Abstract
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) is an autosomal dominant small-vessel disease of the brain caused by mutations in the NOTCH3 receptor. The highly stereotyped nature of the mutations, which alter the number of cysteine residues within the epidermal growth factor-like repeats (EGFR), predicts that all mutations share common mechanisms. Prior in vitro assays and genetic studies in the mouse support the hypothesis that common mutations do not compromise canonical Notch3 function but instead convey a non-physiological and deleterious activity to the receptor through the unpaired cysteine residue. Intriguingly, in vitro studies predict that mutations located in the Delta/Serrate/LAG-2 ligand binding domain-(EGFR10-11) may result in a loss of Notch3 receptor function. However, the in vivo relevance and functional significance of this with respect to the pathogenic mechanisms and clinical expression of the disease remain largely unexplored. To ascertain, in vivo, the functional significance of EGFR10-11 mutations, we generated transgenic mice with one representative mutation (C428S) in EGFR10 of Notch3. These mice, like those with a common R90C mutation, developed characteristic arterial accumulation of Notch3 protein and granular osmiophilic material upon aging. By introducing the mutant C428S transgene into a Notch3 null background, we found that, unlike the R90C mutant protein, the C428S mutant protein has lost wild-type Notch3 activity and exhibited mild dominant-negative activity in three different biological settings. From a large prospectively recruited cohort of 176 CADASIL patients, we identified 10 patients, from five distinct pedigrees carrying a mutation in EGFR10 or 11. These mutations were associated with significantly higher Mini-Mental State Examination and Mattis Dementia Rating Scale scores (P < 0.05), when compared with common mutations. Additionally, we found a strong effect of this genotype on the burden of white matter hyperintensities (P < 0.01). Collectively, these results highlight distinctive functional and phenotypic features of EGFR10-11 mutations relative to the common CADASIL mutations. Our findings are compatible with the hypothesis that EGFR10-11 mutations cause the disease through the same gain of novel function as the common mutations, and lead us to propose that reduced Notch3 signalling acts as a modifier of the CADASIL phenotype.
Keywords: CADASIL, Notch3, transgenic, genotype-phenotype correlations
Introduction
Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy (CADASIL) (MIM 125310) is a small-vessel disease caused by dominant mutations in the NOTCH3 gene (600276) (Joutel et al., 1996). Main clinical manifestations include migraine with aura, recurrent strokes and cognitive decline leading to severe disability and premature death (Chabriat et al., 1995; Dichgans et al., 1998; Opherk et al., 2004). The disease is caused by a unique systemic non-amyloid arteriopathy, although involving primarily brain arteries, characterized by prominent alterations of vascular smooth muscle cells and pathognomonic granular osmiophilic material deposits (Ruchoux et al., 1995; Kalimo et al., 2002).
Notch3 encodes a single pass transmembrane protein belonging to the evolutionary conserved Notch receptor family. The Notch signalling pathway is a signalling mechanism that plays a central role in the development and maturation of most vertebrate organs, with pleiotropic effects depending on dose and context (Gridley, 2007). Notch3 is predominantly expressed in vascular smooth muscle cells and is critically required for the structural and functional integrity of small arteries by controlling the arterial differentiation and maturation of smooth muscle cells (Joutel et al., 2000; Domenga et al., 2004). Notch3 receptor functions at the cell surface as a heterodimer composed of a large extracellular domain (Notch3ECD) containing 34 epidermal growth factor-like repeats (EGFR), non-covalently attached to the membrane tethered intracellular domain (Joutel et al., 2000). Ligand binding initiates a series of proteolytic cleavages that release the Notch intracellular domain, which translocates to the nucleus. Here, the Notch intracellular domain interacts with the transcription factor RBP-Jκ and co-activators and activates the transcription of target genes that remain to be identified (Schweisguth, 2004). The best-known ligands of Notch receptors are transmembrane proteins of the delta-like/Jagged family, which are collectively referred to the canonical DSL (Delta/Serrate/LAG-2) ligands (D'Souza et al., 2008). Additional ligands have been recently identified, including MAGP1 or MAGP2 that are highly expressed in the vessel wall (Sakamoto et al., 2002; Hu et al., 2003; Eiraku et al., 2005; Miyamoto et al., 2006). Yet, the precise ligands of Notch3 in the vessel are unknown.
NOTCH3 has 33 exons but in general CADASIL mutations occur in the epidermal growth factor-like repeats encoded by exons 2–24 and lead to an odd number of cysteine residues (Joutel et al., 1997; Oberstein et al., 1999; Singhal et al., 2004; Peters et al., 2005). The highly stereotyped nature of CADASIL mutations predicts that all mutations cause the disease through the same mechanisms. Recent in vitro and in vivo studies suggest that CADASIL is not due to compromised canonical Notch3 function but instead that mutations convey a non-physiological and deleterious activity to the receptor. Reporter gene assays have shown that the majority of CADASIL-associated NOTCH3 mutant alleles, including those located in the mutational hotspot EGFR2-5, are able to activate RBP-Jκ transcription at wild-type levels (Karlstrom et al., 2002; Joutel et al., 2004; Peters et al., 2004; Haritunians et al., 2005; Low et al., 2006). Genetic studies in the mouse of one representative mutation in EGFR2, R90C, further showed retained receptor function in brain arteries (Monet et al., 2007). Intriguingly, certain naturally occurring mutations are predicted to result, by contrast, in a loss of functional Notch3 receptor. These peculiar mutations are located in EGFR10 and 11, which are required for binding to the canonical DSL ligands, and, two representative mutations were found to abrogate RBP-Jκ signalling in response to this type of ligands in in vitro assays (Joutel et al., 2004; Peters et al., 2004). However, the in vivo relevance of these findings is questionable since it is yet unknown whether, in a physiological context, Notch3 signalling is triggered by a DSL ligand or alternatively by non-canonical ligands that do not require these specific EGFR for binding. Moreover, the significance of a potential loss of function of Notch3 with respect to the pathogenic mechanisms and clinical expression of the disease is largely unknown.
We therefore undertook the present study to ascertain the significance of these peculiar NOTCH3 mutations located in EGFR10-11, with respect to the pathogenic mechanisms and clinical expression of the disease, using genetic studies in mice and humans. Specifically, we generated transgenic mice expressing a human NOTCH3 with the C428S mutation in EGFR10. Transgenic mice were crossed with Notch3 null mice to directly evaluate the impact of this mutation on Notch3 function in brain arteries, with respect to effects on smooth muscle cell arterial identity, structure and RBP-Jκ-mediated activity. We further investigated whether these peculiar NOTCH3 mutations were associated with distinguishing clinical features as compared with common mutations, in CADASIL patients recruited from a large prospective cohort study.
Methods
Experimental mice and genotyping
Transgenic mice expressing human wild-type NOTCH3, under the control of murine SM22α promoter (TghN3(WT)), have been described previously (Monet et al., 2007). Transgenic mice expressing human mutant NOTCH3 with the C428S mutation (TghN3(C428S)) were similarly generated. Nine transgenic founders had germline transmission and two transgenic lines expressing human mutant NOTCH3 were established upon quantification of transgene mRNA expression (see below). Notch3 null mice, SM22α-lacZ mice, Notch pathway Activity Sensor (NAS) mice have been reported elsewhere (Moessler et al., 1996; Domenga et al., 2004; Souilhol et al., 2006). All mice were bred on a C57BL/6J background. The genotyping of mice was performed by PCR using proteinase K-digested tail or toe biopsies as DNA templates and specific primers as follows: human NOTCH3 transgene (sense) 5′-cgatggaatgggtttccact-3′ and (antisense) 5′ aggcaggagcaggaaaagga-3′, Notch3 null allele (sense) 5′-tcgccttcttgacgagttct-3′ and (antisense) 5′-gcgatgcaatttcctcattt-3′; wild-type Notch3 allele (sense) 5′-ccatgaggatgctatctgtgac-3′ and (antisense) 5′-cacattggcacaagaatgagcc-3′, SM22α-lacZ (sense) 5′-ggatcgatctcgccatacagcgcg-3′ and (antisense) 5′-ccagacaccgaagctactctcctt-3′ and TP1-nlacZ (sense) 5′-gatgcgcccatctacacc-3′ and (antisense) 5′-gctctggccttcctgta-3′. All mouse protocols were approved by the Animal Care and Use Committees of Paris, Ile de France.
Real time RT–PCR analysis
Brain arteries, including arteries of the circle of Willis and the medium-sized branches, were dissected under microscope, snap frozen in liquid nitrogen and stored at −80°C. Total RNA was extracted from snap frozen arteries, treated with DNAse I using the SV total RNA isolation system (Promega) and converted into cDNA using the MMLV (Invitrogen). Quantitative PCR was performed using the SYBR Green PCR master mix (BIORAD) on a MyiQTM Single-Color Real-Time PCR detection system (Biorad). Each sample was amplified in duplicate using primers specific for the human NOTCH3 transgene, murine Notch3, GAPDH, β-actin and SM22α cDNA. The hNOTCH3 to mNotch3 mRNA ratios were determined as previously reported using a pBluescript recombinant plasmid containing one copy of the human NOTCH3 transgene and one copy of murine Notch3 cDNA transcript (Monet et al., 2007). In a separate set of analysis, expression levels of hNOTCH3 transgenes were normalized for β-actin and SM22α expression.
β-galactosidase staining and quantification
Whole-mount staining for β-galactosidase activity was performed as described previously (Domenga et al., 2004). Quantitation of arterial LacZ expression, as determined by the conversion of X-gal to the blue-coloured product by β-galactosidase, was performed by computer-assisted morphometrical analysis. Arteries of the circle of Willis and their medium-sized branches were imaged with a Wild Heerbrugg scope equipped with a CCD colour video camera. Digital images were then converted in grey levels and analysed with NIS-Element BR 3.0 image analysis software (Nikon) to determine the mean of intensity values of pixels in arteries. The mean values were averaged per genotype and differences were evaluated by ANOVA and Scheffe's test.
Histology and immunohistochemistry
Histological analyses were carried out on paraffin embedded 7 µm sections stained with haematoxylin and eosin and on Epon E812 embedded semi-thin sections stained with toluidine blue. We used antibodies to human Notch3ECD (clone 1E4) and α-smooth muscle actin (clone 1A4, DAKO) for immunohistochemistry. We performed 1E4 immunostaining on acetone fixed cryostat sections and α-smooth muscle actin immunostaining on paraffin embedded sections. Immunohistochemical signals were revealed using the avidin–biotin–peroxydase system (Vector Laboratories) or TRITC-conjugated antibodies to mouse IgG (Jackson Immunoresearch Laboratories).
High resolution and transmission electron microscopy
Artery and surrounding brain tissue were dissected under microscope, fixed in CARSON solution and embedded in Epon E812 resin. We observed semi-thin sections stained with toluidine blue under a DMR microscope (Leica) and ultrathin sections, stained with lead citrate and uranyl acetate, in a CM100 electron microscope (Philips) as described (Monet et al., 2007).
CADASIL patients, clinical assessment and MRI analysis
Patients were drawn from a prospective cohort study of patients with CADASIL. Study design, clinical and demographic data have been previously reported (Viswanathan et al., 2006). In brief, all enrolled patients had a definite diagnosis of CADASIL as defined by the presence of a pathogenic mutation in the NOTCH3 gene and underwent detailed baseline neurological examination including a Mini-Mental State examination, Mattis Dementia Rate Scale, and degree of disability based on the modified Rankin scale. MRI scans were obtained and processed as described (Viswanathan et al., 2006; Viswanathan et al., 2008). The total volume of white matter hyperintensity and lacunar lesions was normalized to the intracranial cavity. The brain parenchymal fraction was defined as the ratio of brain tissue volume to total intracranial cavity volume. The number of microbleeds as defined in Viswanathan et al. (2006) was recorded. Mean apparent diffusion coefficient was determined as described (Viswanathan et al., 2008). Written, informed consent was obtained from all patients and an independent ethics committee approved the study.
Statistical methods of genotype-phenotype correlation analysis
Each patient with a mutation in EFGR10-11 (case) was matched to two patients with a mutation in EGFR2-5 (controls), randomly selected among those of same age (±2 years) and sex. We used McNemar's Chi-square test to compare the frequency of binary variables between cases and their matched controls and we performed Student's t-test for paired data to compare the mean values of quantitative variables between the two groups. In order to verify the robustness of our findings, we performed a sensitivity analysis on the whole sample. The Wilcoxon–Mann–Withney test was used to compare the median between the two groups (cases versus all other patients) of variables that were not normally distributed.
Results
Generation of TghNotch3(C428S) mice
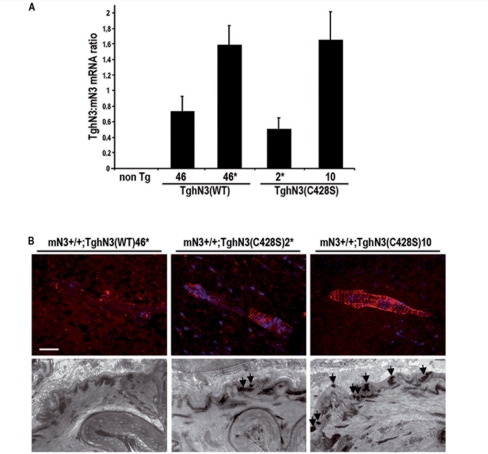
Using cell-based binding studies and gene reporter assays, two of the 34 EGFR of Notch3 (EGFR10 and EGFR11) have been identified as necessary for DSL ligand-binding and DSL ligand-induced signalling (Joutel et al., 2004). To address the mechanisms for the pathogenesis of these peculiar mutations in vivo, we analysed the functional significance of one representative mutation (C428S) in a transgenic mouse model. The C428S mutation is located in EGFR10 and has been identified in two distinct CADASIL pedigrees carrying the c.1282T>A and c.1283G>C nucleotide substitutions in exon 8 (Joutel et al., 2001) and our unpublished data. Our prior in vitro study revealed that the C428S mutation impaired binding to Jagged1 and Jagged1-induced transcriptional activity of a RBP-Jκ responsive luciferase reporter (Joutel et al., 2004). We generated transgenic mice expressing C428S mutant NOTCH3 termed TghN3(C428S). Mutant protein was targeted in arterial smooth muscle cells as previously described (Ruchoux et al., 2003). Two independent lines (2* and 10) were selected for the current study on the basis of the expression level of the transgene in the brain arteries. Line 2*, which has been bred to homozygosity for the transgene, expressed the NOTCH3 transgene at a level approximately half of the endogenous mNotch3 gene. Line 10 expressed higher levels of the transgene, close to 150% of the endogenous mNotch3 gene (Fig. 1A).
Figure 1.
Generation and characterization of TghN3(C428S) mice. (A) Expression level of human NOTCH3 transgenes in isolated brain arteries from wild-type [TghN3(WT)] and mutant [TghN3(C428S)] transgenic mice on a normal Notch3+/+ background, at 1 month of age. Total RNA was prepared from brain arteries collected from 2 to 3 mice. Lines 46 and 46* carry the wild-type transgene at the heterozygous and homozygous state respectively. Lines 2* and 10 are independent lines carrying the mutant transgene at the heterozygous and homozygous state respectively. Shown are mean ratios (±SEM) of human NOTCH3 mRNA to murine Notch3 mRNA measured in 3–4 RNA preparations for each line as determined by real-time RT–PCR. (B) Upper panel, shown are representative brain artery sections from 10-month-old TghN3(WT)46*, 18-month-old TghN3(C428S)2* and 8-month-old TghN3(C428S)10 mice on a Notch 3+/+ background, immunostained with the 1E4 mAb against human NOTCH3ECD (dapi nuclear staining). TghN3(C428S) mice from both lines exhibit the characteristic granular Notch3 immunostaining, indicative of accumulation of aggregated NOTCH3ECD whereas TghN3(WT) mice show no staining in the same experimental conditions. Lower panel, representative electron micrographs of brain arteries from 10-month-old TghN3(WT)46*, 18-month-old TghN3(C428S)2* and 8-month-old TghN3(C428S)10 mice on a Notch 3+/+ background showing granular osmiophilic material deposits (arrows) in TghN3(C428S) arteries whereas granular osmiophilic material deposits are absent in TghN3(WT) artery. Scale bar: 35 µm (upper panel), 0.7 µm (lower panel).
Immunohistochemistry using a human Notch3ECD specific monoclonal antibody demonstrated Notch3ECD aggregation in aged TghN3(C42S8) mice. Robust Notch3ECD aggregation, easily recognizable by its typical granular appearance drawing the outlines of vascular smooth muscle cells, was detected at 8 and 18 months of age in lines 10 and 2*, respectively (Fig. 1B, upper panel). Also, electron microscopy analysis showed granular osmiophilic material deposits in the brain arteries of aged mutant mice (Fig. 1B, lower panel). Therefore, TghN3(C428S) mice exhibit the characteristic arterial lesions of CADASIL upon aging, and age of onset of these latter changes appears to correlate with the expression level of the mutant transgene. However, TghN3(C428S) mice like TghN3(R90C) mice do not develop overt brain parenchyma lesions (data not shown).
Analysis of C428S function in the absence of wild-type Notch3
We previously developed an in vivo assay to measure the functional activity of a human NOTCH3 protein in the mouse, under physiological conditions, without a priori hypotheses on the identity of the Notch3 ligand (Monet et al., 2007). In this assay, a human NOTCH3 transgene is introduced into the Notch3 null background and the ability of the NOTCH3 transgene to rescue the smooth muscle cell defects is examined in the large and medium-sized brain arteries of the circle of Willis. Additionally, Notch3-mediated RBP-Jκ activity in smooth muscle cell of brain arteries is measured by introducing the TP1-nlacZ transgene which is composed of a lacZ reporter gene linked to a well-characterized promoter, which consists of 12 RBP-Jκ binding motifs upstream from a minimal promoter (Souilhol et al., 2006). Our prior data indicated that this assay is semi-quantitative over the range of concentration of human wild-type NOTCH3 transgene examined, that comprised between 20% and 80% of endogenous Notch3 transcript levels, with no restoration of arterial defects and no detectable RBP-Jκ activity at the lowest concentration and complete restoration with almost wild-type RBP-Jκ activity at the highest concentration (Monet et al., 2007). The two transgenic lines (2* and 10) expressing human C428S mutant NOTCH3 were backcrossed to Notch3−/− mice and tested for rescuing and RBP-Jκ activities. All the mice were analysed between 15 and 30 days after birth and at least three mice for each line and genotype were analysed.
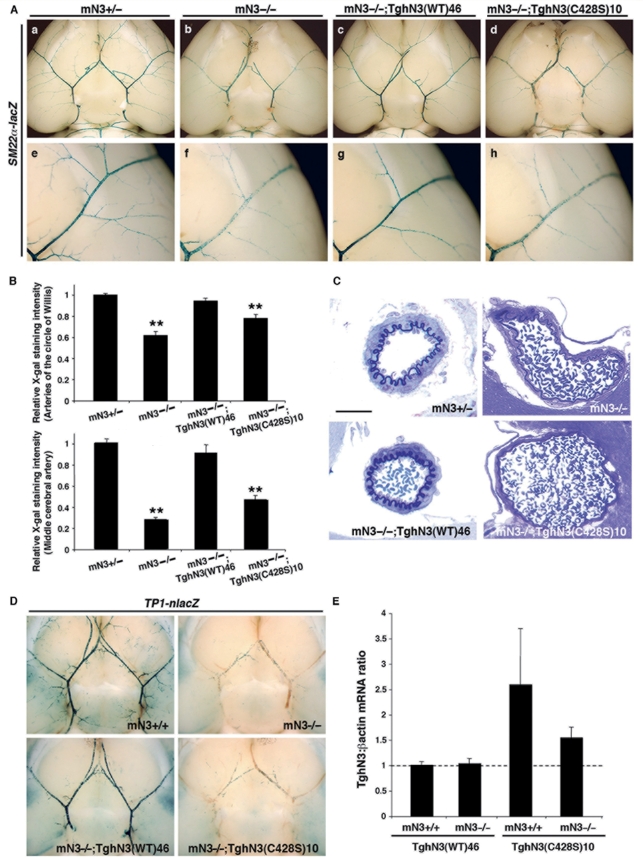
We first analysed the morphology of the brain arteries and the fine structure of smooth muscle cells by high resolution optic microscopy analysis on semi-thin sections and electron microscopy analysis on ultra-thin sections. Figure 2C (and data not shown) shows that arteries of mN3−/−;TghN3(C428S) mice from both lines exhibited defects similar to those of Notch3 null mice with enlarged arteries, flattened elastica lamina, and abnormally thin smooth muscle cells, while brain arteries and smooth muscle cells of mN3−/−;TghN3(WT) mice had a wild-type appearance in morphology and ultrastructure. Thus these results establish that the C428S mutant hNOTCH3 transgene cannot rescue the Notch3−/− arterial defects. Second, in further crosses, the SM22α-lacZ transgene was introduced in addition to the hNOTCH3 transgene in Notch3−/− mice to analyse arterial identity of smooth muscle cells (Domenga et al., 2004; Monet et al., 2007). β-galactosidase expression was similarly impaired in Notch3 null mice and mN3−/−;TghN3(C428S) mice from both lines (Fig. 2A). Specifically, in SM22α-lacZ;mN3−/−;TghN3(C428S)10 mice, X-gal staining was mildly reduced in the arteries of the circle of Willis (78 ± 4%, P < 0.001) and strongly reduced in the medium-sized branches (47 ± 4%, P < 0.001) as compared to control SM22α-lacZ;Notch3+/− and rescued SM22α-lacZ;mN3−/−;TghN3(WT) mice, thus indicating no restoration of arterial identity by the mutant protein (Fig. 2B). Third, we introduced the TP1-nlacZ transgene in addition to the human NOTCH3 transgene in Notch3−/− mice to measure RBP-Jκ activity. X-gal staining showed that RBP-Jκ activity was abrogated in the arteries of the circle of Willis and their medium-sized branches of TP1-nlacZ;mN3−/−;TghN3(C428S) mice from both lines while it was close to wild-type level in those of rescued TP1-nlacZ;mN3−/−;TghN3(WT) mice (Fig. 2D and data not shown).
Figure 2.
The C428S mutation creates a non-functional allele. (A) One-month-old mice of the indicated line and genotype heterozygous for the SM22α-lacZ transgene, were assayed for smooth muscle cell arterial identity with X-gal staining on whole brain. Shown are ventral views of whole brain with the circle of Willis and its main branches (a–d) and higher magnification views of the middle cerebral artery (e–h). mN3−/−,TghN3(WT) rescued mice have comparable X-gal staining in the large and middle size brain arteries (c and g) to that seen in the Notch3+/− control mice (a and e). In contrast, X-gal staining in mN3−/−, TghN3(C428S) mice is mildly reduced in the arteries of the circle of Willis (d) and strongly reduced in the middle cerebral artery (h) as seen in Notch3−/− mice (b and f). (B) Relative SM22α-lacZ transgene expression level in the arteries of the circle of Willis (upper panel) and the middle cerebral artery (MCA) (lower panel) of 1-month-old mice heterozygous for the SM22α-lacZ transgene of the following genotypes: Notch3+/- (n = 15 mice), Notch3−/- (n = 10 mice), mN3−/−;TghN3(WT)46 (n = 5 mice) and mN3−/-;TghN3(C428S)10 (n = 7 mice) in the upper panel and Notch3+/− (n = 25 MCA from 13 mice), Notch3−/− (n = 20 MCA from 12 mice), mN3−/−;TghN3(WT)46 (n = 9 MCA from 5 mice), and mN3−/−;TghN3(C428S)10 (n = 10 MCA from five mice) in the lower panel. Shown are mean intensities of X-gal staining (±SEM) in arteries expressed relative to those in Notch3+/- mice arbitrarily set to a value of 1. Differences were evaluated by ANOVA and Scheffe's test (**P < 0.001 versus Notch3+/- or mN3−/−;TghN3(WT)46 mice). (C) Shown are representative semi-thin sections of brain arteries stained with toluidine blue from mice of the indicated line and genotype, aged one month. Brain artery in mN3−/-; TghN3(C428S) mice is enlarged, with a flattened elastica lamina and abnormally thin smooth muscle cells as seen in Notch3−/− mice. Scale bar: 25 µm. (D) One-month-old mice of the indicated genotype, carrying the TP1-nlacZ transgene at the heterozygous state, were assayed for RBP-Jκ activity, with X-gal staining on dissected whole-brain. Shown are ventral views of whole brains. Robust X-gal staining is detected in arteries of the Notch3+/+ and mN3−/−;TghN3(WT) rescued mice while vascular staining is similarly abrogated in mN3−/−;TghN3(C428S) and Notch3−/− mice. (E) Relative expression levels of human NOTCH3 transgenes in isolated brain arteries (circle of Willis and its main branches) from wild-type TghN3(WT)46 and mutant TghN3(C428S)10 transgenic mice on Notch3+/+ and Notch3−/− backgrounds, at one month of age. Expression level of hNOTCH3 transgenes was normalized for β-actin and expressed relative to the level in mN3+/+;TghN3(WT) arteries and arbitrarily set to a value of 1.
Notably, we controlled for expression level of the hNOTCH3 transgenes when introduced into the Notch3 null background. This is particularly important since the hNOTCH3 transgenes are under the control of the SM22α promoter, and, transcriptional activity of this promoter is influenced by the level of Notch3 activity. Quantitative RT–PCR demonstrated that expression level of the wild-type NOTCH3 transgene, in the same brain arteries than those analysed above, was comparable in Notch3+/+ and Notch3−/− backgrounds, consistent with our finding that re-introduction of the wild-type Notch3 transgene in Notch3−/− arteries restored wild-type Notch3 activity. Conversely, expression level of the C428S mutant NOTCH3 transgene, which exhibited no rescuing activity, was reduced in the Notch3−/− background as compared to the wild-type Notch3 background (Fig. 2E). Nevertheless, it is worth mentioning that TghN3(C428S) mice retained a significant level of NOTCH3 transgene mRNA on the Notch3−/− background, that is ∼1.5-fold the amount expressed by the TghN3(WT) mice. Taken together, these data provide compelling evidence that the C428S mutation yields a non-functional protein in vivo, in the brain arteries in three different biological settings.
Analysis of C428S function in the presence of wild-type Notch3
Notch3 haploinsufficiency in the mouse has been associated with preserved arterial identity of smooth muscle cells as well as normal morphology of brain and peripheral arteries although RBP-Jκ activity is significantly reduced (Monet et al., 2007). Because CADASIL is inherited as a dominant disease, i.e. patients carry a wild-type allele in addition to the mutated allele, this prompted us to ascertain whether the mutant C428S allele is a simple loss of function allele resulting in haploinsufficiency or alternatively creates a dominant-negative allele. In this latter view, abnormal protein derived from the mutant allele interacts and interferes with protein derived from the wild-type allele, culminating in substantial loss of function.
We examined the brain arteries of the TghN3(C428S) mice from both lines on the Notch3+/− background in the three biological settings described above. Figure 3A shows that expression of the SM22α-lacZ transgene was reduced in the arteries of the circle of Willis (79 ± 6%, P < 0.001) and the medium-sized branches (74 ± 7%, P < 0.001) of SM22α-lacZ; mN3+/−;TghN3(C428S)10 mice, as compared to control mice, indicating an impaired arterial identity. High resolution optic microscopy and electron microscopy analysis clearly showed that brain arteries of mN3+/−;TghN3(C428S)10 had structural defects with segments of flattened elastical limina associated with thinning of smooth muscle cells (Fig. 3B, a and b). Morphological defects were further confirmed by electron microscopy analysis, which demonstrated thinning and vacuolization of smooth muscle cells as well as altered cytoskeleton (Fig. 3B; c and d). Finally, RBP-Jκ activity was strongly reduced in the arteries of mN3+/−;TghN3(C428S)10 mice as compared to Notch3+/− mice (Fig. 3C). Consistent with a dominant-negative effect, mN3+/−;TghN3(C428S) mice from the low-expressing line 2* exhibited less attenuated SM22α-lacZ expression and RBP-Jκ activity. Also, attenuation of RBP-Jκ activity in TghN3(C428S)10 mice was lesser on a Notch3+/+ background (Fig. 3C and data not shown). Taken together, our findings strongly suggest that the C428S mutant NOTCH3 can antagonize the function of wild-type Notch3.
Figure 3.
Dominant negative effect of the C428S mutant NOTCH3 protein. One-month-old mN3+/−;TghN3(C428S)10 and control mice were assayed for smooth muscle cell arterial identity (A), brain arterial morphology and smooth muscle cell ultrastructure (B) and Notch3/RBP-Jκ activity (C). (A) Left panel: lateral views of hemisphere from mice carrying the SM22α-lacZ transgene, showing reduced X-gal staining in the middle cerebral artery and its branches in TghN3(C428S) mice on a Notch3+/− background as compared to control Notch3+/− mice. Right panel: Relative β-galactosidase activity in the middle cerebral artery of mN3+/−;TghN3(C428S)10 (n = 6 arteries from three mice) and Notch3+/− mice (n = 25 arteries from 13 mice) heterozygous for the SM22α-lacZ transgene. Shown are mean intensities of blue X-gal staining (±SEM) expressed relative to those in Notch3+/− mice arbitrarily set to a value of 1. (*P < 0.001 versus Notch3+/− mice; unpaired Student's t-test). (B) Shown are representative semi-thin sections of brain arteries stained with toluidine blue (a and b) and representative electron micrographs (c and d) showing focal alterations of the arterial vessel with flattened elastica lamina (e.l.), smooth muscle cell (SMC) thinning and vacuolization (arrowheads) in mN3+/−; TghN3(C428S). Scale bar: 20 µm (a and b), 2.9 µm (c and d). (C) Shown are ventral views of whole brain from mice of the indicated genotype carrying the TP1-nlacZ transgene at the heterozygous state. X-gal staining of brain arteries in TghN3(C428S) mice on a Notch3+/− background is significantly reduced as compared to Notch3+/− control mice and mildly reduced in TghN3(C428S) mice on a Notch3+/+ background as compared to Notch3+/+ control mice.
Frequency of CADASIL-associated NOTCH3 mutations in the DSL-ligand binding
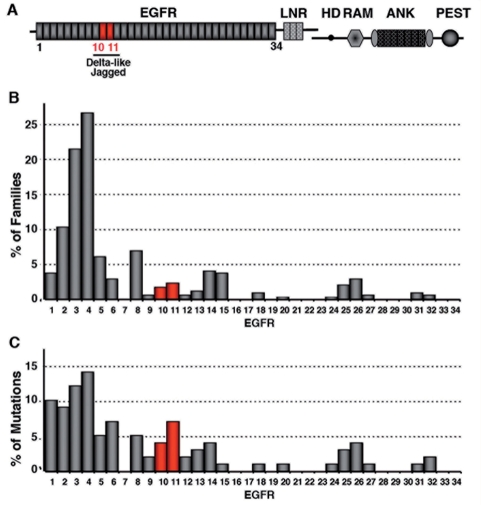
To assess the frequency of mutations occurring in this functional domain, which is encoded by exon 8 and short portions of exons 7 and 9, we performed a retrospective review of the literature. One hundred and fifty seven distinct CADASIL-associated NOTCH3 mutations have been reported in at least 503 different pedigrees (data not shown). Because earlier studies have identified mutational hotspots, notably in exons 3–4, 11 and 18, we suspected that mutations occurring outside these exons, particularly in EGFR10-11 coding exons, might have been underestimated (Joutel et al., 1997; Oberstein et al., 1999; Dichgans et al., 2000). We therefore considered only studies in which mutational scanning of all 34 EGFRs coding exons had been performed in large cohorts of patients (Joutel et al., 1997; Oberstein et al., 1999; Escary et al., 2000; Markus et al., 2002; Santa et al., 2003; Singhal et al., 2004; Dotti et al., 2005; Peters et al., 2005; Lee et al., 2006; Malandrini et al., 2007). Likewise, we excluded CADASIL families from Finland in whom a strong founder effect has been detected (Mykkanen et al., 2004). We selected a total of 350 distinct families, from at least seven different countries, with 99 different CADASIL mutations. Notably, this series included 39 recurrent mutations detected in 290 families, with 10 highly recurrent mutations, each being found in more than 10 distinct families. Overall there were 11 (11%) different mutations in EGFR10 or EGFR11 occurring in 14 (4%) distinct families (Fig. 4). All 11 mutations were single nucleotide substitutions and included eight mutations leading to the loss of a cysteine residue and three mutations to the gain of a cysteine residue (Supplementary Table 1). Of note, there were 40 (40%) different mutations in the mutational hotspot EGFR2-5, occurring in 225 distinct families (78%).
Figure 4.
Distribution of CADASIL-associated NOTCH3 mutations across EGFR1-34. (A) Schematic drawing of the NOTCH3 receptor with the different domains (EGFR, epidermal growth factor-like repeats; LNR, Lin12 repeats; HD, heterodimerization domain; TM, transmembrane domain; RAM domain; ANK, Ankyrin repeats; PEST domain). EGFR10-11 required for DSL-ligand binding domain are underlined. (B and C) Distribution across EGFR1-34 of the 99 CADASIL mutations identified in 350 distinct families, who underwent mutation scanning of exons 2–24 encoding the 34 EGFR. Bars represent the percentage of families harboring a mutation within a given EGFR (B) or the percentage of mutations per EGFR (C).
Genotype–phenotype correlations
The findings above prompted us to investigate whether NOTCH3 mutations in EGFR10-11 were associated with distinguishing phenotypes. Among the 176 patients from a prospective cohort study of CADASIL patients (Viswanathan et al., 2006; Jouvent et al., 2007; O'Sullivan et al., 2007), 10 patients from five distinct families had a mutation in the DSL-ligand binding domain, including six patients with the C428S mutation. These 10 patients were first used for case-control comparisons of disability, cognitive scores and MRI markers with 20 randomly selected patients from this cohort. We restricted the comparison group to the patients having a mutation in the mutational hotspot EFGR 2-5 because prior in vitro and in vivo studies showed that these mutations did not compromise Notch3 function. EGFR10-11 mutations were associated with significantly higher Mini-Mental State Examination and Mattis Dementia Rating Scale scores (P < 0.05) as well as a non-significant trend for a lower modified Rankin score. There was also a significantly higher volume of white matter hyperintensities (P < 0.01) and a non-significant trend to lower volume of lacunar lesions and lower number of microhaemorrhages in cases with EGFR10-11 mutations than in controls with EGFR2-5 mutations (Table 1). Further comparison of clinical and MRI markers between these 10 patients and all other patients with a mutation outside EGFR10-11 gave almost similar results (Supplementary Table 2).
Table 1.
Comparative assessment of neurological disability, cognitive impairment and MRI lesion load between EGFR10-11 and EGFR2-5 mutations carriers
| EGFR10-11 mutations | EGFR2-5 mutations | P-value | |
|---|---|---|---|
| Number of subjects | 10 | 20 | |
| Age, years (age range, mean ± SD) | 36–77, 61 ± 4.3 | 38–77, 60.5 ± 3.0 | n.s. |
| Disability and cognitive scores | |||
| Rankin scale score (% ≥2) | 10.0 | 42.1 | 0.09 |
| Mini-Mental State Examination score, mean ± SD | 28.0 ± 1.3 | 24.2 ± 5.6 | 0.05 |
| Mattis dementia rating scale score, mean ± SD | 141.4 ± 1.7 | 123.9 ± 26.0 | 0.04 |
| MRI markers | |||
| Brain parenchymal fraction, mean ± SD | 81.8 ± 5.4 | 79.5 ± 6.2 | 0.41 |
| Normalized volume of lacunar lesions, mean ± SD | 0.070 ± 0.06 | 0.085 ± 0.09 | 0.66 |
| Normalized volume of white matter hyperintensities, mean ± SD | 12.7 ± 5.8 | 6.48 ± 5.0 | 0.009 |
| Mean Apparent Diffusion Coefficient (10−4 mm2s−1) ± SD | 11.9 ± 0.5 | 12.2 ± 0.4 | 0.65 |
| Number of microbleeds, mean ± SD | 0.11 ± 0.33 | 2.6 ± 5.8 | 0.21 |
Discussion
To investigate the significance of Notch3 mutations located in the ligand binding domain, which account for about 4% of CADASIL families, we conducted genetic studies in mice and humans. Our results highlight distinctive functional and phenotypic features associated with these mutations and lead us to propose that this genotype acts a modifier of the CADASIL phenotype.
A major finding arising from this study is the difference between EGFR10-11 mutations and common mutations with respect to their effect on Notch3 function. Prior studies showed that common mutations, including those located in the mutational hotspot EGFR2-5, did not impair Notch3 signalling (Karlstrom et al., 2002; Joutel et al., 2004; Peters et al., 2004; Haritunians et al., 2005; Low et al., 2006). We further showed that one representative mutation, R90C in EGFR2, retained wild-type function in vivo in brain arteries, with respect to arterial identity (Supplementary Fig. 1), arterial structure and RBP-Jκ activity, even when Notch3ECD accumulated (Monet et al., 2007). The present study demonstrates that, by contrast, a representative mutation lying in EGFR10 (C428S) abrogates Notch3 function in vivo and antagonizes the function of the wild-type Notch3 allele. It is worth mentioning that both mutations were evaluated in the same three biological settings. Importantly, all three assays were conducted without a priori hypothesis on how Notch3 signalling is triggered. The molecular basis for the dominant negative effect of the C428S mutation remains unclear. Data presented here as well as observations from the literature suggest that the location of the mutation, in the DSL ligand binding domain, rather than its nature, altering one of the six cysteine residues of an EGFR, is largely responsible for the dominant negative effect. First, the R90C mutation in EGFR2, which also alters the number of cysteine residues in an EGFR, does not compromise Notch3 function (Monet et al., 2007). Second, the antimorphic effect is unlikely related to Notch3ECD aggregation, since it is observed at one month of age when Notch3ECD aggregation is yet not apparent (data not shown). Third, previous studies in Drosophila showed that the Q491V mutation in the DSL ligand binding domain of Notch receptor behaved genetically as both a Notch antimorphic and a loss of function mutation (de Celis et al., 1993). It is unlikely that C428S mutant Notch3 impedes trafficking of wild-type Notch3 to the cell surface since our previous cell biotinylation experiments showed that this mutant protein was properly localized to the cell surface (Joutel et al., 2004). Also, it is unlikely that mutant Notch3 titrates the ligand since our prior analysis showed that the mutation abrogated binding to Jagged1 (Joutel et al., 2004). Thus, at the present time we can only speculate that the C428S mutant Notch3 titrates wild-type Notch3 receptor into non-functional complexes.
Our genotype-phenotype correlation analysis suggests that EGFR10-11 mutations are associated with distinguishing clinical and MRI features. First, we found that this genotype was associated with relatively preserved cognitive performances in comparison to age-matched patients with common mutations who had lower Mini-Mental State Examination and Mattis Dementia Rating Scale scores. Moreover, there was a non-significant trend for lower disability in the EGFR10-11 mutation carriers as measured by the modified Rankin score, this difference reached statistical significance when the control group was extended to the whole cohort (Supplementary Table 2). Recent studies demonstrated that lacunar lesion volume and brain atrophy are the two strongest independent MRI markers of global cognitive impairment and disability in CADASIL (Jouvent et al., 2007; Viswanathan et al., 2007; Jouvent et al., 2008; Viswanathan et al., 2008). In the present study, EGFR10-11 mutation carriers presented with a slightly lower volume of lacunar infarctions and larger brain parenchymal fraction although the difference with the control group did not reach statistical significance, possibly because of the limited size of our sample. In contrast, we found a highly significant effect of EGFR10-11 mutations on the burden of white matter hyperintensities. Unexpectedly, this genotype was found associated with a larger volume of white matter hyperintensities in all statistical comparisons, although white matter hyperintensities have been reported to increase dramatically with age during the progression of the disease. The discrepancy between the preserved clinical status and larger amount of white matter hyperintensities in patients with EGFR10-11 mutations may be related to the nature of tissue alterations underlying white matter hyperintensities, as visualized by conventional MRI in CADASIL. They reflect largely variable types of tissue lesions and microstructural alterations of variable clinical relevance, ranging from discrete myelin oedema to major loss of white matter structural components (astrocytes, oligodendrocytes, axons and myelin). Accumulating evidence suggests that the extent of white matter hyperintensities does not correlate with the severity of functional or cognitive impairment when other MRI markers such as lacunar infarctions or brain parenchymal fraction are taken into account and thus may have only limited clinical consequences (Peters et al., 2006; Jouvent et al., 2007; Viswanathan et al., 2007, 2008; Jouvent et al., 2008). Moreover, diffusion tensor imaging studies previously showed that microstructural tissue loss underlying white matter hyperintensities was extremely variable among CADASIL patients. The results of these studies also demonstrated that the degree of cerebral tissue damage but not the extent of white matter hyperintensities were actually related to the clinical severity with a possible threshold for appearance of severe disability and/or dependence (Chabriat et al., 1999; Molko et al., 2002; Holtmannspotter et al., 2005). Therefore, one possibility could be that patients with EGFR10-11 mutations exhibit more white matter hyperintensities but with less destructive cerebral tissue lesions (microstructural loss, lacunar infarctions, atrophy). Further studies will be needed to test this hypothesis.
Only two smaller studies have previously examined the effect of this peculiar genotype on the CADASIL phenotype. These data appear in line with our results. In the first study, seven members from a single large Colombian kindred segregating a missense mutation (C455R) in EGFR11 were included (median age 42 years, age range 27–67-years old) (Arboleda-Velasquez et al., 2002). It is remarkable that all seven members had extensive white matter hyperintensities as assessed with the Scheltens score. In addition, although Arboleda-Velasquez et al. (2002) reported an earlier age-of-onset of stroke of more than one decade in this family, the four older patients had relatively well-preserved cognitive and functional status more than two decades after their first stroke. In the second study, three subjects (median age 53 years, age range 37–67 years) from a single family harboring the C440G mutation in EGFR11 were included. They also had a trend for a larger volume of white matter lesions (Singhal et al., 2004).
An important question arising from these findings is whether EGFR10-11 mutations and other common CADASIL mutations cause the disease through distinct mechanisms. The present study and our previous work establish that a change in cysteine residue number, but not the effect of the mutation on signalling, is the common denominator in the CADASIL disease. Our prior work supports the hypothesis that gain of novel function for the mutant protein is a likely mechanism for the more common CADASIL mutations and that this additional function may arise from novel protein–protein interactions. Specifically, the unpaired cysteine residue of the mutant Notch3 receptor may titrate key factors for vascular smooth muscle cell viability and function within the granular osmiophilic material deposits (Monet et al., 2007). Since Notch3ECD aggregation and granular osmiophilic material deposits are important features shared by common mutations and those in EGFR10-11 (this study and our unpublished results), the current data are compatible with a gain of novel function mechanism‚ common to both classes of mutations. Furthermore, our data lead us to propose that reduced Notch3 signaling acts as a modifier of the CADASIL phenotype. It has been proposed that the disease may progress in a two-step process, with a first silent stage of cumulative minor injuries, followed by a second stage of neurological disability when tissue damage becomes more severe and destructive (Chabriat et al., 1999). Based on our findings, we hypothesize that reduced Notch3 signalling associated with EGFR10-11 mutations might favour the occurrence of asymptomatic white matter changes visible on T2-weighted images while delaying the accumulation of severe and clinically relevant lesions, such as lacunar infarctions and brain atrophy.
Our study has limitations. First, our conclusions regarding the functional significance of EGFR10-11 mutations are based on the in vivo analysis of a single representative mutation in this domain. However in vitro studies have shown that another CADASIL-associated NOTCH3 mutation in EGFR 11 (C455R) behaved similarly to the C428S mutation (Joutel et al., 2004; Peters et al., 2004). Second, although this is the largest series of CADASIL patients with a mutation in the DSL ligand-binding domain analysed, genotype–phenotype correlations were obtained in a still limited number of patients and therefore must be interpreted cautiously. Definite proof will need replicating genotype–phenotype correlations in larger populations that will require international multicentre collaborations given the low frequency of this genotype. Also, a mouse model recapitulating brain parenchyma damages of CADASIL, when available, will be an invaluable tool in combination with Notch3 null mice to test the hypothesis that reduced Notch3 signalling is a modifier of the CADASIL phenotype.
In conclusion, this study highlights distinguishing functional and phenotypic features of CADASIL mutations in the Notch3 ligand-binding domain. Additional studies in humans and mice are warranted to further confirm these results and investigate the exciting possibility that reduced Notch3 signalling might attenuate the clinical severity of the CADASIL disease.
Supplementary material
Supplementary material is available at Brain online.
Funding
National Institutes of Health (R01 NS054122 to A.J.); GIS-ANR-Maladies Rares (05-011-02 to A.J.); PHRC (AOR-02-001 to H.C.); fellowship from the French Ministry of Education and Research (to M.M.L.); Contrat d’Interface INSERM-AP-HP (to A.J.).
Supplementary Material
Acknowledgements
We would like to thank Thomas Gridley for critical reading of the manuscript, Francina Langa-Vives and Cécile Goujet at the CIGM and SEAT transgenic mouse facilities, respectively, and Frédérick Baudin for animal maintenance.
Glossary
Abbreviations:
- DSL
Delta/Serrate/LAG-2
- EGFR
Epidermal Growth Factor-like Repeat
- Notch3ECD
Notch3 extracellular domain
References
- Arboleda-Velasquez JF, Lopera F, Lopez E, Frosch MP, Sepulveda-Falla D, Gutierrez JE, et al. C455R notch3 mutation in a Colombian CADASIL kindred with early onset of stroke. Neurology. 2002;59:277–9. doi: 10.1212/wnl.59.2.277. [DOI] [PubMed] [Google Scholar]
- Chabriat H, Pappata S, Poupon C, Clark CA, Vahedi K, Poupon F, et al. Clinical severity in CADASIL related to ultrastructural damage in white matter: in vivo study with diffusion tensor MRI. Stroke. 1999;30:2637–43. doi: 10.1161/01.str.30.12.2637. [DOI] [PubMed] [Google Scholar]
- Chabriat H, Vahedi K, Iba-Zizen MT, Joutel A, Nibbio A, Nagy TG, et al. Clinical spectrum of CADASIL: a study of 7 families. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Lancet. 1995;346:934–9. doi: 10.1016/s0140-6736(95)91557-5. [DOI] [PubMed] [Google Scholar]
- D'Souza B, Miyamoto A, Weinmaster G. The many facets of Notch ligands. Oncogene. 2008;27:5148–67. doi: 10.1038/onc.2008.229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Celis JF, Barrio R, del Arco A, Garcia-Bellido A. Genetic and molecular characterization of a Notch mutation in its Delta- and Serrate-binding domain in Drosophila. Proc Natl Acad Sci USA. 1993;90:4037–41. doi: 10.1073/pnas.90.9.4037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dichgans M, Ludwig H, Muller-Hocker J, Messerschmidt A, Gasser T. Small in-frame deletions and missense mutations in CADASIL: 3D models predict misfolding of Notch3 EGF-like repeat domains. Eur J Hum Genet. 2000;8:280–5. doi: 10.1038/sj.ejhg.5200460. [DOI] [PubMed] [Google Scholar]
- Dichgans M, Mayer M, Uttner I, Bruning R, Muller-Hocker J, Rungger G, et al. The phenotypic spectrum of CADASIL: clinical findings in 102 cases. Ann Neurol. 1998;44:731–9. doi: 10.1002/ana.410440506. [DOI] [PubMed] [Google Scholar]
- Domenga V, Fardoux P, Lacombe P, Monet M, Maciazek J, Krebs LT, et al. Notch3 is required for arterial identity and maturation of vascular smooth muscle cells. Genes Dev. 2004;18:2730–5. doi: 10.1101/gad.308904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dotti MT, Federico A, Mazzei R, Bianchi S, Scali O, Conforti FL, et al. The spectrum of Notch3 mutations in 28 Italian CADASIL families. J Neurol Neurosurg Psychiatry. 2005;76:736–8. doi: 10.1136/jnnp.2004.048207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eiraku M, Tohgo A, Ono K, Kaneko M, Fujishima K, Hirano T, et al. DNER acts as a neuron-specific Notch ligand during Bergmann glial development. Nat Neurosci. 2005;8:873–80. doi: 10.1038/nn1492. [DOI] [PubMed] [Google Scholar]
- Escary JL, Cecillon M, Maciazek J, Lathrop M, Tournier-Lasserve E, Joutel A. Evaluation of DHPLC analysis in mutational scanning of Notch3, a gene with a high G-C content. Hum Mutat. 2000;16:518–26. doi: 10.1002/1098-1004(200012)16:6<518::AID-HUMU9>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Gridley T. Notch signaling in vascular development and physiology. Development. 2007;134:2709–18. doi: 10.1242/dev.004184. [DOI] [PubMed] [Google Scholar]
- Haritunians T, Chow T, De Lange RP, Nichols JT, Ghavimi D, Dorrani N, et al. Functional analysis of a recurrent missense mutation in Notch3 in CADASIL. J Neurol Neurosurg Psychiatry. 2005;76:1242–8. doi: 10.1136/jnnp.2004.051854. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holtmannspotter M, Peters N, Opherk C, Martin D, Herzog J, Bruckmann H, et al. Diffusion magnetic resonance histograms as a surrogate marker and predictor of disease progression in CADASIL: a two-year follow-up study. Stroke. 2005;36:2559–65. doi: 10.1161/01.STR.0000189696.70989.a4. [DOI] [PubMed] [Google Scholar]
- Hu QD, Ang BT, Karsak M, Hu WP, Cui XY, Duka T, et al. F3/contactin acts as a functional ligand for Notch during oligodendrocyte maturation. Cell. 2003;115:163–75. doi: 10.1016/s0092-8674(03)00810-9. [DOI] [PubMed] [Google Scholar]
- Joutel A, Andreux F, Gaulis S, Domenga V, Cecillon M, Battail N, et al. The ectodomain of the Notch3 receptor accumulates within the cerebrovasculature of CADASIL patients. J Clin Invest. 2000;105:597–605. doi: 10.1172/JCI8047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Corpechot C, Ducros A, Vahedi K, Chabriat H, Mouton P, et al. Notch3 mutations in CADASIL, a hereditary adult-onset condition causing stroke and dementia. Nature. 1996;383:707–10. doi: 10.1038/383707a0. [DOI] [PubMed] [Google Scholar]
- Joutel A, Favrole P, Labauge P, Chabriat H, Lescoat C, Andreux F, et al. Skin biopsy immunostaining with a Notch3 monoclonal antibody for CADASIL diagnosis. Lancet. 2001;358:2049–51. doi: 10.1016/S0140-6736(01)07142-2. [DOI] [PubMed] [Google Scholar]
- Joutel A, Monet M, Domenga V, Riant F, Tournier-Lasserve E. Pathogenic mutations associated with cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy differently affect Jagged1 binding and Notch3 activity via the RBP/JK signaling Pathway. Am J Hum Genet. 2004;74:338–47. doi: 10.1086/381506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joutel A, Vahedi K, Corpechot C, Troesch A, Chabriat H, Vayssiere C, et al. Strong clustering and stereotyped nature of Notch3 mutations in CADASIL patients. Lancet. 1997;350:1511–5. doi: 10.1016/S0140-6736(97)08083-5. [DOI] [PubMed] [Google Scholar]
- Jouvent E, Mangin JF, Porcher R, Viswanathan A, O'Sullivan M, Guichard JP, et al. Cortical changes in cerebral small vessel diseases: a 3D MRI study of cortical morphology in CADASIL. Brain. 2008;131:2201–8. doi: 10.1093/brain/awn129. [DOI] [PubMed] [Google Scholar]
- Jouvent E, Viswanathan A, Mangin JF, O'Sullivan M, Guichard JP, Gschwendtner A, et al. Brain atrophy is related to lacunar lesions and tissue microstructural changes in CADASIL. Stroke. 2007;38:1786–90. doi: 10.1161/STROKEAHA.106.478263. [DOI] [PubMed] [Google Scholar]
- Kalimo H, Ruchoux MM, Viitanen M, Kalaria RN. CADASIL: a common form of hereditary arteriopathy causing brain infarcts and dementia. Brain Pathol. 2002;12:371–84. doi: 10.1111/j.1750-3639.2002.tb00451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karlstrom H, Beatus P, Dannaeus K, Chapman G, Lendahl U, Lundkvist J. A CADASIL-mutated Notch 3 receptor exhibits impaired intracellular trafficking and maturation but normal ligand-induced signaling. Proc Natl Acad Sci USA. 2002;99:17119–24. doi: 10.1073/pnas.252624099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YC, Yang AH, Liu HC, Wong WJ, Lu YC, Chang MH, et al. Cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy: two novel mutations in the NOTCH3 gene in Chinese. J Neurol Sci. 2006;246:111–5. doi: 10.1016/j.jns.2006.02.011. [DOI] [PubMed] [Google Scholar]
- Low WC, Santa Y, Takahashi K, Tabira T, Kalaria RN. CADASIL-causing mutations do not alter Notch3 receptor processing and activation. Neuroreport. 2006;17:945–9. doi: 10.1097/01.wnr.0000223394.66951.48. [DOI] [PubMed] [Google Scholar]
- Malandrini A, Gaudiano C, Gambelli S, Berti G, Serni G, Bianchi S, et al. Diagnostic value of ultrastructural skin biopsy studies in CADASIL. Neurology. 2007;68:1430–2. doi: 10.1212/01.wnl.0000264018.46335.c8. [DOI] [PubMed] [Google Scholar]
- Markus HS, Martin RJ, Simpson MA, Dong YB, Ali N, Crosby AH, et al. Diagnostic strategies in CADASIL. Neurology. 2002;59:1134–8. doi: 10.1212/wnl.59.8.1134. [DOI] [PubMed] [Google Scholar]
- Miyamoto A, Lau R, Hein PW, Shipley JM, Weinmaster G. Microfibrillar proteins MAGP-1 and MAGP-2 induce Notch1 extracellular domain dissociation and receptor activation. J Biol Chem. 2006;281:10089–97. doi: 10.1074/jbc.M600298200. [DOI] [PubMed] [Google Scholar]
- Moessler H, Mericskay M, Li Z, Nagl S, Paulin D, Small JV. The SM 22 promoter directs tissue-specific expression in arterial but not in venous or visceral smooth muscle cells in transgenic mice. Development. 1996;122:2415–25. doi: 10.1242/dev.122.8.2415. [DOI] [PubMed] [Google Scholar]
- Molko N, Pappata S, Mangin JF, Poupon F, LeBihan D, Bousser MG, et al. Monitoring disease progression in CADASIL with diffusion magnetic resonance imaging: a study with whole brain histogram analysis. Stroke. 2002;33:2902–8. doi: 10.1161/01.str.0000041681.25514.22. [DOI] [PubMed] [Google Scholar]
- Monet M, Domenga V, Lemaire B, Souilhol C, Langa F, Babinet C, et al. The archetypal R90C CADASIL-NOTCH3 mutation retains NOTCH3 function in vivo. Hum Mol Genet. 2007;16:982–92. doi: 10.1093/hmg/ddm042. [DOI] [PubMed] [Google Scholar]
- Mykkanen K, Savontaus ML, Juvonen V, Sistonen P, Tuisku S, Tuominen S, et al. Detection of the founder effect in Finnish CADASIL families. Eur J Hum Genet. 2004;12:813–9. doi: 10.1038/sj.ejhg.5201221. [DOI] [PubMed] [Google Scholar]
- O'Sullivan M, Ngo E, Viswanathan A, Jouvent E, Gschwendtner A, Saemann PG, et al. Hippocampal volume is an independent predictor of cognitive performance in CADASIL. doi: 10.1016/j.neurobiolaging.2007.09.002. Neurobiol Aging; doi:10.1016/j.neurobiolaging.2007.09.002 (In press) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oberstein SA, Ferrari MD, Bakker E, van Gestel J, Kneppers AL, Frants RR, et al. Diagnostic Notch3 sequence analysis in CADASIL: three new mutations in Dutch patients. Dutch CADASIL Research Group. Neurology. 1999;52:1913–5. doi: 10.1212/wnl.52.9.1913. [DOI] [PubMed] [Google Scholar]
- Opherk C, Peters N, Herzog J, Luedtke R, Dichgans M. Long-term prognosis and causes of death in CADASIL: a retrospective study in 411 patients. Brain. 2004;127:2533–9. doi: 10.1093/brain/awh282. [DOI] [PubMed] [Google Scholar]
- Peters N, Holtmannspotter M, Opherk C, Gschwendtner A, Herzog J, Samann P, et al. Brain volume changes in CADASIL: a serial MRI study in pure subcortical ischemic vascular disease. Neurology. 2006;66:1517–22. doi: 10.1212/01.wnl.0000216271.96364.50. [DOI] [PubMed] [Google Scholar]
- Peters N, Opherk C, Bergmann T, Castro M, Herzog J, Dichgans M. Spectrum of mutations in biopsy-proven CADASIL: implications for diagnostic strategies. Arch Neurol. 2005;62:1091–4. doi: 10.1001/archneur.62.7.1091. [DOI] [PubMed] [Google Scholar]
- Peters N, Opherk C, Zacherle S, Capell A, Gempel P, Dichgans M. CADASIL-associated Notch3 mutations have differential effects both on ligand binding and ligand-induced Notch3 receptor signaling through RBP-Jk. Exp Cell Res. 2004;299:454–64. doi: 10.1016/j.yexcr.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Ruchoux MM, Domenga V, Brulin P, Maciazek J, Limol S, Tournier-Lasserve E, et al. Transgenic mice expressing mutant Notch3 develop vascular alterations characteristic of cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Am J Pathol. 2003;162:329–42. doi: 10.1016/S0002-9440(10)63824-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruchoux MM, Guerouaou D, Vandenhaute B, Pruvo JP, Vermersch P, Leys D. Systemic vascular smooth muscle cell impairment in cerebral autosomal dominant arteriopathy with subcortical infarcts and leukoencephalopathy. Acta Neuropathol (Berl) 1995;89:500–12. doi: 10.1007/BF00571504. [DOI] [PubMed] [Google Scholar]
- Sakamoto K, Yamaguchi S, Ando R, Miyawaki A, Kabasawa Y, Takagi M, et al. The nephroblastoma overexpressed gene (NOV/ccn3) protein associates with Notch1 extracellular domain and inhibits myoblast differentiation via Notch signaling pathway. J Biol Chem. 2002;277:29399–405. doi: 10.1074/jbc.M203727200. [DOI] [PubMed] [Google Scholar]
- Santa Y, Uyama E, Chui DH, Arima M, Kotorii S, Takahashi K, et al. Genetic, clinical and pathological studies of CADASIL in Japan: a partial contribution of Notch3 mutations and implications of smooth muscle cell degeneration for the pathogenesis. J Neurol Sci. 2003;212:79–84. doi: 10.1016/s0022-510x(03)00109-6. [DOI] [PubMed] [Google Scholar]
- Schweisguth F. Regulation of notch signaling activity. Curr Biol. 2004;14:R129–38. [PubMed] [Google Scholar]
- Singhal S, Bevan S, Barrick T, Rich P, Markus HS. The influence of genetic and cardiovascular risk factors on the CADASIL phenotype. Brain. 2004;127:2031–8. doi: 10.1093/brain/awh223. [DOI] [PubMed] [Google Scholar]
- Souilhol C, Cormier S, Monet M, Vandormael-Pournin S, Joutel A, Babinet C, et al. Nas transgenic mouse line allows visualization of Notch pathway activity in vivo. Genesis. 2006;44:277–86. doi: 10.1002/dvg.20208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viswanathan A, Godin O, Jouvent E, O'Sullivan M, Gschwendtner A, Peters N, et al. Impact of MRI markers in subcortical vascular dementia: A multi-modal analysis in CADASIL. doi: 10.1016/j.neurobiolaging.2008.09.001. Neurobiol Aging; doi:10.1016/j.neurobiolaging.2008.09.001 (In press) [DOI] [PubMed] [Google Scholar]
- Viswanathan A, Gschwendtner A, Guichard JP, Buffon F, Cumurciuc R, O'Sullivan M, et al. Lacunar lesions are independently associated with disability and cognitive impairment in CADASIL. Neurology. 2007;69:172–9. doi: 10.1212/01.wnl.0000265221.05610.70. [DOI] [PubMed] [Google Scholar]
- Viswanathan A, Guichard JP, Gschwendtner A, Buffon F, Cumurcuic R, Boutron C, et al. Blood pressure and haemoglobin A1c are associated with microhaemorrhage in CADASIL: a two-centre cohort study. Brain. 2006;129:2375–83. doi: 10.1093/brain/awl177. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.