Summary
Microbial pathogens use environmental cues to trigger the developmental events needed to infect mammalian hosts or transmit to disease-vectors. The parasites causing African sleeping sickness respond to citrate/cis aconitate (CCA) to initiate life-cycle development when transmitted to their tsetse-fly vector. This requires hypersensitization of the parasites to CCA by exposure to low temperature, conditions encountered after tsetse feeding at dusk or dawn. Here we identify a carboxylate-transporter family, PAD (Proteins Associated with Differentiation) required for perception of this differentiation signal. Consistent with predictions for the response of trypanosomes to CCA, PAD proteins are expressed on the surface of the transmission-competent ‘stumpy-form’ parasites in the bloodstream and at least one member is thermoregulated, showing elevated expression and surface-access at low-temperature. Moreover, RNAi-mediated ablation of PAD expression diminishes CCA-induced differentiation and eliminates CCA-hypersensitivity under cold-shock conditions. As well as being molecular transducers of the differentiation signal in these parasites, PAD proteins provide the first surface-marker able to discriminate the transmission-stage of trypanosomes in their mammalian host.
Insect-borne parasites undergo life-cycle differentiation to adapt to rapid changes in temperature1,2, nutritional-availability3 and potential immunological-attack4 as they enter their arthropod vector. The cues that induce such changes are often well characterised, such as temperature reduction and pH changes5 or exposure to arthropod-derived factors6. However, the surface molecules that transmit these signals and initiate intracellular differentiation events in microbial parasites are often not well characterised.
African trypanosomes are protozoan parasites responsible for fatal disease in humans and livestock in sub-Saharan Africa, generating significant restrictions in health and welfare in afflicted regions7. The transmission of these parasites by tsetse flies requires the development of bloodstream ‘stumpy-forms’, a G0-arrested cell-type pre-adapted for transmission8-10. Stumpy-forms arise via quorum-sensing from proliferative ‘slender-forms’ at the peak of each parasitaemia in response to an unidentified parasite-derived signalling factor, stumpy induction factor (SIF)11. Although the morphological extremes of slender and stumpy-forms are easily distinguished, the development from slender- to stumpy-morphology is progressive12, with transitional forms loosely described as ‘intermediates’. To date, no developmentally-regulated surface marker has been identified that discriminates slender and stumpy-cells.
When ingested by tsetse during a bloodmeal, slender-forms are killed whereas stumpy-forms differentiate to procyclic-forms13. Bloodstream trypanosomes can also be induced to differentiate in vitro by the Krebs-cycle intermediates, citrate or cis-aconitate (CCA)14. Recently, it was discovered that temperature reduction from 37°C to 20°C could induce hypersensitivity of stumpy-forms to CCA2, such that differentiation was induced at concentrations found in the tsetse (15.9μM15) or ingested blood-meal (~130μM16). This probably represents a natural condition in the trypanosome life-cycle, since tsetse are exposed to cool conditions when feeding at dusk or early morning2. Nonetheless, the molecule responsible for the transmission of the CCA differentiation signal has remained unidentified.
PAD family identification and expression
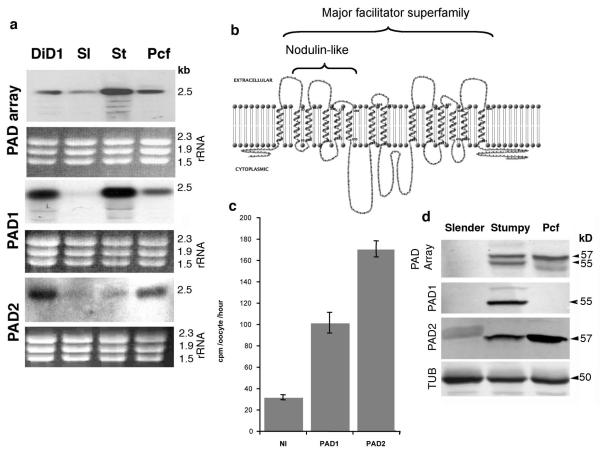
We previously selected a trypanosome line (DiD1, Defective in Differentiation-1) with reduced ability to differentiate to procyclic forms10. The expression profile of DiD1 was compared with its differentiation-competent parent by differential hybridization of labelled cDNA to genomic arrays. This identified 2 adjacent genes on chromosome 7 of the T.brucei genome (Tb927.7.5930 and Tb927.7.5940), there being a stronger signal with DiD1-derived cDNA compared to the parental. These genes comprise the first two genes in an 8-gene array at the end of a unidirectional gene cluster (named the ‘PAD’ gene array, for ‘Proteins Associated with Differentiation’; see below) (Supplementary Fig. 1a). Although no mutation in either gene was detected (Supplementary Fig. 2), northern blotting confirmed the differential expression of both PAD1 and PAD2 genes in the DiD1 line (Figure 1a). Further, PAD1 and PAD2 mRNA showed stage-regulated expression, PAD1 being enriched in stumpy-forms, whereas PAD2 was elevated in procyclic-forms. Neither PAD1 nor PAD2 mRNA was significantly expressed in slender-forms (Figure 1a).
Figure 1. Identification and characteristics of PAD proteins.
- (a) Expression of PAD-mRNAs in DiD-1, parental slender-forms (“Sl”), stumpy-forms (“St”) and procyclic-forms (“Pcf”). rRNAs show loading.
- (b) Predicted transmembrane domains in PAD1.
- (a) [14C]citrate uptake into Xenopus oocytes microinjected with 20 ng cRNA encoding PAD1 or PAD2, or non-injected. The data (from 8-10 oocytes, shown ± s.e.m.) are representative of ten experiments in which oocytes expressing PAD1 exhibited 1.4-3.7-fold increase in [14C]citrate uptake and oocytes expressing PAD2 exhibited 1.6-5.4-fold increase relative to non-injected controls.
- (b) Expression of PAD proteins in slender-, stumpy- and procyclic-forms normalised to α-tubulin. The PAD2-antibody cross-reacts weakly with the 60kDal VSG in slender-forms.
The PAD genes encode closely-related members of a family of 14 transmembrane-spanning proteins of the major facilitator superfamily (Figure 1b; Supplementary Fig. 1b). PSI-BLAST searches revealed a conserved domain in plant nodulin-like proteins (PF06813) and closest overall similarity to carboxylate transporters. Supporting this, Xenopus oocytes microinjected with cRNA encoding either PAD1 or PAD2 showed a marked increase in the uptake of 14C citrate relative to non-injected controls (Figure 1c).
An antibody detecting all members of the PAD protein family reacted with distinct bands in different trypanosome life-cycle stages (Figure 1d); although there was no significant expression in slender-forms, stumpy-forms exhibited two prominent bands at 55kDal and 57kDal, whereas procyclic-forms predominantly expressed the 57kDal band. Neither corresponded to the predicted size of any PAD protein, probably because proteins with extensive transmembrane regions frequently migrate aberrantly. Specific anti-peptide antibodies showed that the 55kDal and 57kDal bands corresponded to PAD1 and PAD2 respectively, and that PAD1 was stumpy-form specific, whereas PAD2 was expressed 6-fold more in procyclic-forms than stumpy-forms (Figure 1d). During synchronous differentiation from stumpy- to procyclic-forms, PAD1 was retained only during the first 24h, whereas PAD2 was strongly induced during this period (up to 17-fold at 24h; Supplementary Fig. 3).
PAD1 marks the transmissible stumpy form
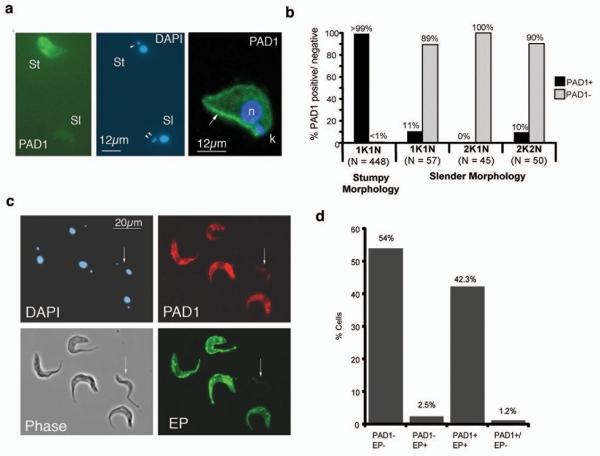
The PAD1 expression profile suggested that it might provide a useful cytological marker for stumpy-forms. To investigate this, the location and expression of PAD1 was analysed in mixed populations of slender and stumpy-forms. Figure 2a (left two panels) shows immunofluorescence images demonstrating stumpy-specific expression of PAD1. Confocal images (Figure 2a, right panel) demonstrated an intense staining at the stumpy-cell periphery, revealing surface membrane labelling. To quantitate the stumpy-specific expression of PAD1, ‘intermediate’ cell populations were assayed with PAD1 and counterstained with DAPI, allowing their cell-cycle position to be determined. In trypanosomes, cells in G1/G0 and S-phase have 1 kinetoplast and 1 nucleus (1K1N), whereas G2/M and post-mitotic cells have a 2K1N or 2K2N DNA configuration, respectively17. Since stumpy-forms are uniformly arrested in G1/G018, 2K1N or 2K2N cells can be unambiguously assigned as slender-forms. Detailed examination of cells within each category of 1K1N, 2K1N or 2K2N (an overall analysis of >10,000 cells) demonstrated that dividing-cells and cells with a slender morphology were overwhelmingly negative for PAD1 (Figure 2b). However, ~10% of 1K1N and 2K2N slender-cells were PAD1-positive: these may represent ‘intermediate’ cells that have committed to stumpy-formation.
Figure 2. PAD1 identifies stumpy forms.
- (a) PAD1-expression on methanol-fixed trypanosomes. The slender-cell (‘sl’; identified by having two kinetoplasts, arrowed in the DAPI panel) is PAD1-negative, the stumpy-cell (‘St’) is PAD1-positive. A confocal image of a paraformaldehyde-fixed stumpy-form is also shown (right); peripheral PAD1-labelling (green) is arrowed; nucleus, ‘n’, and kinetoplast, ‘k’, (both stained blue).
- (b) Quantitation of PAD1 expression on different cell-cycle stages.
- (c) PAD1-positive cells are those competent to differentiate. Slender and stumpy-forms were methanol-fixed 6hr through differentiation. A slender-morphology PAD1-negative, EP-Procyclin-negative cell is arrowed, other cells are PAD1-positive (red), EP-procyclin-positive (green). Phase-contrast and DAPI images are also shown.
- (d) Quantitation of the cells expressing PAD1 and/or EP-procyclin. n=500.
To investigate whether only the PAD1-positive bloodstream trypanosomes were competent to differentiate to procyclic-forms, mixed populations of slender and stumpy-forms were examined 6 hours after exposure to cis-aconitate (CA) for EP-procyclin expression, an early marker for differentiation19. This confirmed a precise correspondence between EP-procyclin and PAD1 expression, with 96% of cells showing matching staining for both markers (Figure 2c,d). Thus, PAD1 identifies stumpy-forms in a bloodstream population as those competent to differentiate to procyclic forms.
PAD2 is thermoregulated
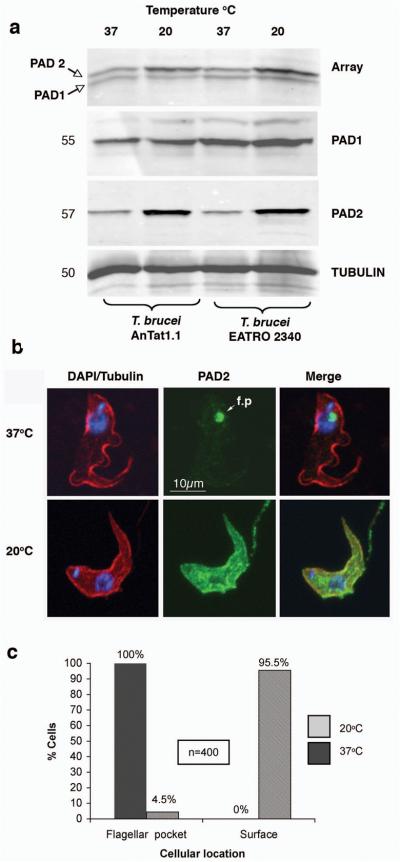
As surface carboxylate transporters expressed on stumpy forms, PAD proteins were good candidates as transducers of the CCA-developmental signal. To evaluate this, we initially tested the inducibility of PAD proteins at 20°C, a predicted requirement for physiological CCA-sensitivity2. This revealed that PAD2 was consistently up-regulated ~4-fold (range 3-32 fold) at 20°C. Analysing two trypanosome strains (T. brucei AnTat 1.1 and T. brucei EATRO 2340) demonstrated 4-fold and 3.8-fold up-regulation of PAD2, respectively, at 20°C when compared to 37°C (Figure 3a). In contrast, PAD1 expression was unaltered. This eliminated the possibility that the elevation of PAD2-expression was due to general enhancement of membrane protein expression at low-temperature. Other stress conditions such as pH or cell concentration did not affect PAD2 expression (Supplementary Fig. 4 and data not shown). Interestingly, at 37°C PAD2 was predominantly at the flagellar pocket, whereas it was surface located at 20°C (Figure 3b, c), this redistribution occurring within 60 min (Supplementary Fig. 5). Thus, PAD2 is a novel example of both a thermoregulated parasite molecule1 and a transmembrane protein exhibiting regulated surface distribution20.
Figure 3. PAD2 is cold-inducible.
- (a) PAD protein expression at 37°C or 20°C in stumpy-forms of T. brucei EATRO 2340 or T. brucei AnTat1.1. Samples were stained for all PAD proteins (“Array”), PAD1 or PAD2. α-tubulin controlled for loading.
- (b) Confocal immunofluorescence-images of paraformaldehyde-fixed stumpy-forms incubated at 37°C or 20°C and co-labelled for α-tubulin (red), PAD2 (green) and DAPI (blue). At 37°C, PAD2 predominantly localised to the flagellar pocket (f.p.; arrowed); at 20°C PAD2 located at the cell surface.
- (c) Quantitation of the PAD2 location at 37°C or 20°C.
PAD proteins convey the CCA differentiation signal
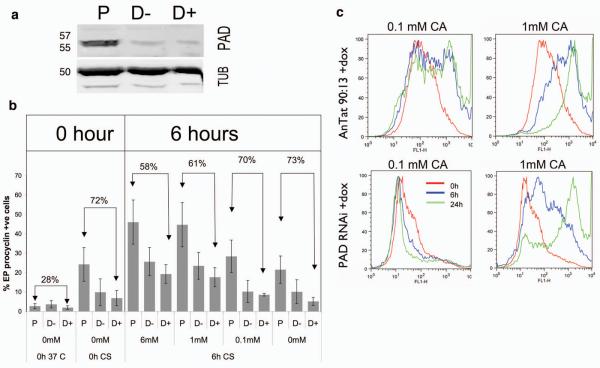
To functionally test PAD proteins in CCA-initiated differentiation, a gene fragment with >95% identity between each PAD gene was targeted by RNAi, enabling simultaneous knock-down of all members. This was performed in the pleomorphic T. brucei AnTat 90:13 line2 that generates stumpy-forms at high frequency, exhibits cold-sensitivity to CCA and has been engineered for doxycycline-inducible RNAi-mediated transcript ablation. The resulting transgenic cell-line, in parallel with the parental line, was grown in mice ±doxycycline induction for 6 days, each producing >80% stumpy populations (Supplementary Fig. 6). Although the RNAi effect was considerably leaky, there was 80% depletion of PAD proteins in the RNAi-induced cells (Figure 4a), demonstrating that significant PAD expression was not required for either stumpy formation or survival (Supplementary Fig. 6). Once the transgenic stumpy-forms were harvested from blood and incubated for 16h at 20°C, they were exposed to different concentrations of CA and differentiation monitored by EP-procyclin expression. Cells incubated at 20°C exhibited the expected cold-induction of EP-procyclin expression2 although this was consistently and inducibly reduced in the RNAi line (Figure 4b, “0h CS” samples). This may indicate some interaction between PAD and EP-procyclin surface expression similar to mammalian monocarboxylate transporters, which co-associate with single-pass membrane proteins for surface access and activity21. Most significantly, however, PAD-depletion reduced the differentiation of cells exposed to CA, with this being more effective at 20°C (F1,44=16.82, p<0.0005 at 6h) than 37°C (F1,44=6.60, p<0.014). Indeed, at 0.1mM CA there was a 70% reduction of EP-procyclin expression compared to the control line at 6h, this expression being further reduced over 24h due to reversal of cold-induction at 27°C (Figure 4C; Supplementary Fig. 7). Matching the response to CA, PAD RNAi also diminished the response of stumpy cells to citrate at 20°C (Supplementary Fig. 8). A cell-line specific differentiation-defect unrelated to PAD RNAi was eliminated by transiently-transfecting pleomorphic cells with the RNAi construct, this recapitulating the differentiation phenotype (Supplementary Fig. 9). Moreover, the RNAi lines differentiated as well as parental cells in response to pronase treatment, an alternative differentiation trigger (Supplementary Fig. 10)15. Hence, ablating PAD mRNAs reduced overall differentiation responses to CCA at all concentrations tested but specifically abrogated the cold-induced CCA-hypersensitivity of stumpy-forms.
Figure 4. RNAi against all PAD genes reduces differentiation.
- (a) PAD-array expression in parental or PAD-RNAi lines (±doxycycline, D+, D) after 16h at 20°C. Tubulin, loading control.
- (b) EP-procyclin in Parental (‘P’) and PAD-RNAi stumpy-forms grown ±doxycycline (D+, D−) after 16h at 20°C, then incubated at 27°C with a CA titration. Means (±s.e.m.) of three experiments are shown, as is the % reduction in EP-procyclin in the PAD-RNAi cells compared with parental cells.
- (c) Flow-cytometry of EP-procyclin expression in PAD-RNAi or parental cells incubated with 0.1mM or 1mM CA. Cold-induced hypersensitivity to CA is ablated in the PAD-RNAi line; at ≥1mM CA both populations differentiate, although this is reduced in the RNAi line. Full flow cytometry data are available in Supplementary Fig. 7.
Conclusions
These experiments demonstrate that the PAD proteins act as transducers of the CCA differentiation signal in Trypanosoma brucei. This conclusion is based on several lines of evidence: (i) PAD proteins are surface molecules expressed on stumpy-forms but absent in transmission-incompetent slender-forms (ii) at the single-cell level, PAD protein expression correlates precisely with the differentiation capacity of bloodstream-form parasites (iii) at least one PAD protein (PAD2) demonstrates cold-regulated expression and localisation, consistent with predictions for the reception of the CCA signal in vivo and (iv) depletion of PAD protein expression in pleomorphic trypanosomes reduces their competence for differentiation and eliminates CCA-responsiveness at physiologically-relevant concentrations. In laboratory-adapted monomorphic slender trypanosomes, which do not express detectable levels of PAD proteins but which can differentiate in response to CCA, signalling is likely driven through other transporters by the high concentrations of CCA required for differentiation in these cell lines14. Consistent with this, incubating monomorphic slender forms, T. brucei AnTat 1.1 90:13 stumpy forms and the PAD RNAi stumpy forms with 14C citrate revealed a clear correlation between PAD expression and cell-associated label (Supplementary Fig. 11). All of these characteristics are compatible with current models for the reception of the signal to differentiation from bloodstream to procyclic-forms8, with the relay of the CCA signal by PAD proteins providing the first molecular insight linking environmental-sensing to trypanosome cell-type differentiation. Notably, this does not exclude an additional or complementary role of proteases in the tsetse midgut, which can act with CCA to promote robust differentiation22.
The identification of PAD1 as a stumpy-specific surface marker protein also provides two key advances. Firstly, the precise correlation between PAD1 expression and differentiation-capacity directly confirms stumpy-forms as the essential transmissible-stage in the bloodstream, supporting classical observations23 but contrasting with some recent models24. Secondly, the discovery of PAD1 permits the quantitative modelling of trypanosome population dynamics in chronic infections25 and the development of bioassays to detect stumpy-formation. This has obvious application for monitoring the activity of the signal for quorum-sensing, SIF, and in high-throughput screens for therapeutic agents that promote stumpy-formation and hence prevent parasite virulence.
Methods Summary
PAD RNAi constructs
PAD protein sequences from GeneDB were analysed using TMHMM26 and displayed using TMRPres2D27 to determine the position of the transmembrane helices. The PAD1 reading frame was amplified from genomic DNA by polymerase chain reaction using 5′-tttaagctttgatcaatgagcgcacccgtcgacaacgtc-3′ and 5′-aaactcgagcatatgtcattgcggagcagcctcacgggc-3′ primers, and cloned into the HindIII-XbaI and XhoI-BamHI cloning sites of pALC1428 to generate the PAD RNAi plasmid.
Parasite growth and transfection
Culturing, transfection, differentiation, and cold-shock assays were performed as described2,29 on T. brucei AnTat1.1 90-132 and selected using 0.5 μg ml−1 puromycin. Stumpy-enriched populations were obtained by DEAE-cellulose purification30 of parasites 6-7 days post-infection into cyclophosphamide-treated mice.
PAD-expression analysis
Peptides specific to PAD1 (H2N-CPKEPTRDAREAAPQ-COOH and H2N-ETCCRREVAE-COOH), PAD2 (H2N-EAEDNQTNAENVC-COOH and H2N-CNADACLEEKAADSSK-COOH) and the entire array (H2N-VETDVDYIAPQFQET-CONH2 and H2N-TQQADKLGQDVCTER-COOH) were used to generate antipeptide antibodies (Eurogentec). Immunofluorescence analysis was performed31 using a Zeiss Axioscope 2 or Leica SP5 confocal microscope and analysed using Volocity software (Improvision). Images were processed using Adobe Photoshop CS. Western blotting was performed by low-voltage SDS-PAGE and wet transfer onto Immobilon-P PVDF (Millipore) according to the manufacturer's instructions; proteins were detected using the LI-COR Odyssey system for quantification against a tubulin loading control. Flow cytometry analysis was performed using the FACS-Calibur flow cytometer (Becton Dickenson)11.
Xenopus oocyte transport assays
PAD genes were cloned into pGHJ and radiolabelled citrate experiments were performed as described32. Uptake measurements were made over 1h in oocytes (5-days post-injection) incubated at pH 9 and 27.5°C.
Statistical analysis
Differentiation data were analysed using a general linear model. Residuals did not conform to a normal distribution and therefore a logarithmic transformation was used. Statistical analysis was carried out using minitab version 15, with P values of p< 0.05 being considered statistically significant.
Methods
Trypanosomes
Trypanosoma brucei brucei AnTat1.1 and Trypanosoma brucei brucei EATRO 2340 were used. Stumpy forms were generated by 5-6 days growth in MF1 mice, treated with cyclophosphamide 24h prior to infection. Slender and monomorphic slender parasites were generated after 3 days growth in rodents.
For pleomorphic transfection, cells from an early-stage parasitaemia (3–4 days, depending on the infection) were purified from the buffy coat and the cells transfected using an AMAXA nucleofector protocol (T-cell nucleofection buffer, programme X001). Drug selection was carried out in cells maintained in HMI-9 media supplemented with 1.1% methyl cellulose at a concentration of approximately 5 × 106 cell ml−1. Transgenic parasites were selected using 0.5 μg ml−1 puromycin.
Macroarray analysis
Expression differences between the DiD1 and parental progenitor were carried out by reverse transcription of 5μg of polyA+ RNA from each cell type, this being labelled using the GE Healthcare Gene Images non-radioactive labelling system. Macroarrays, a gift from Professor Elisabetta Ullu, Yale University, USA, comprised 15,000 plasmid clones of 1-2Kb sheared DNA (~0.8x genome coverage) from Trypanosoma brucei TREU 927/4 cloned into pUC18 and arrayed in 384 microtitre dishes over 39 plates. Bacterial clones were spotted onto 22cm2 nylon membranes in a 4×4 array format. After hybridization, signals were detected by ECL and differential signals, obtained in duplicate, were identified and validated by northern analysis.
Western analysis of PAD proteins
Cells were resuspended at 3 × 106 cells in 10 μl of Laemmli sample buffer containing β-mercaptoethanol at room temperature. Genomic DNA was then sheared by sonication and the sample placed upon ice. 10 μl of protein sample was then resolved on a 10% polyacrylamide gel at 100V at 4°C for approximately 4 hours using chilled buffers. Gels were blotted onto PVDF Immobilon-P (Millipore) at 4°C using a wet blotting system (BioRAD) with chilled buffers. For western blotting, primary antibodies were used at 1:1000 and secondary antibodies were used at 1:5000. Detection after primary antibody incubation used IRDye 680 Goat Anti-Rabbit IgG or IRDye 800CW Goat Anti-Mouse IgG, and was analysed via a Li-COR Odyssey Imager.
Immunofluorescence
For methanol fixation to preserve the overall cell shape, cells were spread onto microscope slides, and air-dried prior to fixation in methanol at −20°C for at least 10 minutes. Cells were rehydrated in PBS for 10 minutes prior to labelling, this being carried out as for paraformaldehyde fixed cells. For paraformaldehyde fixation, 2×106 cells were resuspended in 100μl vPBS (8g/l NaCl; 0.22g/l KCl; 2.27g/l Na2HPO4; 0.41g/l KH2PO4; 15.7g/l sucrose, 1.8g/l glucose; pH7.4), and then an equal volume of 6% paraformaldehyde added. After 10 minutes the suspension was diluted to 5ml in vPBS and settled onto poly-L-lysine coated slides for 20 minutes and then washed with PBS. The cells were then permeabilised in 0.05% Triton X100 for 10 minutes, blocked in 20 % foetal calf serum (FCS) in vPBS for 45 minutes, and stained with the primary antibody (1:100) in 20 % FCS in vPBS for >1 hour.
Subsequent to washing in excess PBS, the cells were stained with the secondary antibody (1:500) in 20 % FCS: vPBS for 1 hour, washed in excess PBS 3 × 5 minutes after which the cellular DNA was stained with a 1 μg ml−1 4′,6-diamidino-2-phenylindole. Cells were mounted in MOWIOL containing phenylene diamine.
Flow cytometry
Between 2 – 5 × 106 cells were fixed in 2 % formaldehyde/ 0.05 % glutaraldehyde for a minimum of 1 hour at 4 °C. Subsequently, the cell suspension was pelleted and resuspended in 200 μl of EP procyclin antibody (Cedar Lane Laboratories) diluted 1:500 in 2 % BSA in PBS. Cells were washed twice prior to being stained with the primary antibody, washed twice and stained with the secondary antibody (1:500). Flow cytometry data were analysed using FlowJO (Tree Star Inc.) software, with unstained cells, and cells stained with only the secondary antibody providing negative controls.
Image acquisition equipment and settings
Immunofluorescence microscopy images (Figure 2c) were captured on a Zeiss axioskop 2 (Carl Zeiss microimaging, Inc.) with a Prior Lumen 200 light source using a QImaging (QImaging, Bucks, UK) Retiga 2000R CCD camera; objectives were either Plan Neofluar 63x (1.25 NA) or Plan Neofluar 100x (1.30 NA). Images were captured via QImage (QImaging, Bucks, UK) and pseudocoloured using Adobe photoshop CS. Confocal imaging (Figure 2a; Figure 3c) used a Leica SP5 confocal laser scanning microscope, using x63 oil immersion objective (N.A.=1.4), with 4.2x zoom. The green channel was imaged using a 488nm argon laser, the red channel was imaged using a 543nm Helium/Neon laser and the image was acquired at 1024/1024 voxels for x/y resolution with sequential optical sections of 0.54μm in z-axis increments. The image was optimised by adjusting laser power and detector sensitivity to minimise bleaching and maintain a digital signal of between 0-255 to avoid signal loss or saturation. The final image was acquired using Volocity™ Software (Improvision Ltd, Coventry, UK) Version 4.4.
Acknowledgements
We thank Elisabetta Ullu (Yale University) for the gift of genomic macroarrays, Markus Engstler (University of Darmstadt, Germany) and Michael Boshart (University of Munich, Germany) for the gift of the AnTat 90:13 line, and Andre Schneider (University of Bern, Switzerland) for pALC14. We thank Deborah Hall, Pamela Davies and Dan Levin for technical assistance, Paula MacGregor for statistical analysis and Athina Paterou and Dianne Murray for Image analysis. This work was supported by a Wellcome Trust project grant and programme grant to KM. SD was supported by a BBSRC studentship, a Wellcome Trust Programme Grant to KM and by a Journal of Cell Science Travelling fellowship for a visit to the laboratory of KK. Support was also provided through a Wellcome Trust strategic award for the Centre for Immunity, Infection and Evolution and a BBSRC REI award for confocal facilities.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Reprints and permissions information is available at npg.nature.com/reprintsandpermissions.
The authors have no competing financial interests.
References
- 1.Fang J, McCutchan TF. Thermoregulation in a parasite's life cycle. Nature. 2002;418(6899):742. doi: 10.1038/418742a. [DOI] [PubMed] [Google Scholar]
- 2.Engstler M, Boshart M. Cold shock and regulation of surface protein trafficking convey sensitization to inducers of stage differentiation in Trypanosoma brucei. Genes Dev. 2004;18(22):2798. doi: 10.1101/gad.323404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee SH, Stephens JL, Paul KS, Englund PT. Fatty acid synthesis by elongases in trypanosomes. Cell. 2006;126(4):691. doi: 10.1016/j.cell.2006.06.045. [DOI] [PubMed] [Google Scholar]
- 4.Hao Z, et al. Tsetse immune responses and trypanosome transmission: implications for the development of tsetse-based strategies to reduce trypanosomiasis. Proc. Natl. Acad. Sci. USA. 2001;98(22):12648. doi: 10.1073/pnas.221363798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zilberstein D, Shapira M. The role of pH and temperature in the development of Leishmania parasites. Ann. Rev. Microbiol. 1994;48:449. doi: 10.1146/annurev.mi.48.100194.002313. [DOI] [PubMed] [Google Scholar]
- 6.Billker O, et al. Identification of xanthurenic acid as the putative inducer of malaria development in the mosquito. Nature. 1998;392(6673):289. doi: 10.1038/32667. [DOI] [PubMed] [Google Scholar]
- 7.Barrett MP, et al. The trypanosomiases. Lancet. 2003;362(9394):1469. doi: 10.1016/S0140-6736(03)14694-6. [DOI] [PubMed] [Google Scholar]
- 8.Fenn K, Matthews KR. The cell biology of Trypanosoma brucei differentiation. Curr. Op. Microbiol. 2007;10(6):539. doi: 10.1016/j.mib.2007.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Szoor B, et al. Protein tyrosine phosphatase TbPTP1: a molecular switch controlling life cycle differentiation in trypanosomes. J. Cell Biol. 2006;175(2):293. doi: 10.1083/jcb.200605090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tasker M, et al. A novel selection regime for differentiation defects demonstrates an essential role for the stumpy form in the life cycle of the African trypanosome. Mol. Biol. Cell. 2000;11(5):1905. doi: 10.1091/mbc.11.5.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vassella E, Reuner B, Yutzy B, Boshart M. Differentiation of African trypanosomes is controlled by a density sensing mechanism which signals cell cycle arrest via the cAMP pathway. J Cell Sci. 1997;110(Pt 21):2661. doi: 10.1242/jcs.110.21.2661. [DOI] [PubMed] [Google Scholar]
- 12.Bruce D, et al. The morphology of the trypanosome causing disease in man in Nyasaland. Proc. Roy. Soc. B. 1912;85:423. [Google Scholar]
- 13.Turner CM, Barry JD, Vickerman K. Loss of variable antigen during transformation of Trypanosoma brucei rhodesiense from bloodstream to procyclic forms in the tsetse fly. Parasitol. Res. 1988;74(6):507. doi: 10.1007/BF00531626. [DOI] [PubMed] [Google Scholar]
- 14.Czichos J, Nonnengaesser C, Overath P. Trypanosoma brucei: cis-aconitate and temperature reduction as triggers of synchronous transformation of bloodstream to procyclic trypomastigotes in vitro. Exp. Parasitol. 1986;62(2):283. doi: 10.1016/0014-4894(86)90033-0. [DOI] [PubMed] [Google Scholar]
- 15.Hunt M, Brun R, Kohler P. Studies on compounds promoting the in vitro transformation of Trypanosoma brucei from bloodstream to procyclic forms. Parasitol. Res. 1994;80(7):600. doi: 10.1007/BF00933009. [DOI] [PubMed] [Google Scholar]
- 16.Jacobs SL, Lee ND. Determination of Citric Acid in Serum and Urine Using Br82. J. Nucl. Med. 1964;5:297. [PubMed] [Google Scholar]
- 17.Sherwin T, Gull K. The cell division cycle of Trypanosoma brucei brucei: timing of event markers and cytoskeletal modulations. Phil. Trans. Roy. Soc. London. 1989;323(1218):573. doi: 10.1098/rstb.1989.0037. [DOI] [PubMed] [Google Scholar]
- 18.Shapiro SZ, Naessen J, Liesegang B, et al. Analysis by flow cytometry of DNA synthesis during the life cycle of African trypanosomes. Acta tropica. 1984;41(4):313. [PubMed] [Google Scholar]
- 19.Matthews KR, Gull K. Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J. Cell Biol. 1994;125(5):1147. doi: 10.1083/jcb.125.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kamsteeg EJ, et al. MAL decreases the internalization of the aquaporin-2 water channel. Proc. Natl. Acad. Sci. USA. 2007;104(42):16696. doi: 10.1073/pnas.0708023104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kirk P, et al. CD147 is tightly associated with lactate transporters MCT1 and MCT4 and facilitates their cell surface expression. EMBO J. 2000;19(15):3896. doi: 10.1093/emboj/19.15.3896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sbicego S, et al. The use of transgenic Trypanosoma brucei to identify compounds inducing the differentiation of bloodstream forms to procyclic forms. Mol. Biochem. Parasitol. 1999;104:311. doi: 10.1016/s0166-6851(99)00157-7. [DOI] [PubMed] [Google Scholar]
- 23.Robertson M. Notes on the polymorphism of Trypanosoma gambiense in the blood and its relation to the exogenous cycvle in Glossina palpalis. Proc. Roy. Soc. B. 1912;85:241. [Google Scholar]
- 24.Bass KE, Wang CC. The in vitro differentiation of pleomorphic Trypanosoma brucei from bloodstream into procyclic form requires neither intermediary nor short-stumpy stage. Mol. Biochem. Parasitol. 1991;44(2):261. doi: 10.1016/0166-6851(91)90012-u. [DOI] [PubMed] [Google Scholar]
- 25.Lythgoe KA, Morrison LJ, Read AF, Barry JD. Parasite-intrinsic factors can explain ordered progression of trypanosome antigenic variation. Proc. Natl. Acad. Sci. USA. 2007;104(19):8095. doi: 10.1073/pnas.0606206104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tusnady GE, Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics (Oxford, England) 2001;17(9):849. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- 27.Spyropoulos IC, Liakopoulos TD, Bagos PG, Hamodrakas SJ. TMRPres2D: high quality visual representation of transmembrane protein models. Bioinformatics (Oxford, England) 2004;20(17):3258. doi: 10.1093/bioinformatics/bth358. [DOI] [PubMed] [Google Scholar]
- 28.Pusnik M, et al. Pentatricopeptide repeat proteins in Trypanosoma brucei function in mitochondrial ribosomes. Mol. Cell. Biol. 2007;27(19):6876. doi: 10.1128/MCB.00708-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.McCulloch R, et al. Transformation of monomorphic and pleomorphic Trypanosoma brucei. Methods in molecular biology (Clifton, N.J. 2004;262:53. doi: 10.1385/1-59259-761-0:053. [DOI] [PubMed] [Google Scholar]
- 30.Lanham SM. Separation of trypanosomes from the blood of infected rats and mice by anion-exchangers. Nature. 1968;218(5148):1273. doi: 10.1038/2181273a0. [DOI] [PubMed] [Google Scholar]
- 31.Field MC, et al. New approaches to the microscopic imaging of Trypanosoma brucei. Microsc Microanal. 2004;10(5):621. doi: 10.1017/S1431927604040942. [DOI] [PubMed] [Google Scholar]
- 32.Saliba KJ, et al. Sodium-dependent uptake of inorganic phosphate by the intracellular malaria parasite. Nature. 2006;443(7111):582. doi: 10.1038/nature05149. [DOI] [PubMed] [Google Scholar]