Abstract
Autism is a common, severe and highly heritable neurodevelopmental disorder. The International Molecular Genetic Study of Autism Consortium (IMGSAC) genome screen for linkage in affected sib-pair families identified a chromosome 7q susceptibility locus (AUTS1), that has subsequently shown evidence of increased sharing in several independent multiplex samples and in two meta-analyses. Taking into account the location of the MET gene under this linkage peak, and the fact that it has recently been reported to be associated with autism, the gene was further analyzed as a promising autism candidate. The gene encodes a transmembrane receptor tyrosine kinase of the hepatocyte growth factor/scatter factor (HGF/SF). MET is best known as an oncogene, but its signalling also participates in immune function, peripheral organ development and repair, and the development of the cerebral cortex and cerebellum (all of which have been observed earlier as being disregulated in individuals with autism). Here we present a family-based association analysis covering the entire MET locus. Significant results were obtained in both single locus and haplotype approaches with a single nucleotide polymorphism in intron 1 (rs38845, P<0.004) and with one intronic haplotype (AAGTG, P<0.009) in 325 multiplex IMGSAC families and 10 IMGSAC trios. Although these results failed to replicate in an independent sample of 82 Italian trios, the association itself was confirmed by a case–control analysis performed using the Italian cohort (P<0.02). The previously reported positive association of rs1858830 failed to replicate in this study. Overall, our findings provide further evidence that MET may play a role in autism susceptibility.
Keywords: autism, hepatocyte growth factor receptor, 7q31.2, nuclear families, case–control study, linkage disequilibrium
Introduction
Autism (Mendelian Inheritance in Man (MIM) 209850) is a severe, complex neurodevelopmental disorder, characterized by impairments in reciprocal social interaction and communication, and restricted and stereotyped patterns of interests and behaviours.1 It predominantly affects males with a sex ratio of approximately 4:1. The latest study in the United Kingdom indicates that the general population prevalence is approximately 38.9 in 10 000, with no apparent systematic geographic or socioeconomic variation.2 Autism spectrum disorders (ASDs) refer to a broader definition of autism, which include classical and atypical autism, Asperger syndrome, and pervasive developmental disorder not otherwise specified and belong to the group of closely related pervasive developmental disorders.3 When the different ASDs are combined, the total prevalence can be as high as 116.1 per 10 000 children.2 Prevalence estimates have risen over the last two decades, likely because of an increased awareness of the disorder and because of broadening and improvement of the diagnostic tools.4 Although the aetiology of autism is not yet understood, it is a highly heritable disorder, with an age of onset usually before 3 years of age, with symptoms persisting throughout life.5
Following the first published genome-wide linkage screen in 19986 several independent studies for autism have been conducted. Evidence for suggestive linkage has been found on almost all chromosomes with little concordance between studies. However, one of the most consistently replicated loci, is located on chromosome 7q. The Autism Susceptibility Locus 1 (AUTS1) region, identified by the International Molecular Genetic Study of Autism Consortium (IMGSAC) on chromosome 7q,6 was further established in two follow-up studies conducted by IMGSAC using additional families and markers.7, 8 It has also subsequently shown consistent positive linkage results in several other studies.9, 10, 11, 12, 13, 14, 15, 16 However, the significance of linkage and precise location of the peak varies between these studies. Thus the peak of linkage in this area remains broad, spanning more than 40 Mb and containing over 200 mapped genes.
The human gene MET (proto-oncogene hepatocyte growth factor receptor; MIM 164860) is located on chromosome 7q31.2, under the linkage peak of interest for IMGSAC families (7q21.3-7q34). MET covers approximately 126 kb and encodes a high-affinity transmembrane receptor tyrosine kinase of the hepatocyte growth factor/scatter factor (HGF/SF). HGF is a mesenchymally derived growth factor that stimulates epithelial cell mitogenesis, motility, invasion and morphogenesis. MET and its ligand are expressed in numerous tissues, although predominantly in cells of epithelial and mesenchymal origin, respectively.17 The receptor comprises an N-terminal α-chain, located outside the membrane, a C-terminal β-chain that contains an extramembrane sequence, a single transmembrane domain and a cytoplasmic tyrosine kinase domain.18, 19 The kinase activity and the receptor's dimerization are essential for the active form of the receptor, and this activation will consequently trigger a number of signalling pathways in target cells.20
MET was primarily identified as a proto-oncogene21 and has a well documented role as a dominant oncogene in tumour development and progression, being overexpressed and/or deregulated in diverse human tumours.22, 23, 24, 25 Furthermore, MET signalling participates in the immune system regulation,26, 27, 28 embryogenesis20 and in the peripheral organ development and repair (such as gastrointestinal29). MET and HGF, both expressed in the developing nervous system, have been implicated in neuronal development,18, 30, 31, 32 more specifically in the cerebral cortex33, 34 and cerebellum.35 Impaired MET/HGF signalling interferes with interneuron migration and disrupts neuronal growth in the cortex,33, 34 and also leads to a decreased proliferation of granule cells, causing a parallel reduction in the size of the cerebellum.35 Interestingly, these features have been observed in the brains of autistic individuals.34, 36, 37, 38
Recently, Campbell et al38 reported genetic and molecular biological evidence pointing to a common functional promoter variant in the MET gene as a contributing risk factor for autism susceptibility. They showed significant overtransmission of the rs1858830 C allele in 204 autistic families (P=0.0005), confirmed in a replication sample of 539 families (P=0.001). Also, the C allele was significantly less prevalent in a group of 189 unrelated controls than in the cases or their parents. Expression studies determined that the C allele results in decreased MET promoter activity and altered binding of specific transcription factor complexes, suggesting a link between reduced gene expression and autism susceptibility.38 MET protein levels were also significantly decreased in ASD cases compared with controls and this was accompanied in ASD brains by increased mRNA expression for proteins involved in regulating MET signalling activity.39
In this study, the role of the MET gene in autism susceptibility was analyzed in the IMGSAC family collection following previous reports of association. The entire known variation within the MET locus was covered to evaluate whether any specific gene variants or haplotypes were associated with autism, thus seeking a better understanding of potential mechanisms underlying the previous positive findings for autism in the 7q region.
Materials and methods
Subjects
The collection and identification of families, assessment methods and inclusion criteria used by IMGSAC are described elsewhere.7 Briefly, after an initial screen, parents undertook the Autism Diagnostic Interview-Revised40 and the Vineland Adaptive Behaviour Scales.41 Probands were administered the Autism Diagnostic Observation Schedule42 and a medical examination was performed to exclude recognizable medical causes of autism, particularly tuberous sclerosis and neurofibromatosis. Karyotyping was undertaken where possible on all affected individuals and molecular genetic testing for Fragile X syndrome performed on one case per family. DNA was extracted from blood samples, buccal swabs or cell lines by use of a DNA purification kit (Nucleon, Manchester, UK) and standard techniques. Written informed consent was given by all parents/guardians and, where possible, by affected individuals. The study has been approved by the relevant ethical committees. For this experiment, a total of 1621 Caucasian individuals from 325 multiplex IMGSAC families and 10 IMGSAC trios were genotyped. The male:female ratio of the affected individuals is 3.3:1. Also, 82 Italian trios from the IMGSAC collection were used in the replication study.
A sample of 185 UK controls from ECACC (European Collection of Cell Cultures) and 88 Italian controls randomly chosen were used for the case–control study. The Italian cohort (samples and controls) used in this study is entirely independent from the Campbell et al study.38
Single nucleotide polymorphism (SNP) genotyping
SNP selection
With the intention of capturing the maximum amount of genetic variation in MET, genotyping data from CEPH (Centre D'étude du Polymorphisme Humain) individuals was downloaded from the HapMap phase II (release 21), for the 7q31.2 region, including 5 kb upstream and downstream of the gene (chromosome 7: 115890847–116029719). Thirty-one haplotype-tagging SNPs were selected using Tagger from Haploview v4.043 (r2>0.8 and minor allele frequency >0.05, aggressive tagging), covering the selected region. In addition, four non-synonymous SNPs and rs1858830 (the promoter variant reported earlier by Campbell et al38) were also chosen (Figure 1). Thirty-four of these were genotyped using the Sequenom MALDI-TOF iPLEX platform (Sequenom, San Diego, CA, USA) and two by a restriction fragment length polymorphism assay.
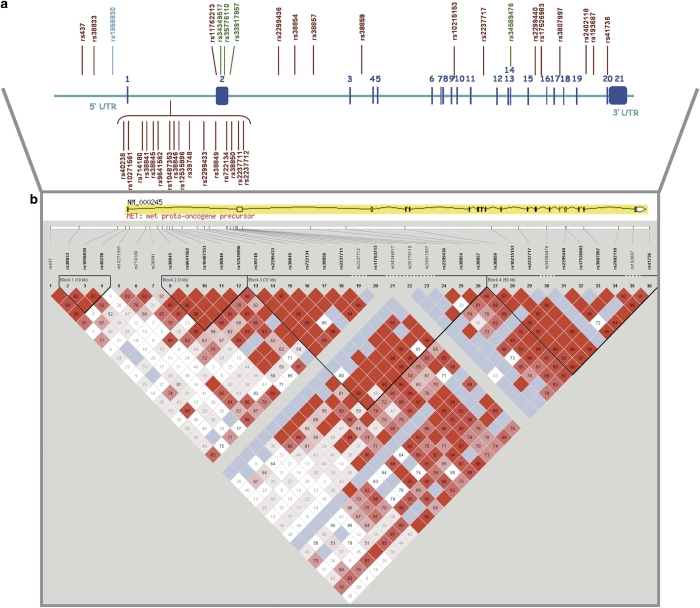
Figure 1.
(a) Schematic representation of the 36 SNPs chosen to cover the MET locus and their respective locations. The 21 exons are indicated by solid blue boxes and numbered. The SNPs represented include the 31 haplotype-tagging SNPs, 4 non-synonymous variants (in green) and rs1858830 (promoter variant in light blue). (b) The graphical output from Haploview for the MET gene, including the markers tested and the haplotype blocks constructed. Analysis of the markers selected to cover the genetic variation within the MET locus showed four different LD blocks. D′ values are given (bright red corresponds to D′=1, with the colour tending towards white as D′ tends towards 0).
Sequenom iPLEX assay
The genotyping assay was designed using MassARRAY design software (Sequenom) and genotypes were obtained using the MassARRAYTyper system (version 3.1.4.0). The 34 SNPs were genotyped with sample and SNP genotyping success rates of ∼99 and 100%, respectively. The Picogreen dsDNA Quantitation Kit (Invitrogen, OR, USA) was used to confirm the concentration of all the DNA samples and 40 ng of DNA per sample was used in the genotyping assay. Two control individuals were included on each plate as genotyping controls for inter-plate reproducibility. Genotype Analyzer (Sequenom) was used to check the quality of genotypes and to assign alleles where possible. The data were exported and uploaded into the integrated genotyping system44—an in-house database.
Restriction digest
The restriction fragment length polymorphism assay was selected for genotyping three markers: rs1858830, rs2237711 in IMGSAC cases (as they were incompatible with the Sequenom system) and rs38845 in the Italian cohort and in the ECACC controls. Primers were designed using Primer3 (http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi) and BLAT (Blast-like alignment tool) analysis was performed (using the University of California – Santa Cruz (UCSC) database) to confirm their specificity. Primer sequences used are as follows: forward (f)-GATTTCCCTCTGGGTGGTGC/reverse (r)-CAAGCCCCATTCTAGTTTCG for rs1858830, (f)-GGGCTATGTCCCATTTCTCA/(r)-GGCAGGGTAGTAGGTTGGAA for rs2237711 and (f)-CTGATTCTGCCCTCACTTCAG/(r)-AAGTATGTGTTAGTGAGACCGAAA for rs38845.
For rs1858830, the KOD Hot Start DNA Polymerase kit (Novagen, Osaka, Japan) was used owing to the high GC content in this region (∼85%). A touchdown-PCR (95 °C for 30 s, 65 °C for 30 s and 72 °C for 30 s, followed by 10 cycles at decreasing annealing temperatures in decrements of 0.5 °C per cycle, then 25 cycles of 95 °C for 30 s, 60 °C for 30 s and 72 °C for 30 s) was performed in 15 μl reactions containing 50 ng of DNA and a final concentration of 2.0 m MgSO4, 0.33 μ primer, 0.2 m dNTPs, 5% DMSO and 0.05 U KOD Hot Start DNA polymerase (Novagen). The PCR for rs2237711 (35 cycles at 90 °C for 30 s, 62 °C for 30 s and 72 °C for 45 s) was performed in 15 μl reactions containing 50 ng of DNA and a final concentration of 1.25 m MgCl2, 0.4 μ primer, 0.2 m dNTPs and 0.25 U BIOTaq DNA polymerase (Bioline, Bath, UK). The touchdown PCR for rs38845 (95 °C for 60 s, 63 °C for 60 s and 72 °C for 30 s, followed by 13 cycles at decreasing annealing temperatures in decrements of 0.5 °C per cycle, then 28 cycles of 95 °C for 60 s, 56 °C for 30 s and 72 °C for 60 s) was performed in 15 μl reactions containing 50 ng of DNA and a final concentration of 2 m MgCl2, 0.3 μ primer, 0.2 m dNTPs and 0.25 U AmpliTaq Gold DNA polymerase (Applied Biosystem, Foster City, USA). In total 5 μl of PCR products of rs1858830, rs2237711 and rs38845 were separated on 2% agarose gels to check the size and quantity of product and the remaining 10 μl incubated with EagI, HhaI and Hpy188I, respectively (5 U of EagI and 1 × NEB buffer 3, in a total volume of 20 μl; 1 U of HhaI and 1 × NEB buffer 4, supplemented with 1 × BSA in a total volume of 20 μl; 2 U of Hpy188I and 1 × NEB buffer 4, in a total volume of 15 μl) at 37 °C for 3 h and visualized with SYBR-safe (Invitrogen) on a 3% agarose gel. Genotypes were identified and entered into integrated genotyping system. As with the iPLEX assay, two control individuals were included on each plate as genotyping controls for inter-plate reproducibility.
Statistical analysis
Error corrections
Mendelian consistency of SNP genotype data in family pedigrees was checked using PedCheck45 and any inconsistent genotypes were removed. Genotypes flanking double recombinants were detected using Merlin46 and disregarded if ambiguous, as these could be indicative of genotyping errors. All the SNPs were tested for Hardy–Weinberg equilibrium using a χ2-test in founders and probands separately.
Association analysis
The pairwise linkage disequilibrium (LD) map for MET was constructed from genotypes using the Haploview software. Association was assessed using the transmission disequilibrium test (TDT).47 TDT uses genotype data from nuclear families consisting of two parents and one or more affected individuals, and is robust to population stratification. A version robust to non-independent siblings was implemented using the DGCgenetics R library (http://www-gene.cimr.cam.ac.uk/clayton/software), allowing for multiple affected offspring from the same nuclear family to be used in this study. Allele frequencies were reported for all the parents, as well as allele transmission frequencies from parents to offspring. Parental transmissions were also examined for each SNP to consider parent-of-origin effects. A haplotype-based TDT analysis was performed using Transmit48 methodology implemented within the tdthap R library (http://cran.r-project.org). For each haplotype, risk estimates and their 95% confidence regions were estimated using a Bayesian method.47, 48
Logistic regression was applied in the analysis of both the alleles and genotypes in the case–control study. We assessed the robustness of results through permutation approaches by sampling and analyzing multiple (50 000) datasets consisting of one affected case per family from the multiplex families.
Correction for multiple testing in family-based study
Performing multiple statistical tests leads to an inflation in the occurrence of false positives and it is necessary to adjust the P-value significance threshold (usually 5%) to account for the number of independent tests. Because of the high LD within the region, a Bonferroni correction using the total number of SNPs would be too conservative.48 By considering the LD pattern, which identified four independent haplotype blocks, it is reasonable to interpret a P<0.0125 to be statistically significant. A separate permutation approach found a similar threshold, established for the principal analyses. However, secondary tests, such as assessing parent-of-origin effect, which we know require larger sample sizes to be meaningful, are not reflected in this threshold.
Bioinformatic analysis
A search for transcription factor binding sites was conducted using the online tools MatInspector49 and TFSearch (http://www.cbrc.jp/research/db/TFSEARCH.html). These software programmes utilize a large library of matrix descriptions for transcription factor binding sites to locate matches in DNA sequences.50
Results
Family-based studies
SNP genotyping and association analysis
A total of 36 SNPs were successfully genotyped across the entire MET locus, all of which were in Hardy–Weinberg equilibrium in both probands and founders (P>0.01). To test the potential association between MET and autism, a family-based analysis was performed using TDT. The P-values for all SNPs tested are reported in Table 1. One SNP (rs38845) showed statistically significant (P<0.01) transmission disequilibrium, having a preferential transmission of the A allele to the affected offspring (P=0.0035, odds ratio =1.3 (95% confidence interval: 1.09, 1.54) – Table 1). This odds ratio of 1.3 indicates that the condition is more likely in the individuals who carry the A allele (conversely, carrying the G allele is protective). Also, parental transmissions were examined for each SNP, but no evidence of parent-of-origin effects was found (P>0.12). Moreover, none of the four non-synonymous SNPs genotyped (rs34349517, rs35776110, rs33917957 and rs34589476) showed association to autism, having very low-heterozygosity content (in particular rs34349517, which was monomorphic in this population). There was 100% concordance with the two control individuals genotyped to control for inter-plate reproducibility.
Table 1. TDT results for the SNPs genotyped across MET.
| SNP ID | Position on 7qa (Mb) | Allele | Frequency | TR | NT | OR | (95% CI) | P-value |
|---|---|---|---|---|---|---|---|---|
| rs437 | 116084132 | C | 0.279 | 182 | 214 | 0.85 | (0.70, 1.04) | 0.107 |
| T | 0.721 | |||||||
| rs38833 | 116090787 | C | 0.840 | 1.00 | (0.59, 1.70) | 1.000 | ||
| T | 0.160 | 27 | 27 | |||||
| rs1858830 | 116099675 | C | 0.556 | 1.06 | (1.26, 0.67) | 0.555 | ||
| G | 0.444 | 261 | 246 | |||||
| rs40238 | 116101563 | A | 0.205 | 33 | 49 | 0.67 | (0.43, 1.05) | 0.070 |
| C | 0.795 | |||||||
| rs10271561 | 116102441 | C | 0.114 | 105 | 117 | 0.90 | (0.69, 1.17) | 0.363 |
| T | 0.886 | |||||||
| rs714180 | 116106238 | A | 0.492 | 258 | 290 | 0.89 | (0.75, 1.05) | 0.166 |
| G | 0.508 | |||||||
| rs38841 | 116107162 | A | 0.612 | 0.90 | (0.75, 1.08) | 0.233 | ||
| G | 0.388 | 219 | 244 | |||||
| rs38845 | 116109038 | A | 0.463 | 289 | 223 | 1.30 | (1.09, 1.54) | 0.0035 |
| G | 0.537 | |||||||
| rs9641562 | 116110027 | A | 0.942 | 0.83 | (0.56, 1.21) | 0.345 | ||
| C | 0.058 | 48 | 58 | |||||
| rs10487353 | 116111172 | G | 0.939 | 0.77 | (0.53, 1.13) | 0.209 | ||
| T | 0.061 | 48 | 62 | |||||
| rs38846 | 116111767 | C | 0.209 | 169 | 178 | 0.95 | (0.77, 1.17) | 0.656 |
| T | 0.790 | |||||||
| rs12535996 | 116112385 | A | 0.057 | 49 | 58 | 0.85 | (0.58, 1.24) | 0.401 |
| G | 0.943 | |||||||
| rs39748 | 116114666 | C | 0.543 | 0.93 | (0.63, 1.36) | 0.719 | ||
| G | 0.457 | 49 | 53 | |||||
| rs2299433 | 116118114 | C | 0.746 | 1.06 | (0.85, 1.33) | 0.634 | ||
| T | 0.254 | 155 | 146 | |||||
| rs38849 | 116119775 | C | 0.270 | 202 | 214 | 0.94 | (0.78, 1.14) | 0.589 |
| G | 0.730 | |||||||
| rs722134 | 116120945 | C | 0.126 | 34 | 27 | 1.26 | (0.76, 2.09) | 0.362 |
| T | 0.874 | |||||||
| rs38850 | 116124885 | A | 0.293 | 240 | 221 | 1.09 | (0.90, 1.30) | 0.374 |
| G | 0.707 | |||||||
| rs2237711 | 116125233 | A | 0.073 | 67 | 72 | 0.93 | (0.67, 1.30) | 0.686 |
| C | 0.927 | |||||||
| rs2237712 | 116125894 | A | 0.965 | 0.53 | (0.24, 1.13) | 0.117 | ||
| G | 0.035 | 10 | 19 | |||||
| rs11762213 | 116126518 | A | 0.067 | 57 | 64 | 0.89 | (0.62, 1.27) | 0.518 |
| G | 0.933 | |||||||
| rs35776110 | 116127333 | C | 0.997 | 0.33 | (0.03, 3.20) | 0.317 | ||
| T | 0.003 | 1 | 3 | |||||
| rs33917957 | 116127498 | A | 0.997 | 0.64 | (0.34, 1.20) | 0.150 | ||
| G | 0.003 | 16 | 25 | |||||
| rs2299436 | 116128619 | A | 0.787 | 1.28 | (0.83, 1.98) | 0.275 | ||
| G | 0.213 | 46 | 36 | |||||
| rs38854 | 116144447 | T | 0.448 | 52 | 53 | 0.98 | (0.67, 1.44) | 0.928 |
| C | 0.552 | |||||||
| rs38857 | 116152649 | C | 0.689 | 0.94 | (0.76, 1.17) | 0.626 | ||
| T | 0.311 | 158 | 168 | |||||
| rs38859 | 116166330 | C | 0.537 | 0.97 | (0.78, 1.19) | 0.757 | ||
| T | 0.463 | 173 | 179 | |||||
| rs10215153 | 116186367 | A | 0.321 | 217 | 236 | 0.92 | (0.76, 1.11) | 0.420 |
| G | 0.679 | |||||||
| rs2237717 | 116192623 | C | 0.557 | 0.95 | (0.66, 1.37) | 0.790 | ||
| T | 0.443 | 55 | 58 | |||||
| rs34589476 | 116199159 | C | 0.994 | 0.43 | (0.11, 1.66) | 0.157 | ||
| T | 0.006 | 3 | 7 | |||||
| rs2299440 | 116203563 | C | 0.816 | 1.17 | (0.90, 1.52) | 0.269 | ||
| T | 0.184 | 118 | 101 | |||||
| rs17526983 | 116203659 | A | 0.070 | 33 | 50 | 0.66 | (0.43, 1.02) | 0.062 |
| G | 0.930 | |||||||
| rs3807997 | 116207794 | C | 0.832 | 1.39 | (0.87, 2.20) | 0.174 | ||
| A | 0.168 | 43 | 31 | |||||
| rs2402118 | 116215809 | A | 0.360 | 263 | 248 | 1.06 | (0.89, 1.26) | 0.532 |
| C | 0.640 | |||||||
| rs193687 | 116219242 | C | 0.960 | 0.85 | (0.53, 1.35) | 0.485 | ||
| T | 0.040 | 33 | 39 | |||||
| rs41736 | 116223004 | C | 0.559 | 1.00 | (0.80, 1.24) | 1 | ||
| T | 0.441 | 160 | 160 |
Abbreviations: CI, confidence interval; NT, non-transmitted; OR, odds ratio; SNP, single nucleotide polymorphism; TDT, transmission disequilibrium test; TR, transmitted.
The position of the SNPs is according to the UCSC Genome Browser March 2006 Assembly.
Significant results are shown in bold.
LD patterns and haplotype analysis
Analysis of the LD patterns across MET (using 31 haplotype-tagging SNPs, along with the 4 non-synonymous SNPs and the rs1858830 promoter variant), defined 4 LD blocks of 10, 3, 37 and 56 kb (Figure 1). The determination of haplotypes of SNPs in LD can offer more power to detect association than testing SNPs individually. Haplotype analysis was performed to further characterize the transmission disequilibrium within the LD blocks. Transmission of haplotypes, including all markers within each block, were tested using Transmit48 (Table 2). Haplotype A–A–G–T–G for markers rs38845–rs9641562–rs10487353–rs38846–rs12535996 was overtransmitted to the affected offspring (P=0.007, odds ratio=1.34 (95% confidence interval: 1.08, 1.66)), revealing a significant transmission distortion in LD block 2. This suggests that this specific haplotype (in intron 1) could potentially increase susceptibility to autism. Statistical significance was not increased compared with single markers and so the association obtained with the five-marker haplotype may be driven by the single-locus association to rs38845.
Table 2. Haplotype transmission disequilibrium results within each LD block for MET haplotypesa and for the haplotype combinations from Haploview, selected by Tagger.
| LD block/SNPs | Haplotype | TR | NT | OR | (95% CI) | P-value |
|---|---|---|---|---|---|---|
| 1st block (3 SNP haplotype) | CCA | 5 | 6 | 0.83 | (0.31, 2.23) | 1.000 |
| rs38833–rs1858830–rs40238 | CCC | 47 | 44 | 1.07 | (0.76, 1.51) | 0.834 |
| CGC | 41 | 35 | 1.17 | (0.80, 1.71) | 0.567 | |
| TCA | 12 | 20 | 0.60 | (0.33, 1.08) | 0.215 | |
| 2nd block (5 SNP haplotype) | AAGCG | 151 | 154 | 0.98 | (0.81, 1.18) | 0.909 |
| rs38845–rs9641562–rs10487353–rs38846–rs12535996 | AAGTG | 199 | 148 | 1.34 | (1.13, 1.61) | 0.007 |
| GAGTG | 207 | 238 | 0.87 | (0.74, 1.02) | 0.155 | |
| GCTTA | 41 | 54 | 0.76 | (0.54, 1.07) | 0.218 | |
| 3rd block (13 SNP haplotype) | CCCTGAAGCAACT | 14 | 27 | 0.52 | (0.30–0.88) | 0.060 |
| rs39748–rs2299433–rs38849–rs722134–rs38850– | CCCTGCAACAACT | 9 | 7 | 1.29 | (0.57, 2.96) | 0.804 |
| rs2237711–rs2237712–rs11762213–rs35776110– | CCCTGCAGCAACT | 30 | 22 | 1.36 | (0.86, 2.17) | 0.332 |
| rs33917957–rs2299436–rs38854–rs38857 | CTGTGCAGCAGCC | 30 | 35 | 0.86 | (0.57, 1.29) | 0.620 |
| CTGTGCAGCAGCT | 10 | 7 | 1.43 | (0.64, 3.24) | 0.629 | |
| GCGCACAGCAATC | 29 | 21 | 1.38 | (0.87, 2.22) | 0.322 | |
| GCGTACAGCAATC | 29 | 32 | 0.91 | (0.59, 1.38) | 0.798 | |
| GCGTGCAGCAATC | 27 | 25 | 1.08 | (0.68, 1.70) | 0.890 | |
| 4th block (10 SNP haplotype) rs38859–rs10215153–rs2237717–rs34589476– rs2299440–rs17526983–rs3807997–rs2402118– rs193687–rs41736 | CACCCGCCCC | 48 | 39 | 1.23 | (0.86, 1.76) | 0.391 |
| CACCCGCCTC | 9 | 10 | 0.90 | (0.42, 1.91) | 1.000 | |
| CGCCCGCCCC | 6 | 12 | 0.50 | (0.22, 1.11) | 0.238 | |
| CGCCTGACCC | 33 | 25 | 1.32 | (0.86, 2.05) | 0.358 | |
| TGTCCACACT | 12 | 18 | 0.67 | (0.36, 1.22) | 0.362 | |
| TGTCCGCACT | 34 | 33 | 1.03 | (0.69, 1.54) | 1.000 | |
| TGTCCGCCCT | 13 | 15 | 0.87 | (0.46, 1.61) | 0.851 |
| Haplotype combinations selected by Tagger | Tagged SNP | Haplotype | TR | NT | OR | (95% CI) | P-value |
|---|---|---|---|---|---|---|---|
| rs38845, rs38846, rs38849 | rs39747 | ATG | 198 | 149 | 1.33 | (1.11, 1.59) | 0.010 |
| rs10215153, rs38859, rs38854 | rs10243024 | GCC | 66 | 54 | 1.22 | (0.90, 1.65) | 0.320 |
| rs38854, rs38849, rs2237717 | rs7798983, rs2237710 | CGC | 80 | 68 | 1.18 | (0.90, 1.65 ) | 0.370 |
| rs2299440, rs2237717, rs3807997 | rs1621, rs41741 | CCC | 64 | 60 | 1.07 | (0.79, 1.43) | 0.788 |
| rs2299436, rs9641562 | rs17138943, rs12539654, rs17138948 | GC | 68 | 75 | 0.91 | (0.69, 1.19) | 0.616 |
Abbreviations: CI, confidence interval; LD, linkage disequilibrium; NT, non-transmitted; OR, odds ratio; SNP, single nucleotide polymorphism; TR, transmitted.
Haplotypes only reported if there are more than 10 informative transmissions in total.
Significant results are shown in bold.
Given that the haplotype-tagging SNPs were chosen using aggressive tagging in Haploview v4.043, the multi-marker haplotype combinations from the Tagger output were also tested (Table 2). The haplotype ATG with markers rs38845–rs38846–rs38849 was significant (P=0.01, odds ratio=1.33 (95% confidence interval: 1.11, 1.59)). This haplotype again contains the significant single marker and the A allele is overtransmitted to the cases, confirming once more the direction of association. This particular combination of markers is tagging a fourth SNP (rs39747). An overall view of the LD structure across the region reveals that this variant is in the haplotype–block 2 boundary, just before rs38845, but is not in complete LD with the latter (D′=95; r2=0.7).
TDT replication study
The SNP showing evidence for association (rs38845) and the promoter variant rs1858830 were further studied in an independent sample of 82 Italian trios. TDT analysis of the two SNPs did not provide significant results (Table 3).
Table 3. TDT results for rs38845 and rs1858830 using the 82 Italian trios. The allele frequencies shown are for the parents only.
| SNP ID | Allele | Frequency | TR | NT | OR | (95% CI) | P-value |
|---|---|---|---|---|---|---|---|
| rs38845 | A | 0.583 | 44 | 43 | 0.98 | (0.64,1.49) | 0.915 |
| G | 0.417 | ||||||
| rs1858830 | C | 0.593 | 1.49 | (0.43, 1.05) | 0.067 | ||
| G | 0.407 | 49 | 33 |
Abbreviations: CI, confidence interval; NT, non-transmitted; OR, odds ratio; TDT, transmission disequilibrium test; TR, transmitted.
Case–control study
A case–control study was also conducted among patients with autism against unselected controls, using allele and genotype data from our SNP of interest (rs38845). The genotypes of 185 ECACC and 88 Italian controls were determined by restriction enzyme digestion. Case–control analysis was performed using logistic regression in IMGSAC probands against ECACC controls and Italian cases against Italian controls. Allelic tests of association applied to the Italian cohort indicated an increased risk of the A allele compared with the G (P=0.017 (95% confidence interval: 1.10, 2.59)). A genotype analysis suggested an increased risk for the AA/GA genotypes compared with GG (a dominant effect of A) of nearly 2.5 times (P=0.02 (95% confidence interval: 1.16, 5.33) (Table 4)). No significant allele or genotype frequency difference was detected in IMGSAC cases vs the sample of ECACC controls.
Table 4. Case–control studies for the rs38845 variant in the United Kingdom and Italian populations.
| OR | OR | |||||||
|---|---|---|---|---|---|---|---|---|
| A vs G | AA/GA vs GG | |||||||
| Cases/controls | Cases n | Case Allele A (%) | Controls n | Control Allele A (%) | (95% CI) | P-value | (95% CI) | P-value |
| IMGSACa/ECACC | 333 | 0.522 | 185 | 0.478 | 1.19 | 0.189 | 1.35 | 0.164 |
| (0.92–1.54) | (0.89, 2.04) | |||||||
| Italian/Italian | 82 | 0.584 | 88 | 0.455 | 1.69 | 0.017 | 2.48 | 0.020 |
| (1.10–2.59) | (1.16, 5.33) |
Abbreviations: CI, confidence interval; IMGSAC, the International Molecular Genetic Study of Autism Consortium; OR, odds ratio.
There are a total of 666 probands, but there are 333 sib-pairs, so we generated 50 000 permutations of unrelated probands and the median (empirical 95% CI): (i) allele A frequency is 0.522 (0.500, 0.543) (ii) OR (A vs G) is 1.19 (1.09, 1.29) and (iii) OR (AA/GA vs GG) is 1.34 (1.16, 1.57).
Bioinformatic analysis of rs38845
The results from MatInspector49 predicted the creation of a binding site for an interferon-stimulated response element (interferon regulatory factor 1 (IRF1) (MIM *147575)) in the region only when the A allele of rs38845 is present. This prediction was confirmed by TFSearch.50 In addition, the TFSearch output showed that the protein C/EBP (CCAAT/enhancer-binding protein) is predicted to bind the same region only when the A allele is present. The binding of these elements could be of importance in the regulation of MET expression.
Discussion
MET has been implicated in brain circuit development and in peripheral functions (such as gastrointestinal repair and immune function),34, 38 which are consistent with observations in a proportion of autistic patients. As the gene is also located in a key area of linkage on chromosome 7, MET was examined as an appealing candidate gene for autism by performing a thorough analysis aiming to cover all its genetic variation. However, the earlier reported genetic association of the common C allele of rs1858830 in the promoter region38 failed to replicate in this study. A family-based analysis was carried out using TDT and Transmit, but there was no evidence of significant association for this marker in the family collection tested. Several explanations can be proposed for this incongruity. The lack of association found with the pre-ascertained variant might be because of the differences in the sample collection, such as the inclusion/exclusion criteria, the different size of family collections and the use of different analytical approaches. Furthermore, although our sample was mostly composed of multiplex families, and Campbell et al reported their association to be stronger in such families, heterogeneity in autism cohorts could still be a potential source of variation. The TDT approach used is robust to population stratification and although it only uses heterozygous parent transmissions, should be an efficient method to fine map susceptibility loci in a region of linkage such as the one on chromosome 7.
The family-based association analysis carried out indicated that there is a genetic association between the marker rs38845 and autism. The A allele is transmitted to the affected offspring more often than expected by chance, suggesting that it increases the risk that carriers of this allele will develop the disorder. In addition, the haplotype analysis revealed that the risk haplotype A–A–G–T–G for the combination of markers within LD block 2 (rs38845–rs9641562–rs10487353–rs38846–rs12535996) and the haplotype ATG (rs38845–rs38846–rs38849) were overtransmitted to the affected offspring. Therefore, these findings indicate that the A allele of rs38845 increases the risk of developing autism, possibly interacting with other susceptibility genes (not yet known), and with still undefined environmental factors. It is noteworthy that rs38845 is not in strong LD with the promoter SNP rs1858830 (Figure 1). We attempted to replicate this finding in an independent sample collection of 82 Italian trios and 88 Italian sex-matched controls. Using TDT, no evidence of association was detected with either rs38845 or rs1858830. Specifically for rs1858830, this result is not completely surprising as the association was first found in multiplex families, while the Italian cohort is composed of simplex families. However, case–control analysis in the Italian cohort revealed a positive association of rs38845, in the same direction. Although the sample size for the Italian case–control sample is arguably small, this result can be taken as a confirmation of the positive result obtained earlier by family-based analysis in the IMGSAC sample set.
Finding different associated variants within the same region (such as ours and Campbell et al, 2006) is often reported.51 When an association is replicated, it is frequently with a different phenotype of the disorder, with different polymorphisms in the same gene and even with different alleles of the same variant, such as examples reported in asthma.52 The lack of concordance between our association studies reflects once more the complexity in studying such a heterogeneous disorder as autism, and should not be taken to mean that the results are false positives. There are now several studies associating MET with psychiatric disorders, especially within the area upstream of the coding region.38, 39 Regulatory mechanisms not yet identified may be acting in this region and be responsible for alterations in gene expression.
Multiple upstream and downstream proteins regulate MET signalling, such as the availability of the HGF ligand and MET activating coreceptors. For instance, the CD44 element that forms a multimeric complex with MET and HGF and is required to activate signalling through the MET receptor tyrosine kinase.39 As the rs38845 polymorphism is located in intron 1, it may have a regulatory role in gene expression, affecting, for example, transcription rate or splicing. Alterations in MET expression may contribute to autism susceptibility. Bioinformatic analyses of this variant predicted that the nuclear factor IRF1 only binds the target DNA sequence when the A allele is present. As the A allele was associated with autism susceptibility in our families, this warrants further investigation.
IRF1 is a nuclear factor that binds the promoter region of both IFN-α and IFN-β genes53, and IRF1 functions as a transcriptional activator for the type I IFN genes54 (in which IFN-α and IFN-β are included). Additionally, an earlier study showed that in human hepatocytes IFN-α upregulates 44 genes by 100% and downregulates 9 genes by 50% (including MET). Downregulation of MET is caused by IFN-α suppression of the MET promoter activity (through downregulation of Spl), which results in an attenuation of HGF/MET-induced signals and cell proliferation.55 Recent literature has suggested that a reduced expression of the MET receptor tyrosine kinase could play a crucial role in the pathobiology of autism.38 Thus, we hypothesize that when the A allele is present at rs38845, IRF1 binds directly to the MET promoter DNA sequence contributing to increased deregulation of HGF/MET-induced signalling in autistic patients by mechanisms not yet known.
In disorders with a complex genetic basis there has been a growing interest in LD, mainly because of the belief that the association studies offer considerable power for mapping common disease genes.56 Nonetheless, the pursuit of common variants might not have been solely the most suitable to identify the susceptibility variants in this specific region, as we should perhaps be looking for rare variants in parallel. The repeated occurrence of non-replication in the story of the genetics of ASDs seems to be a constant dilemma, and reflects that going from a broad peak of linkage to a functional genetic variant continues to be a challenge for autism today.
Overall, these results provide further evidence that MET may play a role in autism susceptibility; however, further studies will be essential to better clarify its part in the pathology of the disorder.
Acknowledgments
We thank all the families who have participated in the study and the professionals who made this study possible. We thank Tom Scerri for help in statistical and bioinformatic analysis, Chris Allan for technical support, and Alistair Pagnamenta, Susana Campino and Kirsty Wing for helpful discussions and critical comments. This work was funded by the Nancy Lurie Marks Family Foundation, the Simons Foundation, the EC 6th FP AUTISM MOLGEN, and the Fundacao para a Ciencia e Tecnologia (FCT-Portugal). Taane Clark is funded by the Bill and Melinda Gates Foundation, Wellcome Trust and Medical Research Council UK.
References
- Lord C, Cook EH, Leventhal BL, Amaral DG. Autism spectrum disorders. Neuron. 2000;28:355–363. doi: 10.1016/s0896-6273(00)00115-x. [DOI] [PubMed] [Google Scholar]
- Baird G, Simonoff E, Pickles A, et al. Prevalence of disorders of the autism spectrum in a population cohort of children in South Thames: the Special Needs and Autism Project (SNAP) Lancet. 2006;368:210–215. doi: 10.1016/S0140-6736(06)69041-7. [DOI] [PubMed] [Google Scholar]
- Volkmar FR, Paul R, Klin A, Cohen D. Handbook of Autism and Pervasive Developmental Disorders. Hoboken, NJ: John Wiley & Sons; 2005. [Google Scholar]
- Rutter M. Incidence of autism spectrum disorders: changes over time and their meaning. Acta Paediatr. 2005;94:2–15. doi: 10.1111/j.1651-2227.2005.tb01779.x. [DOI] [PubMed] [Google Scholar]
- Bailey A, Le Couteur A, Gottesman I, et al. Autism as a strongly genetic disorder: evidence from a British twin study. Psychol Med. 1995;25:63–77. doi: 10.1017/s0033291700028099. [DOI] [PubMed] [Google Scholar]
- IMGSAC A full genome screen for autism with evidence for linkage to a region on chromosome 7q. Hum Mol Genet. 1998;7:571–578. doi: 10.1093/hmg/7.3.571. [DOI] [PubMed] [Google Scholar]
- IMGSAC A genomewide screen for autism: strong evidence for linkage to chromosomes 2q, 7q, and 16p. Am J Hum Genet. 2001;69:570–581. doi: 10.1086/323264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb JA, Barnby G, Bonora E, et al. Analysis of IMGSAC autism susceptibility loci: evidence for sex limited and parent of origin specific effects. J Med Genet. 2005;42:132–137. doi: 10.1136/jmg.2004.025668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett S, Beck JC, Bernier R, et al. An autosomal genomic screen for autism. Collaborative linkage study of autism. Am J Med Genet. 1999;88:609–615. doi: 10.1002/(sici)1096-8628(19991215)88:6<609::aid-ajmg7>3.3.co;2-c. [DOI] [PubMed] [Google Scholar]
- Philippe A, Martinez M, Guilloud-Bataille M, et al. Genome-wide scan for autism susceptibility genes. Paris Autism Research International Sibpair Study. Hum Mol Genet. 1999;8:805–812. doi: 10.1093/hmg/8.5.805. [DOI] [PubMed] [Google Scholar]
- Risch N, Spiker D, Lotspeich L, et al. A genomic screen of autism: evidence for a multilocus etiology. Am J Hum Genet. 1999;65:493–507. doi: 10.1086/302497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao Y, Wolpert CM, Raiford KL, et al. Genomic screen and follow-up analysis for autistic disorder. Am J Med Genet. 2002;114:99–105. doi: 10.1002/ajmg.10153. [DOI] [PubMed] [Google Scholar]
- Alarcon M, Cantor RM, Liu J, Gilliam TC, Geschwind DH. Evidence for a language quantitative trait locus on chromosome 7q in multiplex autism families. Am J Hum Genet. 2002;70:60–71. doi: 10.1086/338241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molloy CA, Keddache M, Martin LJ. Evidence for linkage on 21q and 7q in a subset of autism characterized by developmental regression. Mol Psychiatry. 2005;10:741–746. doi: 10.1038/sj.mp.4001691. [DOI] [PubMed] [Google Scholar]
- Badner JA, Gershon ES. Regional meta-analysis of published data supports linkage of autism with markers on chromosome 7. Mol Psychiatry. 2002;7:56–66. doi: 10.1038/sj.mp.4000922. [DOI] [PubMed] [Google Scholar]
- Trikalinos TA, Karvouni A, Zintzaras E, et al. A heterogeneity-based genome search meta-analysis for autism-spectrum disorders. Mol Psychiatry. 2006;11:29–36. doi: 10.1038/sj.mp.4001750. [DOI] [PubMed] [Google Scholar]
- Lin JC, Naujokas M, Zhu H, Nolet S, Park M. Intron-exon structure of the MET gene and cloning of an alternatively-spliced Met isoform reveals frequent exon-skipping of a single large internal exon. Oncogene. 1998;16:833–842. doi: 10.1038/sj.onc.1201599. [DOI] [PubMed] [Google Scholar]
- Birchmeier C, Gherardi E. Developmental roles of HGF/SF and its receptor, the c-Met tyrosine kinase. Trends Cell Biol. 1998;8:404–410. doi: 10.1016/s0962-8924(98)01359-2. [DOI] [PubMed] [Google Scholar]
- Rodrigues GA, Naujokas MA, Park M. Alternative splicing generates isoforms of the met receptor tyrosine kinase which undergo differential processing. Mol Cell Biol. 1991;11:2962–2970. doi: 10.1128/mcb.11.6.2962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulasne D, Foveau B. The shadow of death on the MET tyrosine kinase receptor. Cell Death Differ. 2008;15:427–434. doi: 10.1038/sj.cdd.4402229. [DOI] [PubMed] [Google Scholar]
- Park M, Dean M, Cooper CS, et al. Mechanism of met oncogene activation. Cell. 1986;45:895–904. doi: 10.1016/0092-8674(86)90564-7. [DOI] [PubMed] [Google Scholar]
- Di Renzo MF, Olivero M, Ferro S, et al. Overexpression of the c-MET/HGF receptor gene in human thyroid carcinomas. Oncogene. 1992;7:2549–2553. [PubMed] [Google Scholar]
- Liu C, Park M, Tsao MS. Overexpression of c-met proto-oncogene but not epidermal growth factor receptor or c-erbB-2 in primary human colorectal carcinomas. Oncogene. 1992;7:181–185. [PubMed] [Google Scholar]
- Scotlandi K, Baldini N, Oliviero M, et al. Expression of Met/hepatocyte growth factor receptor gene and malignant behavior of musculoskeletal tumors. Am J Pathol. 1996;149:1209–1219. [PMC free article] [PubMed] [Google Scholar]
- Kaji M, Yonemura Y, Harada S, Liu X, Terada I, Yamamoto H. Participation of c-met in the progression of human gastric cancers: anti-c-met oligonucleotides inhibit proliferation or invasiveness of gastric cancer cells. Cancer Gene Ther. 1996;3:393–404. [PubMed] [Google Scholar]
- Okunishi K, Dohi M, Nakagome K, et al. A novel role of hepatocyte growth factor as an immune regulator through suppressing dendritic cell function. J Immunol. 2005;175:4745–4753. doi: 10.4049/jimmunol.175.7.4745. [DOI] [PubMed] [Google Scholar]
- Beilmann M, Vande Woude GF, Dienes HP, Schirmacher P. Hepatocyte growth factor-stimulated invasiveness of monocytes. Blood. 2000;95:3964–3969. [PubMed] [Google Scholar]
- Beilmann M, Odenthal M, Jung W, Vande Woude GF, Dienes HP, Schirmacher P. Neoexpression of the c-met/hepatocyte growth factor-scatter factor receptor gene in activated monocytes. Blood. 1997;90:4450–4458. [PubMed] [Google Scholar]
- Ido A, Numata M, Kodama M, Tsubouchi H. Mucosal repair and growth factors: recombinant human hepatocyte growth factor as an innovative therapy for inflammatory bowel disease. J Gastroenterol. 2005;40:925–931. doi: 10.1007/s00535-005-1705-x. [DOI] [PubMed] [Google Scholar]
- Yang XM, Park M. Expression of the met/hepatocyte growth factor/scatter factor receptor and its ligand during differentiation of murine P19 embryonal carcinoma cells. Dev Biol. 1993;157:308–320. doi: 10.1006/dbio.1993.1137. [DOI] [PubMed] [Google Scholar]
- Streit AC, Stern CD.Competence for neural induction: HGF/SF, HGF1/MSP and the c-Met receptor Ciba Found Symp 1997212155–165.discussion 165–158. [DOI] [PubMed] [Google Scholar]
- Krasnoselsky A, Massay MJ, DeFrances MC, Michalopoulos G, Zarnegar R, Ratner N. Hepatocyte growth factor is a mitogen for Schwann cells and is present in neurofibromas. J Neurosci. 1994;14:7284–7290. doi: 10.1523/JNEUROSCI.14-12-07284.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powell EM, Mars WM, Levitt P. Hepatocyte growth factor/scatter factor is a motogen for interneurons migrating from the ventral to dorsal telencephalon. Neuron. 2001;30:79–89. doi: 10.1016/s0896-6273(01)00264-1. [DOI] [PubMed] [Google Scholar]
- Levitt P, Eagleson KL, Powell EM. Regulation of neocortical interneuron development and the implications for neurodevelopmental disorders. Trends Neurosci. 2004;27:400–406. doi: 10.1016/j.tins.2004.05.008. [DOI] [PubMed] [Google Scholar]
- Ieraci A, Forni PE, Ponzetto C. Viable hypomorphic signaling mutant of the Met receptor reveals a role for hepatocyte growth factor in postnatal cerebellar development. Proc Natl Acad Sci U S A. 2002;99:15200–15205. doi: 10.1073/pnas.222362099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Casanova MF, Buxhoeveden DP, Switala AE, Roy E. Minicolumnar pathology in autism. Neurology. 2002;58:428–432. doi: 10.1212/wnl.58.3.428. [DOI] [PubMed] [Google Scholar]
- Palmen SJ, van Engeland H, Hof PR, Schmitz C. Neuropathological findings in autism. Brain. 2004;127:2572–2583. doi: 10.1093/brain/awh287. [DOI] [PubMed] [Google Scholar]
- Campbell DB, Sutcliffe JS, Ebert PJ, et al. A genetic variant that disrupts MET transcription is associated with autism. Proc Natl Acad Sci U S A. 2006;103:16834–16839. doi: 10.1073/pnas.0605296103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell DB, D′Oronzio R, Garbett K, et al. Disruption of cerebral cortex MET signaling in autism spectrum disorder. Ann Neurol. 2007;62:243–250. doi: 10.1002/ana.21180. [DOI] [PubMed] [Google Scholar]
- Lord C, Rutter M, Le Couteur A. Autism Diagnostic Interview-Revised: a revised version of a diagnostic interview for caregivers of individuals with possible pervasive developmental disorders. J Autism Dev Disord. 1994;24:659–685. doi: 10.1007/BF02172145. [DOI] [PubMed] [Google Scholar]
- Sparrow S, Balla D, Cicchetti D. Vineland Adaptive Behaviour Scales. Circle Pines, American Guidance Service: MN; 1984. [Google Scholar]
- Lord C, Risi S, Lambrecht L, et al. The autism diagnostic observation schedule-generic: a standard measure of social and communication deficits associated with the spectrum of autism. J Autism Dev Disord. 2000;30:205–223. [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Fiddy S, Cattermole D, Xie D, Duan XY, Mott R. An integrated system for genetic analysis. BMC Bioinformatics. 2006;7:210. doi: 10.1186/1471-2105-7-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O′Connell JR, Weeks DE. PedCheck: a program for identification of genotype incompatibilities in linkage analysis. Am J Hum Genet. 1998;63:259–266. doi: 10.1086/301904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abecasis GR, Cherny SS, Cookson WO, Cardon LR. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat Genet. 2002;30:97–101. doi: 10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- Spielman RS, McGinnis RE, Ewens WJ. Transmission test for linkage disequilibrium: the insulin gene region and insulin-dependent diabetes mellitus (IDDM) Am J Hum Genet. 1993;52:506–516. [PMC free article] [PubMed] [Google Scholar]
- Clayton D, Jones H. Transmission/disequilibrium tests for extended marker haplotypes. Am J Hum Genet. 1999;65:1161–1169. doi: 10.1086/302566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quandt K, Frech K, Karas H, Wingender E, Werner T. MatInd and MatInspector: new fast and versatile tools for detection of consensus matches in nucleotide sequence data. Nucleic Acids Res. 1995;23:4878–4884. doi: 10.1093/nar/23.23.4878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarke GM, Carter KW, Palmer LJ, Morris AP, Cardon LR. Fine mapping versus replication in whole-genome association studies. Am J Hum Genet. 2007;81:995–1005. doi: 10.1086/521952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ober C. Perspectives on the past decade of asthma genetics. J Allergy Clin Immunol. 2005;116:274–278. doi: 10.1016/j.jaci.2005.04.039. [DOI] [PubMed] [Google Scholar]
- Lengyel P. Tumor-suppressor genes: news about the interferon connection. Proc Natl Acad Sci U S A. 1993;90:5893–5895. doi: 10.1073/pnas.90.13.5893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harada H, Willison K, Sakakibara J, Miyamoto M, Fujita T, Taniguchi T. Absence of the type I IFN system in EC cells: transcriptional activator (IRF-1) and repressor (IRF-2) genes are developmentally regulated. Cell. 1990;63:303–312. doi: 10.1016/0092-8674(90)90163-9. [DOI] [PubMed] [Google Scholar]
- Radaeva S, Jaruga B, Hong F, et al. Interferon-alpha activates multiple STAT signals and down-regulates c-Met in primary human hepatocytes. Gastroenterology. 2002;122:1020–1034. doi: 10.1053/gast.2002.32388. [DOI] [PubMed] [Google Scholar]
- Ardlie KG, Kruglyak L, Seielstad M. Patterns of linkage disequilibrium in the human genome. Nat Rev Genet. 2002;3:299–309. doi: 10.1038/nrg777. [DOI] [PubMed] [Google Scholar]