, we identified three mRNAs that were also predicted by various miRNA target prediction programs to be evolutionarily conserved targets of miR-126 (Supplemental Table 2): Sprouty-related protein-1 (Spred-1), VCAM-1 and integrin alpha-6. Indeed, VCAM-1 mRNA was recently shown to be a target for repression by miR-126 in vitro (Harris et al., 2008).
Intriguingly, Spred-1 has been shown to function as a negative regulator of the Ras/MAP kinase pathway (Wakioka et al., 2001). Given the ability of miR-126 to enhance MAP kinase signaling in response to VEGF and FGF, and the diminished angiogenic growth factor signaling in the absence of miR-126, Spred-1 seemed a likely mediator of the angiogenic actions of miR-126. The predicted energy of the miR-126/Spred-1 interaction is ~-17.9 kcal/mol. Most importantly, the “seed” region (nucleotides 1–7) of miR-126 is completely complementary to the sequence of the Spred-1 3’UTR, and the complementary sequences of miR-126 and the Spred-1 3’ UTR are conserved from amphibians to mammals (Fig. 6C). Consistent with the conclusion that Spred-1 mRNA is a target for repression by miR-126, Spred-1 protein expression was increased in yolk sac from miR-126−/− mice compared to wild-type littermates (Fig. 6D). In contrast, CRK, which is predicted by several miRNA target prediction programs to be a miR-126 target, was unchanged, as was GAPDH, as a control.
When the Spred-1 3’ UTR was fused to a luciferase reporter and tested for repression by miR-126 in transfected cells, miR-126 strongly repressed expression of the Spred-1 3’ UTR luciferase reporter (Fig. 6E). Mutation of six nucleotides in the miR-126 “seed” region (miR-126m) or its complementary sequence in the Spred-1 3’ UTR (Spred-1m UTR) relieved the repressive effect of miR-126 (Fig. 6E). Infection of HUVEC cells with an adenovirus expressing miR-126 also repressed expression of Spred-1 mRNA by about 2-fold (Fig. 6F). Conversely, a miR-126 antisense RNA elevated the expression of Spred-1 mRNA in HAEC cells (Fig. 6F). The efficiency of miR-126 overexpression or knockdown was monitored by Northern blot analysis with miR-126 probe (data not shown).
To establish whether miR-126 is necessary to repress Spred-1 expression, ECs were isolated from the kidneys of miR-126−/− and wild-type adult mice. The identity of ECs was monitored by the uptake of DiI-labeled acetylated low density lipoprotein (DiI- Ac-LDL) and staining with an antibody against von Willebrand factor (data not shown). As expected, Spred-1 mRNA was significantly up-regulated in miR-126−/− ECs compared to wild-type ECs (Fig. 6G), confirming the microarray results.
Further support for the involvement of Spred-1 in inhibiting miR-126 modulated EC migration and angiogenesis, was provided by the aortic ring assay in which retrovirus-mediated overexpression of Spred-1 diminished EC outgrowth (Fig. 6H), whereas knockdown of Spred-1 expression with a small interfering RNA enhanced endothelial outgrowth in explants from miR-126−/− mice (Fig. 6H). Finally, in a scratch-wound assay in vitro, miR-126 antisense RNA dramatically impaired HUVEC migration, whereas Spred-1 siRNA restored migratory activity to cells expressing miR-126 antisense RNA (Fig. 6I). These results support the conclusion that miR-126 augments angiogenic signaling by diminishing the inhibitory influence of Spred-1 on the MAP kinase pathway.
DISCUSSION
The results of this study reveal an essential role for miR-126 in angiogenesis and maintenance of vascular integrity in vivo. The actions of miR-126 appear to reflect, at least in part, its potentiation of MAP kinase signaling downstream of VEGF and FGF, which act as potent inducers of angiogenesis (Fig. 7). Spred-1, an intracellular inhibitor of the Ras/MAP kinase pathway, serves as a target for repression by miR-126. Thus, in the absence of miR-126, Spred-1 expression is elevated, resulting in repression of angiogenic signaling. Conversely, miR-126 overexpression relieves the repressive influence of Spred-1 on the signaling pathways activated by VEGF and FGF, favoring angiogenesis. Consistent with these findings, over-expression of Spred-1 in ECs impairs angiogenesis and cell migration, mimicking the miR-126 loss-of-function phenotype, whereas knockdown of Spred-1 expression enhances angiogenesis and rescues the miR-126 loss-of-function phenotype in cultured ECs.
Figure 7. A model for the function of miR-126 in angiogenesis.
Binding of VEGF and FGF to their receptors on ECs leads to activation of the MAP kinase signaling pathway, which culminates in the nucleus to stimulate the transcription of genes involved in angiogenesis. miR-126 represses the expression of Spred-1, a negative regulator of Ras/MAP kinase signaling. Thus, loss of miR-126 function diminishes MAP kinase signaling in response to VEGF and FGF, whereas gain of miR-126 function enhances angiogenic signaling.
It is intriguing that only a subset of miR-126−/− embryos succumbs to embryonic lethality from vascular rupture, whereas others survive to adulthood. We propose that miR-126 stochastically modulates critical angiogenic signaling events during a temporal window in embryogenesis, perhaps because of a specific threshold of angiogenic signaling at this stage. If embryos are able to pass this developmental time point, the functions of miR-126 may become less critical for maintenance of the vasculature. The partial embryonic lethality of miR-126 mutant mice suggests that this miRNA acts to modulate gene expression programs, rather than functioning as an “onoff” switch for angiogenesis.
In addition to its requirement in normal vascular development during embryogeneis, the functions of miR-126 appear to be important following MI, when injured vessels at the site of the infarct initiate neoangiogenesis to restore blood flow to the injured myocardial wall. Under conditions of stress, as in the heart following MI, the actions of miR-126 may acquire heightened importance due to the requirement of angiogenic signaling for neovascularization. In this regard, VEGF and FGF expression increases in response to myocardial ischemia and is critical for the development of collateral vessels in the ischemic myocardium (Semenza, 2003). Cardiac injury, in addition to activating the migration and proliferation of nearby ECs, results in the homing of circulating hematopoietic progenitor cells to sites of ischemia and their contribution to cardiac repair (Kocher et al., 2001; Takahashi et al., 1999). miR-126 is expressed in hematopoietic stem cells, and might therefore contribute to the regenerative functions of this cell population (Garzon et al., 2006; Landgraf et al., 2007).
Control of Angiogenesis by miR-126
Angiogenic growth factors, such as VEGF and FGF, modulate EC proliferation, migration and adhesion by activating the MAP kinase pathway, which culminates in the nucleus to enhance the expression of genes required for angiogenesis and vascular integrity. The abnormalities associated with miR-126 loss-of-function are similar to the vascular defects resulting from the inhibition of MAP kinase signaling in ECs (Hayashi et al., 2004).
Consistent with the conclusion that miR-126 promotes angiogenesis by dampening the expression of Spred-1, Spred-1 inhibits cell motility and Rho-mediated actin reorganization (Miyoshi et al. 2004), processes important for angiogenesis. Spred-1, and other members of the Spred family, function as membrane-associated suppressors of growth factor-induced ERK activation and block cell proliferation and migration in response to growth factor signaling (Wakioka et al., 2001). The inhibitory actions of Spred proteins are mediated by interference of phosphorylation and activation of Raf, an upstream activator of the MAP kinase pathway. Among the three Spred proteins, only Spred-1 contains a predicted target sequence for miR-126. While our results are consistent with the conclusion that Spred-1 plays a major role as a mediator of the pro-angiogenic actions of miR-126, it is likely that the actions of miR-126 reflect the combined functions of multiple target proteins that modulate angiogenesis and vascular integrity.
Biogenesis of miR-126
Based on the co-expression of miR-126 and Egfl7 mRNA, as well as our finding that miR-126 is generated from a retained intron in a subset of Egfl7 pre-mRNAs (unpublished results), we conclude that miR-126 originates from the Egfl7 pre-mRNA. While there are intronic miRNAs that are transcribed independently of their host genes, in all cases to date, these miRNAs are transcribed on the opposite strand of the mRNA, in contrast to miR-126 and Egfl7 mRNA. Moreover, Ets binding sites are required for endothelial-specific expression and no such sites are present in intron 7 of the Egfl7 gene.
Recently, Egfl7 knockout mice were reported to display vascular abnormalities remarkably similar to those of miR-126 null mice (Schmidt et al., 2007). The deletion mutation in those mice was reported to result in the absence of an Egfl7 transcript, suggesting that miR-126 expression is also eliminated. However, miR-126 expression was not examined. Thus, the possibility that the phenotype of those mutant mice actually reflects the loss-of-function of miR-126 warrants consideration.
It is becoming increasingly apparent that the integration of miRNAs into introns of protein coding genes represents a common mechanism for coordinating the expression and regulatory functions of miRNAs with protein coding genes. As another example of this form of co-regulation, we showed previously that miR-208, which is encoded by an intron of the alpha-myosin heavy chain (MHC) gene, functions within a regulatory network to control cardiac stress response (van Rooij et al., 2007). Incorporation of a miRNA into an intron of a tissue-specific gene provides an efficient mechanism for ensuring the co-regulation of the miRNA with the gene programs it regulates.
Therapeutic Implications
The endothelium plays myriad roles in cardiovascular homeostasis and remodeling during disease, including the control of vascular tone and permeability, smooth muscle cell growth and proliferation, leukocyte adhesion, coagulation, and thrombosis. miRNAs have been implicated in regulating EC gene expression and function in vitro (Kuehbacher et al., 2007; Suarez et al., 2007)), but the functions of miRNAs in EC biology in vivo have not been explored. The discovery that miR-126 is required for vascular integrity and angiogenesis, as well as survival post-MI, suggests that strategies to elevate miR-126 in the ischemic myocardium could enhance cardiac repair. Conversely, diminishing miR-126 expression may be efficacious in settings of pathological vascularization, such as cancer, atherosclerosis, retinopathy and stroke. Recently, miR-126 was reported to inhibit tumorigenesis and to be down-regulated in metastatic breast tumors, although the specific cell type in which it was down-regulated and the targets of miR-126 that might mediate these actions were not defined (Tavazoie et al., 2008). We speculate that miR-126 and other miRNAs will be found to play key roles in tissue remodeling and diseases.
EXPERIMENTAL PROCEDURES
Generation of miR-126 Mutant Mice
Methods to generate miR-126 null mice are described in Supplemental Text.
Surgical Procedure
Mice were subjected to MI by coronary artery ligation as previously described (van Rooij et al., 2004). A detailed version of the procedure is described in Supplemental Data. Animal surgical procedures were approved by the UT Southwestern IACUC.
miRNA Northern Blot
Generation and Analysis of Transgenic Mice
Transgenes were generated by cloning DNA fragments from the Egfl7 5’ flanking region into the hsp68 basal promoter upstream of a lacZ reporter gene (Kothary et al., 1989). These reporter constructs were injected into fertilized oocytes from B6C3F mice and implanted into pseudopregnant ICR mice. Embryos were collected and stained for β -galactosidase activity. Transgenic embryos were identified by PCR analysis with lacZ primer pairs.
Histology, BrdU Labeling, TUNEL assay and Immunohistochemistry
Histology was performed as described (Chang et al., 2006). For BrdU labeling, animals were injected intraperitoneally with 100 ug BrdU/g 4 hours prior to sacrifice. For whole-mount immunostaining, embryos or P2 retinas were fixed in 4% paraformaldehyde for 2 hours, and processed for staining with PECAM using standard procedures. For section immunohistochemistry, embryos or mouse hearts were fixed in 4% paraformadehyde overnight, and processed for cryo-section and single or double immunostaining using standard procedures. Apoptosis was determined by the TUNEL assay using an In Situ Cell Death Detection Kit, TMR red (Roche).
Cell Culture
Mouse EC isolation was performed as described in the Supplemental Data. HAEC (Clonetics) and HUVEC (ATCC) cells were grown in EC growth medium (EGM) (Clonetics/Cambrex). For FGF-2 or VEGF treatment, ECs were starved with EC basal medium (EBM-2) with 0.1% FBS for 24 hours, and then treated with growth factors for the indicated periods of time. Adenovirus expressing miR-126 or lacZ was generated and cells were infected as described (Wang et al., 2008). Retrovirus-expressing Spred-1 or GFP was generated as described (Nonami et al., 2004). For miR-126-3p inhibitor transfection, HAEC cells were transfected with 2’-O-methyl-miR-126 antisense oligonucleotide (Ambion) and/or human Spred-1 siRNA pool, or a control oligonucleotide at a concentration of 50 nM using Liptofectamine 2000 (Invitrogen). Forty-eight hours after transfection, the cells were starved, treated with VEGF-A, and harvested for protein analyses. miR-126 expression was determined by Northern blot analysis with miR-126-3p starfire™ probe. For aortic ring virus infection, Spred-1 or control GFP retrovirus was added to the cultured aortic rings. Transfection of the mouse Spred-1 siRNA pool into miR-126 null aortic rings was performed as described above. After overnight transfection or infection, the aortic rings were cultured in fresh medium for 4–6 days to monitor the aortic ring sprouting. The expression of Spred-1 was determined by Western blot and Realtime PCR analysis.
Reporter Assays
The 0.5 kb region 1 enhancer of pEgfl7/miR-126, and the ETS DNA binding site deletion mutant of the fragment generated by site directed mutagenesis, was cloned into the pGL3 vector upstream of an engineered ANF basal promoter. COS-7 cells in 24-well plates were transfected with 50 ng of reporter plasmids in the presence or absence of increasing amount of Ets1 or Ets1 DNA binding mutant expression plasmid (John et al., 2008).
Spred-1 3’ UTR and Spred-1 mutant 3’ UTR generated by mutagenesis were directionally cloned into the pMIR-REPORT vector (Ambion). miR-126 genomic DNA fragment and miR-126m DNA fragment, were cloned into the pCMV-Myc vector. Spred-1 or Spred1m UTR construct was then co-transfected with miR-126 or miR-126m expression plasmid into COS-7 cells. Reporter assays were performed as described (Chang et al., 2005).
Electron Microscopy
Electron microscopy was performed at Children’s Medical Center of Dallas as described in Supplemental Data.
Aortic Ring Assay
Four-well culture dishes (Nunclon™ Surface, Nunc™) were covered with 250 µl of matrigel (Chemicon) and allowed to gel for 15 min at 37°C, 5% CO2. Thoracic aortas were excised from 4–6 week-old mice. Fibro-adipose tissue was dissected away from the aortas, which were then cut into 1-mm rings, rinsed with EGM-2 (Cambrex), placed on matrigel coated wells and covered with additional Matrigel. The aortic rings were cultured in EGM-2 (Cambrex) plus 3% mouse serum (Taconic).
In Vivo Matrigel Plug Assay
Growth factor reduced-Matrigel (BD Bioscience) was mixed with heparin (60 units/ml), and FGF-2 (250 ng/ml, R&D) or PBS as control. Matrigel (0.5ml) was injected subcutaneously into the ventral area of anesthetized mice. The animals were euthanized after 7 days, and the Matrigel plugs were carefully dissected away from the host tissue, and processed for frozen section and immuno-stained for PECAM1.
Scratch-wound assay
The scratch-wound assay was performed using HUVEC cells as described (Wang et al., 2008).
RNA, Western blot analysis and in situ hybridization
Standard procedures were used and described in the Supplemental Data.
Statistics
Statistics was carried out using 2 way t-test. P values less than 0.05 were considered to be significant.
Supplementary Material
ACKNOWLEDGEMENTS
We are grateful to Eva van Rooij for scientific input and insightful comments on the manuscript. We thank Jose Cabrera for graphics; Jennifer Brown for editorial assistance; Xiumin Li, and John M. Shelton for technical help. We thank Dr. Garrett-sinha for providing Ets1 and Ets1DBmut plasmids, Dr. A. Yoshimura for Spred-1 retrovirus, Dr. Robert Gerard for miR-126 adenovirus, and Dr. Kai Schuh for Spred-1 antibody. This work was supported by grants from the National Institutes of Health, the Donald W. Reynolds Clinical Cardiovascular Research Center, the Sandler Foundation for Asthma Research, and the Robert A. Welch Foundation to ENO. S.W. was supported by a fellowship grant from the American Heart Association
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- Campagnolo L, Leahy A, Chitnis S, Koschnick S, Fitch MJ, Fallon JT, Loskutoff D, Taubman MB, Stuhlmann H. EGFL7 is a chemoattractant for endothelial cells and is up-regulated in angiogenesis and arterial injury. Am. J. Pathol. 2005;167:275–284. doi: 10.1016/S0002-9440(10)62972-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in health and disease. Nat. med. 2003;9:653–660. doi: 10.1038/nm0603-653. [DOI] [PubMed] [Google Scholar]
- Chang SR, Bezprozvannaya S, Li SJ, Olson EN. An expression screen reveals modulators of class II histone deacetylase phosphorylation. Proc. Natl. Acad. Sci. U S A. 2005;102:8120–8125. doi: 10.1073/pnas.0503275102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SR, Young BD, Li SJ, Qi XX, Richardson JA, Olson EN. Histone deacetylase 7 maintains vascular integrity by repressing matrix metalloproteinase 10. Cell. 2006;126:321–334. doi: 10.1016/j.cell.2006.05.040. [DOI] [PubMed] [Google Scholar]
- Cross MJ, Claesson-Welsh L. FGF and VEGF function in angiogenesis: signalling pathways, biological responses and therapeutic inhibition. Trends Pharmacol. Sci. 2001;22:201–207. doi: 10.1016/s0165-6147(00)01676-x. [DOI] [PubMed] [Google Scholar]
- Eliceiri BP, Klemke R, Stromblad S, Cheresh DA. Integrin alpha v beta 3 requirement for sustained mitogen-activated protein kinase activity during angiogenesis. J. Cell Biol. 1998;140:1255–1263. doi: 10.1083/jcb.140.5.1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitch MJ, Campagnolo L, Kuhnert F, Stuhlmann H. Egfl7, a novel epidermal growth factor-domain gene expressed in endothelial cells. Dev Dyn. 2004;230:316–324. doi: 10.1002/dvdy.20063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garzon R, Pichiorri F, Palumbo T, Iuliano R, Cimmino A, Aqeilan R, Volinia S, Bhatt D, Alder H, Marcucci G, et al. MicroRNA fingerprints during human megakaryocytopoiesis. Proc. Natl. Acad. Sci. U S A. 2006;103:5078–5083. doi: 10.1073/pnas.0600587103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giroux S, Tremblay M, Bernard D, Cadrin-Girard JF, Aubry S, Larouche L, Rousseau S, Huot J, Landry J, Jeannotte L, et al. Embryonic death of Mek1-deficient mice reveals a role for this kinase in angiogenesis in the labyrinthine region of the placenta. Curr Biol. 1999;9:369–372. doi: 10.1016/s0960-9822(99)80164-x. [DOI] [PubMed] [Google Scholar]
- Harris TA, Yamakuchi M, Ferlito M, Mendell JT, Lowenstein CJ. MicroRNA-126 regulates endothelial expression of vascular cell adhesion molecule 1. Proc. Natl. Acad. Sci. U S A. 2008;105:1516–1521. doi: 10.1073/pnas.0707493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi M, Kim SW, Imanaka-Yoshida K, Yoshida T, Abel ED, Eliceiri B, Yang Y, Ulevitch RJ, Lee JD. Targeted deletion of BMK1/ERK5 in adult mice perturbs vascular integrity and leads to endothelial failure. J. Clin. Invest. 2004;113:1138–1148. doi: 10.1172/JCI19890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He L, Hannon GJ. MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 2004;5:522–531. doi: 10.1038/nrg1379. [DOI] [PubMed] [Google Scholar]
- Hood JD, Bednarski M, Frausto R, Guccione S, Reisfeld RA, Xiang R, Cheresh DA. Tumor regression by targeted gene delivery to the neovasculature. Science. 2002;296:2404–2407. doi: 10.1126/science.1070200. [DOI] [PubMed] [Google Scholar]
- Jackson RJ, Standart N. How do microRNAs regulate gene expression? Sci STKE. 2007;367:1–13. doi: 10.1126/stke.3672007re1. [DOI] [PubMed] [Google Scholar]
- John SA, Clements JL, Russell LM, Garrett-Sinha LA. Ets-1 regulates plasma cell differentiation by interfering with the activity of the transcription factor Blimp-1. J. Biol. Chem. 2008;283:951–962. doi: 10.1074/jbc.M705262200. [DOI] [PubMed] [Google Scholar]
- Kocher AA, Schuster MD, Szabolcs MJ, Takuma S, Burkhoff D, Wang J, Homma S, Edwards NM, Itescu S. Neovascularization of ischemic myocardium by human bone-marrow-derived angioblasts prevents cardiomyocyte apoptosis, reduces remodeling and improves cardiac function. Nat. Med. 2001;7:430–436. doi: 10.1038/86498. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible Expression of an Hsp68-Lacz Hybrid Gene in Transgenic Mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Kuehbacher A, Urbich C, Zeiher AM, Dimmeler S. Role of Dicer and Drosha for endothelial microRNA expression and angiogenesis. Circ. Res. 2007;101:59–68. doi: 10.1161/CIRCRESAHA.107.153916. [DOI] [PubMed] [Google Scholar]
- Kutryk MJ, Stewart DJ. Angiogenesis of the heart. Microsc. Res. Tech. 2003;60:138–158. doi: 10.1002/jemt.10252. [DOI] [PubMed] [Google Scholar]
- Lagos-Quintana M, Rauhut R, Yalcin A, Meyer J, Lendeckel W, Tuschl T. Identification of tissue-specific microRNAs from mouse. Curr Biol. 2002;12:735–739. doi: 10.1016/s0960-9822(02)00809-6. [DOI] [PubMed] [Google Scholar]
- Landgraf P, Rusu M, Sheridan R, Sewer A, Iovino N, Aravin A, Pfeffer S, Rice A, Kamphorst AO, Landthaler M, et al. A mammalian microRNA expression atlas based on small RNA library sequencing. Cell. 2007;129:1401–1414. doi: 10.1016/j.cell.2007.04.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latronico MVG, Catalucci D, Condorelli G. Emerging role of MicroRNAs in cardiovascular biology. Circ. Res. 2007;101:1225–1236. doi: 10.1161/CIRCRESAHA.107.163147. [DOI] [PubMed] [Google Scholar]
- Lelievre E, Lionneton F, Soncin F. Role of the ETS transcription factors in the control of endothelial-specific gene expression and in angiogenesis. Bull. Cancer. 2001;88:137–142. [PubMed] [Google Scholar]
- Miyoshi K, Wakioka T, Nishinakamura H, Kamio M, Yang L, Inoue M, Hasegawa M, Yonemitsu Y, Komiya S, Yoshimura A. The Sprouty-related protein, Spred, inhibits cell motility, metastasis, and Rho-mediated actin reorganization. Oncogene. 2004;23:5567–5576. doi: 10.1038/sj.onc.1207759. [DOI] [PubMed] [Google Scholar]
- Musiyenko A, Bitko V, Barik S. Ectopic expression of miR-126*, an intronic product of the vascular endothelial EGF-like 7 gene, regulates prostein translation and invasiveness of prostate cancer LNCaP cells. J. Mol. Med. 2008;86:313–322. doi: 10.1007/s00109-007-0296-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panka DJ, Atkins MB, Mier JW. Targeting the mitogen-activated protein kinase pathway in the treatment of malignant melanoma. Clin. Cancer Res. 2006;12:2371s–2375s. doi: 10.1158/1078-0432.CCR-05-2539. [DOI] [PubMed] [Google Scholar]
- Parker LH, Schmidt M, Jin SW, Gray AM, Beis D, Pham T, Frantz G, Palmieri S, Hillan K, Stainier DY, et al. The endothelial-cell-derived secreted factor Egfl7 regulates vascular tube formation. Nature. 2004;428:754–758. doi: 10.1038/nature02416. [DOI] [PubMed] [Google Scholar]
- Scheinowitz M, Abramov D, Eldar M. The role of insulin-like and basic fibroblast growth factors on ischemic and infarcted myocardium: A mini review. Int. J. Cardiol. 1997;59:1–5. doi: 10.1016/s0167-5273(96)02902-6. [DOI] [PubMed] [Google Scholar]
- Schmidt M, Paes K, De Maziere A, Smyczek T, Yang S, Gray A, French D, Kasman I, Klumperman J, Rice DS, et al. EGFL7 regulates the collective migration of endothelial cells by restricting their spatial distribution. Development. 2007;134:2913–2923. doi: 10.1242/dev.002576. [DOI] [PubMed] [Google Scholar]
- Semenza GL. Angiogenesis in ischemic and neoplastic disorders. Annu. Rev. Med. 2003;54:17–28. doi: 10.1146/annurev.med.54.101601.152418. [DOI] [PubMed] [Google Scholar]
- Soncin F, Mattot V, Lionneton F, Spruyt N, Lepretre F, Begue A, Stehelin D. VE-statin, an endothelial repressor of smooth muscle cell migration. EMBO J. 2003;22:5700–5711. doi: 10.1093/emboj/cdg549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suarez Y, Fernandez-Hernando C, Pober JS, Sessa WC. Dicer dependent microRNAs regulate gene expression and functions in human endothelial cells. Circ. Res. 2007;100:1164–1173. doi: 10.1161/01.RES.0000265065.26744.17. [DOI] [PubMed] [Google Scholar]
- Syed IS, Sanborn TA, Rosengart TK. Therapeutic angiogenesis: A biologic bypass. Cardiology. 2004;101:131–143. doi: 10.1159/000075994. [DOI] [PubMed] [Google Scholar]
- Takahashi T, Kalka C, Masuda H, Chen D, Silver M, Kearney M, Magner M, Isner JM, Asahara T. Ischemia- and cytokine-induced mobilization of bone marrow-derived endothelial progenitor cells for neovascularization. Nat. Med. 1999;5:434–438. doi: 10.1038/7434. [DOI] [PubMed] [Google Scholar]
- Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang QQ, Bos PD, Gerald WL, Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Olson EN. MicroRNAs: powerful new regulators of heart disease and provocative therapeutic targets. J. Clin. Invest. 2007;117:2369–2376. doi: 10.1172/JCI33099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microRNA. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- Wakioka T, Sasaki A, Kato R, Shouda T, Matsumoto A, Miyoshi K, Tsuneoka M, Komiya S, Baron R, Yoshimura A. Spred is a Sprouty-related suppressor of Ras signalling. Nature. 2001;412:647–651. doi: 10.1038/35088082. [DOI] [PubMed] [Google Scholar]
- Wang S, Li X, Parra M, Verdin E, Bassel-Duby R, Olson EN. Control of endothelial cell proliferation and migration by VEGF signaling to histone deacetylase 7. Proc. Natl. Acad. Sci. U S A. 2008;105:7738–7743. doi: 10.1073/pnas.0802857105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wienholds E, Kloosterman WP, Miska E, Alvarez-Saavedra E, Berezikov E, de Bruijn E, Horvitz HR, Kauppinen S, Plasterk RH. MicroRNA expression in zebrafish embryonic development. Science. 2005;309:310–311. doi: 10.1126/science.1114519. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.