Summary
Mouse embryonic stem (ES) cells derived from pluripotent early epiblast contribute functionally differentiated progeny to all foetal lineages of chimaeras. By contrast, epistem cell (EpiSC) lines from post-implantation epithelialised epiblast are unable to colonise the embryo even though they express the core pluripotency genes Oct4, Sox2 and Nanog. We examined interconversion between these two cell types. ES cells can readily become EpiSCs in response to growth factor cues. By contrast, EpiSCs do not change into ES cells. We exploited PiggyBac transposition to introduce a single reprogramming factor, Klf4, into EpiSCs. No effect was apparent in EpiSC culture conditions, but in ground state ES cell conditions a fraction of cells formed undifferentiated colonies. These EpiSC-derived induced pluripotent stem (Epi-iPS) cells activated expression of ES cell-specific transcripts including endogenous Klf4, and downregulated markers of lineage specification. X chromosome silencing in female cells, a feature of the EpiSC state, was erased in Epi-iPS cells. They produced high-contribution chimaeras that yielded germline transmission. These properties were maintained after Cre-mediated deletion of the Klf4 transgene, formally demonstrating complete and stable reprogramming of developmental phenotype. Thus, re-expression of Klf4 in an appropriate environment can regenerate the naïve ground state from EpiSCs. Reprogramming is dependent on suppression of extrinsic growth factor stimuli and proceeds to completion in less than 1% of cells. This substantiates the argument that EpiSCs are developmentally, epigenetically and functionally differentiated from ES cells. However, because a single transgene is the minimum requirement to attain the ground state, EpiSCs offer an attractive opportunity for screening for unknown components of the reprogramming process.
Keywords: Induced pluripotent stem (iPS) cell, Chimaera, Leukaemia inhibitory factor (Lif), Reprogramming, Mitogen-activated protein kinase (Erk) kinase (Mek/Mkk), Embryonic stem (ES) cell
INTRODUCTION
In the mouse, pluripotent stem cell lines can be established from two distinct phases of early development (Rossant, 2008). Embryonic stem (ES) cells are obtained from naïve epiblast in pre-implantation blastocysts (Batlle-Morera et al., 2008; Brook and Gardner, 1997; Evans and Kaufman, 1981; Martin, 1981). Epistem cells (EpiSCs) are derived from columnar epithelial epiblast of the early post-implantation embryo (Brons et al., 2007; Tesar et al., 2007). ES cells retain the character of early epiblast and can be incorporated into the host embryo when injected into blastocysts (Gardner and Rossant, 1979). They subsequently contribute to all lineages of the developing and adult mouse (Beddington and Robertson, 1989; Bradley et al., 1984; Gardner and Rossant, 1979; Nagy et al., 1993). By contrast, neither freshly isolated post-implantation epiblast cells nor EpiSCs are capable of functional colonisation of a host blastocyst (Rossant et al., 1978; Tesar et al., 2007).
Both ES cells and EpiSCs are capable of multilineage differentiation in vitro and can form teratomas when grafted into adult mice (Brons et al., 2007; Tesar et al., 2007). Both cell types express the three transcriptional regulators, Oct4 (Pou5f1 - Mouse Genome Informatics), Sox2 and Nanog, that are generally considered to constitute the core pluripotency network (Boyer et al., 2005; Loh et al., 2006; Wang et al., 2006). However, there are significant differences in gene expression between ES cells and EpiSCs (Tesar et al., 2007). Furthermore, the culture conditions for maintaining the two cell types are quite distinct. ES cells self-renew in response to the cytokine leukaemia inhibitory factor (Lif) (Smith et al., 1988; Williams et al., 1988) and either serum, bone morphogenetic protein, or the inhibition of Mek/Erk signalling (Burdon et al., 1999; Ying et al., 2003; Ying et al., 2008). They are driven into differentiation by FGF/Erk signalling (Kunath et al., 2007; Stavridis et al., 2007). EpiSCs, by contrast, are maintained by FGF and activin (Brons et al., 2007).
The molecular basis for restriction of egg cylinder epiblast and EpiSCs as compared with naïve epiblast and ES cells is presently unclear. This issue acquires added significance in light of evidence that human embryo-derived stem cells are more akin to EpiSCs than to ground state ES cells (Brons et al., 2007; Rossant, 2008; Tesar et al., 2007). Here, we report on requirements for the interconversion of mouse ES cells and EpiSCs.
MATERIALS AND METHODS
EpiSC derivation and culture
EpiSCs were derived from E5.75 mouse embryos using activin A (20 ng/ml) and Fgf2 (12 ng/ml) essentially as described (Brons et al., 2007), except that we employed N2B27 medium (Ying and Smith, 2003). OE cell lines were derived from F1 embryos carrying an Oct4GiP (eGFPiresPuro) transgene (Ying et al., 2002). EpiSCs were also derived from non-transgenic strain 129 embryos. Cells were used between 10 and 25 passages. Differentiated cells could be eliminated as required from OE cultures by puromycin (1 μg/ml) selection for expression of the Oct4GiP transgene.
Embryonic stem cell and induced pluripotent stem (iPS) cell culture
2i/Lif comprises the Mek inhibitor PD0325901 (1 μM), the Gsk3 inhibitor CHIR99021 (3 μM), and leukaemia inhibitory factor (Lif, 100 U/ml) in N2B27 medium (Ying et al., 2008). Cultured cells were expanded by dissociation with trypsin and replating every 3 days.
ES cells overexpressing Klf4 were generated by electroporation of Oct4βgeo reporter cells (IOUD2) (Burdon et al., 1999) with a pPyCAGKlf4iP construct followed by puromycin selection (1 μg/ml). For ES cell to EpiSC differentiation, cells were plated at a density of 1-2×104 per cm2 in fibronectin-coated plates. Twenty-four hours after plating, the medium was changed to N2B27 containing activin A and Fgf2. Thereafter, cells were maintained in EpiSC culture conditions and passaged every 2-3 days. For colony formation, 1000 cells were plated per well in fibronectin-coated 6-well plates in activin A/Fgf2, or in laminin-coated plates in 2i/Lif. After 6 days, colonies were fixed and stained for alkaline phosphatase. Colonies were scored using ImageJ software.
PiggyBac vector transfection
To establish PiggyBac (PB) transgenic EpiSC lines, 1×106 cells were co-transfected using Lipofectamine 2000 (Invitrogen) with 1 μg of pGG137Klf4 or control pGG131 vector plus 2-3 μg of the PBase-expressing vector pCAGPBase (Wang et al., 2008). Transfection efficiency was evaluated by flow cytometry for DsRed expression. To select for stable transfectants, hygromycin (200 μg/ml) was applied for at least 5 days. To delete transgenes, 1×105 cells were transfected with 1 μg of Cre expression plasmid using Lipofectamine 2000. Five days after transfection, DsRed-negative cells were purified and individually deposited into a 96-well plate using a MoFlo high-performance cell sorter (DakoCytomation). After expansion, genomic PCR was employed to identify revertants lacking the Klf4 transgene. RT-PCR was used to confirm the lack of Klf4 transgene and of DsRed expression.
iPS cell induction and propagation
EpiSCs, either stable transfectants isolated after hygromycin selection or cells immediately after transfection, were plated at a density of 1×104, 5×104 and 1×105 cells per well of 6-well tissue culture plates in EpiSC culture condition. After 24 hours, medium was replaced with that containing 2i/Lif and subsequently refreshed every other day. The number of Oct4-GFP-positive clones was manually counted using fluorescence microscopy. ES cell-like clones were picked after 14 days in 2i/Lif and subsequently expanded by Accutase (PAA Laboratories) dissociation and replating every 3-4 days.
RT-PCR
Total RNA was prepared using the RNeasy Mini Kit (Qiagen) with DNaseI treatment. First-strand cDNA was synthesised using Superscript III reverse transcriptase (Invitrogen). Unless specified otherwise, real-time PCR was performed using Taqman Gene Expression Assays (Applied Biosystems). Gene expression was determined relative to Gapdh using the ΔCt method. Expression of the Klf4 transgene and of DsRed was determined by standard curve calibration. All quantitative PCR (qPCR) reactions were performed in a 7900HT Fast Real-Time PCR System (Applied Biosystems).
Taqman probes
Oct4, Mm00658129_gH; Klf4, Mm00516104_m1; Klf2, Mm01244979_g1; Klf5, Mm00456521_m1; Nanog, Mm02384862_g1; Rex1, Mm03053975_g; Fgf5, Mm00438615_m1; Lefty, Mm00438615_m1; brachyury (T), Mm01318252_m1; Nr0b1, Mm00431729_m1; Stella (Dppa3), Mm00836373_g1; Gapdh, 4352339E; β-actin (Actb), 4352341E.
Chimaera production
Term chimaeras were produced by microinjection into C57BL/6 blastocysts. Selected female chimaeras were mated with C57BL/6J black males. Germline transmission from cultured cell-derived oocytes manifests in agouti offspring.
RESULTS AND DISCUSSION
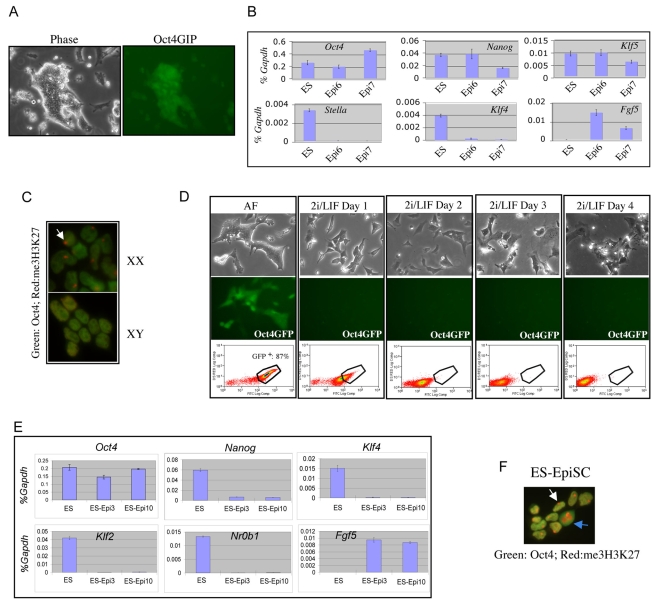
We derived EpiSCs from E5.75 mouse embryos carrying the Oct4GiP transgene (Ying et al., 2002). Cell lines were established and maintained without feeders in serum-free N2B27 medium (Ying and Smith, 2003) supplemented with activin A and Fgf2 (bFGF) (Brons et al., 2007). They grew as monolayers of closely apposed cells on a fibronectin substrate (Fig. 1A). The majority of cells expressed the Oct4-GFP reporter (Fig. 1A). Consistent with the original descriptions (Brons et al., 2007; Tesar et al., 2007), the EpiSCs we derived expressed the pluripotency markers Oct4 and Nanog, but not the early epiblast marker Stella (Dppa3) (Fig. 1B). EpiSCs also differ from ES cells by upregulation of the post-implantation markers Fgf5, T (brachyury) and Lefty (see Fig. S1 in the supplementary material). We established both male and female EpiSC lines. Immunofluorescence revealed a prominent body of nuclear staining for the repressive histone modification trimethylated H3 lysine 27 (me3H3K27) in the female line (Fig. 1C). This is diagnostic of a silent X chromosome (Silva et al., 2003). Thus, an emphatic epigenetic distinction between early and late epiblast is conserved in ES cells and EpiSCs, respectively. This is reflected in a differential ability to colonise chimaeric embryos (Tesar et al., 2007). We found that after morula aggregation, Oct4GiP EpiSCs could mix with inner cell mass (ICM) cells in blastocysts, but that they quickly downregulated GFP. Consistent with this, no contribution was detectable in egg cylinders after embryo transfer (see Fig. S6 in the supplementary material).
Fig. 1.
EpiSCs are distinct from, and do not spontaneously convert to, ES cells. (A) Phase contrast and fluorescence images of established EpiSC line. (B) qRT-PCR analysis of marker gene expression in ES cells and EpiSCs. ES, ES cells in 2i/Lif. Epi6 and Epi7 are two independent EpiSC lines. y-axis, relative expression normalised to Gapdh. (C) Immunostaining of male and female EpiSCs for me3H3K27 and Oct4. White arrow indicates focus of staining diagnostic of an inactive X chromosome. (D) EpiSCs lose Oct4 expression and differentiate or die in 2i/Lif. AF, EpiSC cultured in activin A plus Fgf2. (E) qRT-PCR analysis of ES cell differentiation into EpiSCs upon culture in Fgf2 and activin. Epi3 and Epi10 indicate cells cultured in Fgf2 and activin for three and ten passages, respectively. y-axis, relative expression normalised to Gapdh. (F) Oct4 and me3H3K27 immunostaining of female ES cell-derived EpiSCs. EpiSCs both express Oct4 and exhibit a nuclear body indicative of the inactive X (white arrow). Blue arrow indicates a dividing cell.
EpiSCs also lose expression of Oct4 and differentiate when transferred to conventional mouse ES cell culture conditions (Brons et al., 2007). Recently, however, it has been established that small molecules that selectively inhibit the Mek/Erk MAP kinase signalling cascade and glycogen synthase kinase 3 (Gsk3) provide, in combination with Lif, an optimal environment for derivation and propagation of ES cells from different rodent backgrounds in serum-free medium (Buehr et al., 2008; Ying et al., 2008) (J.N., unpublished). The combination of two inhibitors with Lif (2i/Lif) also promotes the generation of iPS cells (Silva et al., 2008). We therefore tested whether EpiSCs cultured in 2i/Lif might acquire features of ground state pluripotency. However, after transfer into 2i/Lif, EpiSCs underwent massive differentiation and death such that Oct4-GFP-expressing cells were entirely eliminated by 3 days (Fig. 1D). Some differentiated cells persisted, but in multiple platings of 1×107 EpiSCs not a single Oct4-GFP-expressing colony was obtained. Since genetic background has a strong influence on the derivation and propagation of ES cells and on iPS cell generation (Batlle-Morera et al., 2008; Silva et al., 2008), we also examined EpiSCs from the permissive 129 strain. These EpiSCs also failed to survive in 2i/Lif (data not shown). We conclude that the EpiSC represents a stable cell state that does not naturally revert to naïve pluripotent status.
The origin of ES cells and EpiSCs from early and late epiblast, respectively, suggests that ES cells might be capable of becoming EpiSCs. Indeed, ES cells transferred into EpiSC culture conditions continued to proliferate. After passaging, cultures became relatively homogenous and EpiSC-like. Thereafter, they displayed the marker profile of EpiSCs rather than of ES cells, with maintained Oct4, reduced Nanog and downregulated Rex1 (Zfp42 - Mouse Genome Informatics), Nr0b1 and Klf4 (Fig. 1E; see Fig. S2 in the supplementary material). Furthermore, EpiSCs derived from female ES cells showed a coincidence of Oct4 expression and X chromosome inactivation (Fig. 1F). This signature distinguishes EpiSCs from ES cells and differentiated somatic cell types. To confirm that this ES cell-derived EpiSC state was truly differentiated, we transferred cells back into 2i/Lif. Occasional ES cell-like colonies could initially be recovered, but not after four or more passages in activin plus Fgf2 (data not shown). We conclude that ES cells differentiate into EpiSCs, although a minority of undifferentiated cells persists for a while, as is commonly observed in other in vitro ES cell differentiation schema (Lowell et al., 2006; Smith, 2001).
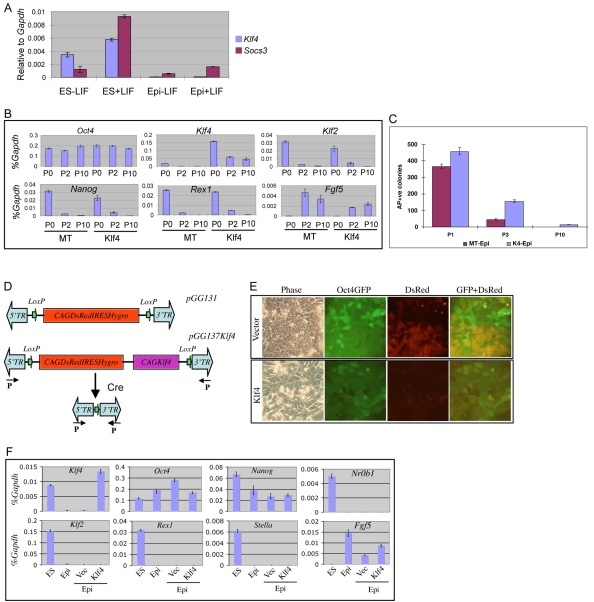
One of the genes prominently downregulated during differentiation of ES cells into EpiSCs is Klf4. Klf4 has been implicated in ES cell self-renewal (Jiang et al., 2008; Li et al., 2005). Klf4 is induced by Lif/Stat 3 signalling in ES cells, but not in EpiSCs (Fig. 2A). To test whether Klf4 might regulate the ES cell to EpiSC transition, we stably transfected ES cells with a Klf4 expression plasmid. These cells show greatly reduced dependency on Lif for self-renewal, as previously reported (Li et al., 2005). On transfer to EpiSC culture conditions, however, they responded similarly to parental ES cells, growing as a monolayer and downregulating ES cell-specific marker expression while maintaining Oct4 (Fig. 2B). This indicates that forced expression of Klf4 does not prevent conversion into EpiSCs. However, even after ten passages in activin and Fgf2, ES cell colonies were obtained at low frequency upon transfer to 2i/Lif (Fig. 2C). Therefore, constitutive Klf4 either allows long-term persistence of a small fraction of undifferentiated ES cells, or enables a fraction of EpiSCs to dedifferentiate and regain the ground state.
Fig. 2.
Klf4 neither prevents ES cell differentiation into EpiSCs nor converts an EpiSC population into ES cells in the presence of activin and FGF. (A) qRT-PCR analysis of Lif induction of Klf4 in ES cells but not in EpiSCs. Cells were stimulated with Lif (+LIF) for 1 hour. (B) ES cells constitutively expressing Klf4 acquire an EpiSC marker profile in Fgf2 plus activin A. MT, empty vector transfectants. P0, P2 and P10 indicate passage numbers in Fgf2/activin. (C) Constitutive Klf4 expression permits continued recovery of ES cell colonies after culture in activin and Fgf2. One thousand cells were plated for each sample in triplicate at the indicated passage (P) number. MT, empty vector transfectants; K4, Klf4 transfectants. (D) PiggyBac vector for expression of Klf4 (pGG137Klf4), and control PiggyBac vector (pGG131). Arrows (P) indicate PCR primers used to amplify the PB LTR fragment after Cre-mediated recombination. (E) Hygromycin-selected Klf4 and control vector-transfected EpiSCs. (F) qRT-PCR analysis showing that forced Klf4 expression does not induce ES cell marker gene expression in EpiSC culture. ES, ES cells; Epi, EpiSCs; Vec, EpiSC transfected with control vector pGG131; Klf4, EpiSCs transfected with pGG137Klf4. y-axis, relative expression normalised to Gapdh.
To distinguish between these possibilities, we investigated whether forced expression of Klf4 in embryo-derived EpiSCs could induce ground state pluripotency. We used PiggyBac (PB) vector-chromosome transposition to achieve high efficiency stable transfection (Wang et al., 2008). The PB vector contains independent CAG promoter units directing expression of the Klf4 open reading frame and of a DsRed reporter with a linked hygromycin resistance gene (Fig. 2D). LoxP sites adjacent to the PB terminal repeats allow for excision of both expression units. Fewer DsRed-expressing cells were obtained after Klf4 transfection than following control vector transfection (data not shown) and the level of DsRed expression was reduced. Overexpression of Klf4 might therefore be toxic to EpiSCs. Nonetheless, Klf4/DsRed-expressing EpiSCs were isolated following hygromycin selection (Fig. 2E). In activin and Fgf2 they did not upregulate ES cell-specific genes (Fig. 2F) and female cells maintained an inactive X chromosome as judged by me3H3K27 staining (see Fig. S3 in the supplementary material). We conclude that the expression of Klf4 at a similar RNA level to that present in ES cells is not alone sufficient to reset EpiSCs and instate full pluripotency in cells maintained in activin and Fgf2.
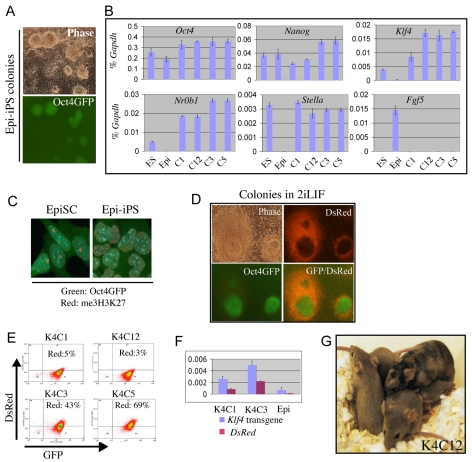
We then examined the effect of Klf4 transfection in 2i/Lif conditions, which have been shown to promote the final stages of iPS cell generation from neural stem cells and fibroblasts (Silva et al., 2008). Klf4-transfected EpiSCs were transferred into 2i/Lif 48 or 72 hours after transfection. They exhibited a wave of differentiation and cell death, similar to non-transfected EpiSCs. After 4 days in 2i/Lif, however, multiple Oct4-GFP-positive colonies emerged (Fig. 3A). These colonies had the tightly packed three-dimensional aggregate form typical of ES cells in 2i. They arose at a frequency of 0.1-0.2% of cells surviving transfection (21 colonies from 1×104 cells in one typical experiment). GFP-positive colonies were never obtained in 2i/Lif from multiple control vector transfections. We then tested whether stably transfected EpiSCs propagated in activin and Fgf2 could convert in 2i/Lif. Klf4 transfectants generated colonies in 2i/Lif with similar kinetics to cells transferred directly after transfection (see Fig. S4 in the supplementary material). The yield was higher, at ∼1%. This could point to some element of reprogramming proceeding in the stable transfectants, but could also be explained by elimination of non-transfectants and cells with toxic overexpression of Klf4. Significantly, the frequency did not noticeably increase with passaging of the stable transfectants, indicating that if there is any partial reprogramming this does not accumulate in the cultures.
Fig. 3.
EpiSCs transfected with Klf4 can convert to ground state pluripotency. (A) Oct4-positive colonies obtained by transfection with Klf4 and transfer to 2i/Lif after 72 hours. Images were taken after 9 days in 2i/Lif. (B) qRT-PCR analysis of marker gene expression in ES cells, EpiSCs and derivative Epi-iPS cells isolated in 2i/Lif. y-axis, relative expression normalised to Gapdh. (C) me3H3K27 staining of female EpiSCs and derivative Epi-iPS cells. (D) Images of Epi-iPS colonies after 10 days in 2i/Lif, showing mutually exclusive expression of DsRed and Oct4-GFP. (E) Flow cytometry analysis of four expanded Epi-iPS clones. Two clones retain weak but detectable red fluorescence. (F) qRT-PCR analysis of Klf4 transgene and DsRed expression in Epi-iPS cell clones and parental EpiSC line. y-axis, relative expression normalised to Gapdh. (G) Chimeric mouse produced from the K4C12 Epi-iPS clone and agouti germline offspring.
Ten out of 12 colonies picked from a transfection with transfer to 2i/Lif after 72 hours expanded with undifferentiated morphology and stable expression of Oct4-GFP. qRT-PCR analysis showed the marker profile of ES cells, with upregulation of Stella and Klf2. Conversely, Fgf5 and brachyury mRNAs were lost (Fig. 3B; see Fig. S5 in the supplementary material). We examined me3H3K27 immunostaining and found that the nuclear body corresponding to the inactive X chromosome was lost in Oct4-GFP-positive cells after transfer to 2i/Lif (Fig. 3C). A potential disadvantage of using the PB vector and CAG promoter is that transgenes might not be silenced. However, in each of the GFP-positive clones we observed partial or complete loss of visible DsRed expression (Fig. 3D,E), although qRT-PCR analysis revealed that the transgenes were not completely silenced (Fig. 3F).
We injected cells without visible DsRed expression into C57BL/6 blastocysts. Healthy chimaeras were obtained with extensive agouti coat colour contributions (Fig. 3G). Female chimaeras mated with C57BL/6 males produced agouti offspring, indicating transmission of the cultured cell genome. This confirms that the developmental capacity has been fully derestricted and the authentic pluripotent state established. These cells should therefore be considered as EpiSC-derived iPS cells, or Epi-iPS cells.
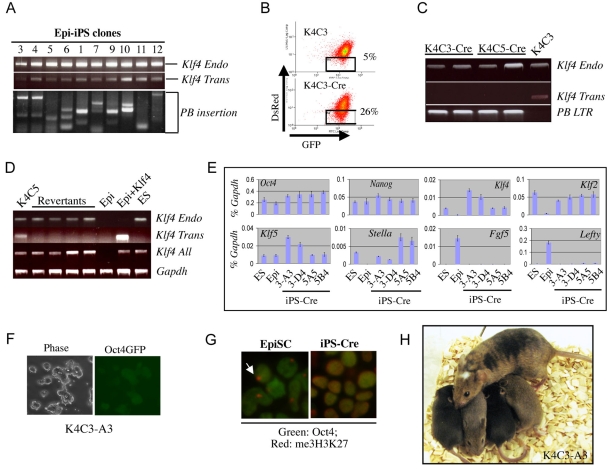
We examined the copy number of PB integrations by genomic PCR analysis of ten Epi-iPS cell clones. Each Epi-iPS cell line carried 1-3 PB insertions (Fig. 4A). To determine whether the low, but still detectable, Klf4 transgene expression might play a role in maintaining the induced phenotype we excised the transgene copies. We chose two DsRed-positive clones and transfected each with a Cre expression plasmid. After 5 days, cells that no longer expressed DsRed were isolated using flow cytometry with single-cell deposition into 96-well plates (Fig. 4B). Resulting clones were screened by genomic PCR for absence of the Klf4 transgene and presence of a reverted PB fragment (Fig. 4C). Two thirds of the expanded clones retained only the PB terminal repeats. RT-PCR analysis failed to detect expression of the Klf4 transgene or DsRed from these revertants (Fig. 4D). They retained ES cell morphology, Oct4-GFP expression and ES cell marker profile (Fig. 4E,F). The X chromosome silencing mark, me3H3K27, was undetectable (Fig. 4G). These cells incorporated efficiently into the ICM and subsequently the egg cylinder after morula aggregation (see Fig. S6 in the supplementary material). We injected transgene-deleted cells into blastocysts and obtained viable high-contribution chimaeras (see Table S1 in the supplementary material). Female chimaeras from two out of three clones produced agouti offspring in their first litter (Fig. 4H), indicative of transmission of iPS cell-derived oocytes. Therefore, complete removal of the Klf4 transgene does not destabilise the induced ground state. This establishes that reprogramming has been finalised and does not depend upon ongoing transgene expression or insertional mutagenesis.
Fig. 4.
Retention of ground state pluripotency after transgene excision. (A) Splinkerette-PCR reveals 1-3 PB insertions in each iPS clone. (B) Flow cytometry showing the DsRed-negative population in the K4C3 line before and after Cre transfection. (C) Genomic PCR showing loss of the Klf4 transgene and gain of the PB-LTR fragment in two revertant clones. (D) RT-PCR analysis showing the lack of Klf4 transgene and DsRed expression in expanded Cre-reverted cells. (E) Marker gene expression in Cre-reverted Epi-iPS cells (iPS-Cre) compared with ES cells and EpiSCs. Data are for two independent Cre-revertant clones derived from each of Epi-iPS cell lines K4C3 and K4C5. y-axis, relative expression normalised to Gapdh. (F) Maintained morphology and Oct4-GFP expression in a Cre-reverted Epi-iPS cell line. (G) me3H3K27 staining of Klf4 transgene-deleted iPS cells as compared with parental EpiSCs. (H) Chimeric mouse made with revertant K4C3-A3 cells, and agouti offspring denoting germline transmission.
The restricted potency of EpiSCs as compared with ES cells appears to mirror the developmental progression from naïve pre-implantation epiblast to epithelialised egg cylinder (Rossant, 2008). ES cells in vitro recapitulate this conversion with stable alterations in gene expression, growth factor dependence and epigenetic status. These differentiation changes may be completely reversed by re-expression of a single gene that is normally downregulated in EpiSCs. Klf4 in combination with culture in 2i/Lif reverts EpiSCs to the naïve ground state. This is mediated by transcriptional resetting accompanied by activation of contrasting epigenetic processes that restrict expression of exogenous DNA sequences and erase X chromosome inactivation. Interestingly, although colonies with features of iPS cells are detectable within 7 days of Klf4 transfection, all clones recovered showed stable integration of at least one copy of the transgene, suggesting a requirement for sustained expression to effect reprogramming. However, the combination of PB-mediated low copy number integration and Cre-mediated excision facilitated formal proof that once attained, the Epi-iPS cell state does not require the continuous presence of introduced DNA elements (Okita et al., 2008; Stadtfeld et al., 2008).
It is apparent that the exogenous transcription factor requirements for inducing pluripotency vary with the starting cell type as do the efficiency and kinetics of molecular reprogramming (Aoi et al., 2008; Kim et al., 2008; Silva et al., 2008). It is striking, however, that even though other core components of the pluripotent network are already present, only ∼1% of Klf4-expressing EpiSCs become iPS cells. This emphasises that even though there are transcriptional similarities, EpiSCs are truly differentiated from ground state ES cells. Their reprogramming efficiency is limited in an analogous manner to that of somatic cells by currently unknown parameters. However, because a single transgene is sufficient, we suggest that EpiSCs might provide an attractive system in which to screen for new components of the reprogramming process.
Finally, continuous expression of Klf4 does not prevent ES cell differentiation into EpiSCs when exposed to inductive extrinsic factors. Nonetheless, downregulation of Klf4 may help to ensure developmental restriction of epithelialised epiblast in the embryo and safeguard against dedifferentiation to a naïve and teratogenic condition. We suggest that the creation of iPS cells might be intimately related, mechanistically, to the molecular transitions through which ground state pluripotency is generated and then restricted in the early phases of mammalian embryogenesis.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/7/1063/DC1
Supplementary Material
We thank Jose Silva for discussion and comments on the manuscript. Mary Chol generated the morula aggregation chimaeras, Marko Hyvonen provided recombinant activin A and Fgf2, and Rachel Walker provided flow cytometry assistance. We thank Adrian Woodhouse, Samuel Jameson and colleagues for mouse husbandry. This research was supported by the Medical Research Council, The Wellcome Trust and the EC Project EuroSystem. G.G. is a Medical Research Council Stem Cell Career Development Fellow and A.S. is a Medical Research Council Professor. Deposited in PMC for release after 6 months.
References
- Aoi, T., Yae, K., Nakagawa, M., Ichisaka, T., Okita, K., Takahashi, K., Chiba, T. and Yamanaka, S. (2008). Generation of pluripotent stem cells from adult mouse liver and stomach cells. Science 321, 699-702. [DOI] [PubMed] [Google Scholar]
- Batlle-Morera, L., Smith, A. G. and Nichols, J. (2008). Parameters influencing derivation of embryonic stem cells from murine embryos Genesis 46, 758-767. [DOI] [PubMed] [Google Scholar]
- Beddington, R. S. P. and Robertson, E. J. (1989). An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development 105, 733-737. [DOI] [PubMed] [Google Scholar]
- Boyer, L. A., Lee, T. I., Cole, M. F., Johnstone, S. E., Levine, S. S., Zucker, J. P., Guenther, M. G., Kumar, R. M., Murray, H. L., Jenner, R. G. et al. (2005). Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947-956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bradley, A., Evans, M. J., Kaufman, M. H. and Robertson, E. (1984). Formation of germ-line chimaeras from embryo-derived teratocarcinoma cell lines. Nature 309, 255-256. [DOI] [PubMed] [Google Scholar]
- Brons, I. G., Smithers, L. E., Trotter, M. W., Rugg-Gunn, P., Sun, B., Chuva de Sousa Lopes, S. M., Howlett, S. K., Clarkson, A., Ahrlund-Richter, L., Pedersen, R. A. et al. (2007). Derivation of pluripotent epiblast stem cells from mammalian embryos. Nature 448, 191-195. [DOI] [PubMed] [Google Scholar]
- Brook, F. A. and Gardner, R. L. (1997). The origin and efficient derivation of embryonic stem cells in the mouse. Proc. Natl. Acad. Sci. USA 94, 5709-5712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buehr, M., Meek, S., Blair, K., Yang, J., Ure, J., Silva, J., McLay, R., Hall, J., Ying, Q. L. and Smith, A. (2008). Capture of authentic embryonic stem cells from rat blastocysts. Cell 135, 1287-1298. [DOI] [PubMed] [Google Scholar]
- Burdon, T., Stracey, C., Chambers, I., Nichols, J. and Smith, A. (1999). Suppression of SHP-2 and ERK signalling promotes self-renewal of mouse embryonic stem cells. Dev. Biol. 210, 30-43. [DOI] [PubMed] [Google Scholar]
- Evans, M. J. and Kaufman, M. H. (1981). Establishment in culture of pluripotential cells from mouse embryos. Nature 292, 154-156. [DOI] [PubMed] [Google Scholar]
- Gardner, R. L. and Rossant, J. (1979). Investigation of the fate of 4-5 day post-coitum mouse inner cell mass cells by blastocyst injection. J. Embryol. Exp. Morphol. 52, 141-152. [PubMed] [Google Scholar]
- Jiang, J., Chan, Y. S., Loh, Y. H., Cai, J., Tong, G. Q., Lim, C. A., Robson, P., Zhong, S. and Ng, H. H. (2008). A core Klf circuitry regulates self-renewal of embryonic stem cells. Nat. Cell Biol. 10, 353-360. [DOI] [PubMed] [Google Scholar]
- Kim, J. B., Zaehres, H., Wu, G., Gentile, L., Ko, K., Sebastiano, V., Arauzo-Bravo, M. J., Ruau, D., Han, D. W., Zenke, M. et al. (2008). Pluripotent stem cells induced from adult neural stem cells by reprogramming with two factors. Nature 454, 646-650. [DOI] [PubMed] [Google Scholar]
- Kunath, T., Saba-El-Leil, M. K., Almousailleakh, M., Wray, J., Meloche, S. and Smith, A. (2007). FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 134, 2895-2902. [DOI] [PubMed] [Google Scholar]
- Li, Y., McClintick, J., Zhong, L., Edenberg, H. J., Yoder, M. C. and Chan, R. J. (2005). Murine embryonic stem cell differentiation is promoted by SOCS-3 and inhibited by the zinc finger transcription factor Klf4. Blood 105, 635-637. [DOI] [PubMed] [Google Scholar]
- Loh, Y. H., Wu, Q., Chew, J. L., Vega, V. B., Zhang, W., Chen, X., Bourque, G., George, J., Leong, B., Liu, J. et al. (2006). The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat. Genet. 38, 431-440. [DOI] [PubMed] [Google Scholar]
- Lowell, S., Benchoua, A., Heavey, B. and Smith, A. G. (2006). Notch promotes neural lineage entry by pluripotent embryonic stem cells. PLoS Biol. 4, 805-818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin, G. R. (1981). Isolation of a pluripotent cell line from early mouse embryos cultured in medium conditioned by teratocarcinoma stem cells. Proc. Natl. Acad. Sci. USA 78, 7634-7638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy, A., Rossant, J., Nagy, R., Abramow-Newerly, W. and Roder, J. C. (1993). Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA 90, 8424-8428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okita, K., Nakagawa, M., Hyenjong, H., Ichisaka, T. and Yamanaka, S. (2008). Generation of mouse induced pluripotent stem cells without viral vectors. Science 322, 949-953. [DOI] [PubMed] [Google Scholar]
- Rossant, J. (2008). Stem cells and early lineage development. Cell 132, 527-531. [DOI] [PubMed] [Google Scholar]
- Rossant, J., Gardner, R. L. and Alexandre, H. L. (1978). Investigation of the potency of cells from the postimplantation mouse embryo by blastocyst injection: a preliminary report. J. Embryol. Exp. Morphol. 48, 239-247. [PubMed] [Google Scholar]
- Silva, J., Mak, W., Zvetkova, I., Appanah, R., Nesterova, T. B., Webster, Z., Peters, A. H., Jenuwein, T., Otte, A. P. and Brockdorff, N. (2003). Establishment of histone H3 methylation on the inactive X chromosome requires transient recruitment of Eed-Enx1 polycomb group complexes. Dev. Cell 4, 481-495. [DOI] [PubMed] [Google Scholar]
- Silva, J., Barrandon, O., Nichols, J., Kawaguchi, J., Theunissen, T. W. and Smith, A. (2008). Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 6, 2237-2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith, A. G. (2001). Embryo-derived stem cells: of mice and men. Annu. Rev. Cell Dev. Biol. 17, 435-462. [DOI] [PubMed] [Google Scholar]
- Smith, A. G., Heath, J. K., Donaldson, D. D., Wong, G. G., Moreau, J., Stahl, M. and Rogers, D. (1988). Inhibition of pluripotential embryonic stem cell differentiation by purified polypeptides. Nature 336, 688-690. [DOI] [PubMed] [Google Scholar]
- Stadtfeld, M., Nagaya, M., Utikal, J., Weir, G. and Hochedlinger, K. (2008). Induced pluripotent stem cells generated without viral integration. Science 322, 945-949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stavridis, M. P., Lunn, J. S., Collins, B. J. and Storey, K. G. (2007). A discrete period of FGF-induced Erk1/2 signalling is required for vertebrate neural specification. Development 134, 2889-2894. [DOI] [PubMed] [Google Scholar]
- Tesar, P. J., Chenoweth, J. G., Brook, F. A., Davies, T. J., Evans, E. P., Mack, D. L., Gardner, R. L. and McKay, R. D. (2007). New cell lines from mouse epiblast share defining features with human embryonic stem cells. Nature 448, 196-199. [DOI] [PubMed] [Google Scholar]
- Wang, J., Rao, S., Chu, J., Shen, X., Levasseur, D. N., Theunissen, T. W. and Orkin, S. H. (2006). A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364-368. [DOI] [PubMed] [Google Scholar]
- Wang, W., Lin, C., Lu, D., Ning, Z., Cox, T., Melvin, D., Wang, X., Bradley, A. and Liu, P. (2008). Chromosomal transposition of PiggyBac in mouse embryonic stem cells. Proc. Natl. Acad. Sci. USA 105, 9290-9295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, R. L., Hilton, D. J., Pease, S., Willson, T. A., Stewart, C. L., Gearing, D. P., Wagner, E. F., Metcalf, D., Nicola, N. A. and Gough, N. M. (1988). Myeloid leukaemia inhibitory factor maintains the developmental potential of embryonic stem cells. Nature 336, 684-687. [DOI] [PubMed] [Google Scholar]
- Ying, Q. L. and Smith, A. G. (2003). Defined conditions for neural commitment and differentiation. Methods Enzymol. 365, 327-341. [DOI] [PubMed] [Google Scholar]
- Ying, Q. L., Nichols, J., Evans, E. P. and Smith, A. G. (2002). Changing potency by spontaneous fusion. Nature 416, 545-548. [DOI] [PubMed] [Google Scholar]
- Ying, Q. L., Nichols, J., Chambers, I. and Smith, A. (2003). BMP induction of Id proteins suppresses differentiation and sustains embryonic stem cell self-renewal in collaboration with STAT 3. Cell 115, 281-292. [DOI] [PubMed] [Google Scholar]
- Ying, Q. L., Wray, J., Nichols, J., Batlle-Morera, L., Doble, B., Woodgett, J., Cohen, P. and Smith, A. (2008). The ground state of embryonic stem cell self-renewal. Nature 453, 519-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.