Summary
The Bicoid (Bcd) protein gradient is generally believed to be established in pre-blastoderm Drosophila embryos by the diffusion of Bcd protein after translation of maternal mRNA, which serves as a strictly localized source of Bcd at the anterior pole. However, we previously published evidence that the Bcd gradient is preceded by a bcd mRNA gradient. Here, we have revisited and extended this observation by showing that the bcd mRNA and Bcd protein gradient profiles are virtually identical at all times. This confirms our previous conclusion that the Bcd gradient is produced by a bcd mRNA gradient rather than by diffusion. Based on our observation that bcd mRNA colocalizes with Staufen (Stau), we propose that the bcd mRNA gradient forms by a novel mechanism involving quasi-random active transport of a Stau-bcd mRNA complex through a nonpolar microtubular network, which confines the bcd mRNA to the cortex of the embryo.
Keywords: bicoid, Morphogenetic gradient, Staufen, ARTS model
INTRODUCTION
The bicoid (bcd) gene is a well-known maternal patterning gene that was discovered in a large-scale screen for female-sterile mutants in Drosophila. Its function is required maternally for the development of the larval head and thorax, and its activity was suggested to decline sharply with increasing distance from the anterior pole of a cleavage-stage embryo (Frohnhöfer and Nüsslein-Volhard, 1986). In textbooks, the Bcd protein serves as paradigm as the first identified morphogen, the concentration gradient of which provides the initial positional information in the anterior half of the embryo where it differentially activates segmentation genes, particularly gap genes. The activity of the morphogen is expressed in a spectacular gradient of the Bcd protein, the concentration of which approximates an exponential decline with distance from the anterior pole in the nuclei of syncytial-blastoderm embryos (Driever and Nüsslein-Volhard, 1988; Houchmandzadeh et al., 2002). This gradient and its activity provided the first experimental proof of the French Flag model of pattern formation proposed by Wolpert (Wolpert, 1969). In this model, Wolpert reasoned that the spatial concentration gradient of a morphogen, with its maximum at one end of a uniaxial field of cells, could provide the positional information that uniquely specifies cellular fates.
The bcd gene was first isolated and characterized at the molecular level in an initial test of the gene network hypothesis, which states that genes belonging to a network with an integrated function are structurally linked by a relatively small number of protein-coding and cis-regulatory domains that are characteristic of the network (Frigerio et al., 1986) (reviewed by Noll, 1993). Two crucial properties of bcd were identified: (1) bcd encodes a transcription factor that includes a homeodomain, and (2) its maternal mRNA forms a concentration gradient along the anteroposterior axis of the embryo at stages preceding cellular blastoderm (Frigerio et al., 1986). These properties explained, for the first time, the ability of the bcd product to act as a maternal morphogen. Changes in the spatial distribution of maternal bcd mRNA, visualized in tissue sections by in situ hybridization with a 3H-labeled bcd cDNA, were documented at three stages: (1) bcd transcripts accumulate at the anterior margin of oocytes in the female abdomen; (2) after fertilization transcripts move posteriorly, forming a concentration gradient along the anteroposterior axis in cleavage-stage embryos; and (3) during nuclear cycle 14, transcripts display an extended gradient along the apical cortex of the syncytial-blastoderm embryo (Frigerio et al., 1986).
Later, another paper appeared that showed the bcd mRNA patterns of an early cleavage-stage and a syncytial-blastoderm embryo (Berleth et al., 1988). Although both patterns were entirely consistent with those published earlier (Frigerio et al., 1986), this paper mentioned only that bcd transcripts were “becoming concentrated in the cortical cytoplasm in the form of an anterior cap”, but disregarded the presence of the mRNA gradient. Soon thereafter, the first publication appeared demonstrating that Bcd protein forms an exponential concentration gradient with its maximum at the anterior pole, reaching background levels in the posterior third of embryos at early nuclear cycle 14 (Driever and Nüsslein-Volhard, 1988). The authors proposed that the exponential gradient is generated from a bcd mRNA source, localized in the anterior-most portion of the embryo, by diffusion and dispersed degradation of the Bcd protein. However, they provided no evidence to support a crucial feature of their model, namely their contention that bcd mRNA, the source of Bcd protein, remains localized at the anterior pole. In a later attempt to compensate for this shortcoming, St Johnston et al. (St Johnston et al., 1989) described the spatial distribution of bcd mRNA. They indicated that it became localized to the periphery of the embryo in a movement that “sometimes resulted in a slight posteriorwards shift in the RNA distribution”. Still, the Bcd protein diffusion model was not questioned.
Since then, this model has become anchored in textbooks as a fundamental paradigm of developmental biology. Its importance and implications have been described in a review (Ephrussi and St Johnston, 2004), supplemented with personal perspectives of the authors who proposed it (Driever, 2004; Nüsslein-Volhard, 2004). Recently, however, this SDD model - the naming of which refers to the localized synthesis, diffusion and spatially uniform degradation of the Bcd protein - has been subjected to a critical test (Gregor et al., 2007). The model assumes a Bcd protein source that is fed by translation of the bcd mRNA localized at the anterior pole, diffusion of Bcd away from its source, and spatially uniform degradation by a first-order reaction (Gregor et al., 2007), which predicts an exponential decay of the Bcd concentration with increasing distance from the anterior pole (Wolpert, 1969). Although this prediction was verified, a series of ingenious experiments that measured the diffusion constant of Bcd in the cortex of embryos uncovered a serious difficulty with the model: the diffusion constant was two orders of magnitude too low to explain the observation that the steady state of the Bcd gradient profile is reached within 1.5 hours (Gregor et al., 2007).
Here, we propose a simple solution to this dilemma by reinforcing and extending results that documented a bcd mRNA gradient, in agreement with the first molecular characterization of the bcd gene (Frigerio et al., 1986). Revisiting our published results and using a sensitive fluorescent in situ hybridization (FISH) method and confocal microscopy, we demonstrate that (1) a bcd mRNA concentration gradient is formed along the cortex of the embryo by nuclear cycle 10 or the beginning of syncytial blastoderm; (2) the gradient falls off exponentially with distance from the anterior pole and persists unchanged during nuclear cycles 10-13; (3) bcd mRNA is transported to the apical nuclear periplasm during syncytial blastoderm; and (4) bcd transcripts are degraded rapidly during the first third of nuclear cycle 14. In addition, we show that the bcd mRNA and protein patterns behave very similarly. These results exclude the SDD model, which needs to be replaced by a new model in which the Bcd protein gradient is dictated by the bcd mRNA gradient and its subsequent translation. In contrast to the SDD model, this ARTS (active RNA transport and synthesis) model explains the formation of the Bcd protein gradient by an active transport of its mRNA on microtubules, followed by synthesis of the protein.
MATERIALS AND METHODS
FISH and staining by immunohistochemistry
For in situ hybridization, a full-length bcd cDNA, c53.46.6, which encodes the longest Bcd protein isoform, was used (Frigerio et al., 1986; Berleth et al., 1988). This cDNA was used in the first demonstration of a bcd mRNA gradient (Frigerio et al., 1986), and served as the source of Bcd antigen for the preparation of an anti-Bcd antiserum in the first publication demonstrating a Bcd protein gradient (Driever and Nüsslein-Volhard, 1988) as well as in subsequent publications (Kosman et al., 1998). In situ hybridization of digoxigenin (DIG)-labeled probes and their immunochemical detection by alkaline phosphatase or horseradish peroxidase (Fig. 1D-F) followed standard protocols (Tautz and Pfeifle, 1989).
Fig. 1.
Visualization of the bcd mRNA gradient by established in situ hybridization methods. (A,B) Dark- (A) and bright- (B) field photomicrographs of a frozen midsagittal section through a Drosophila embryo at nuclear cycle 14, hybridized in situ with a 3H-labeled bcd cDNA probe [fig. 5e,f of Frigerio et al. (Frigerio et al., 1986), reproduced with permission]. The inset in B is a magnified view of the anterior region, as marked with a bracket, that shows the silver grains (black) in the apical periplasm of nuclei. (C) The bcd mRNA gradient. Silver grains in the apical periplasm of each dorsal nucleus in B were counted under a microscope and plotted against the position of the nucleus along the anteroposterior axis [nucleus 1 at the anterior tip corresponds to 0% egg length (EL)]. Irregularities and small gaps at nuclei 10-20 were caused by shearing forces during sectioning. (D-F) Transcripts detected after in situ hybridization with a DIG-labeled probes of a bcd cDNA (D,E) or of nos and bcd cDNAs (F) by alkaline phosphatase and exposure to its substrate for 30 minutes (D,F) or by horseradish peroxidase and exposure to its substrate for 2 minutes (E) in embryos at nuclear cycle 10 (D), nuclear cycle 14 (E), or during early cleavage stage (F). The inset in E is a magnified view of nuclei close to the anterior tip of the embryo, where localization of bcd mRNA in the apical periplasm is clearly visible. (G-I) ImageJ scans along the anteroposterior axis (between the dashed red lines) of embryos in D-F, demonstrating the bcd mRNA gradients. The bcd mRNA profiles shown in G-I are derived from the embryos in D-F, respectively. All embryos in this figure are oriented with their dorsal side up and anterior to the left.
To optimize the sensitivity of FISH, we modified a standard in situ hybridization protocol (Tautz and Pfeifle, 1989) (detailed protocol available on request). Care was taken to keep the parameters in different in situ hybridization experiments constant. To ensure that the apical periplasm of embryos was not affected during devitellinization and hybridization, embryos were fixed up to 1 day before devitellinization. During devitellinization, only short and mild vortexing was applied. To maintain the integrity of the apical periplasm during hybridization, embryos were fixed for 1 hour in 1% formaldehyde before treatment with 10 μg/ml proteinase K for 5 minutes, which is five times shorter than recommended by current protocols. Hybridized probes were detected with an anti-DIG monoclonal antibody (Roche) diluted 1:200, and Alexa 555- (Invitrogen) or Cy3-coupled goat anti-mouse secondary antibodies diluted 1:1500. To determine the developmental stage, embryos were counterstained with DAPI or TOTO3 (Invitrogen).
An anti-Bcd antiserum, raised in rabbits against amino acids 112-396 of Bcd (including the homeodomain), was affinity-purified and used at 1:20 dilution. To visualize bcd mRNA as well as Bcd protein, Bcd protein was detected by the sequential use of anti-Bcd and Alexa 555-coupled goat anti-rabbit antisera, followed by FISH and the detection of bcd mRNA by the sequential use of an anti-DIG monoclonal antibody and Alexa 647-coupled goat anti-mouse antiserum. Alexa 555 (Bcd protein) and Alexa 647 (bcd mRNA) were chosen as fluorochrome coupled to secondary antibodies to minimize their cross-channel activity. Anti-Stau antibodies (a generous gift of Daniel St Johnston) were used at 1:2500 dilution.
When only proteins (Stau or Bcd) were visualized, consistently better fluorescent signal intensities (extremely low background combined with a wide range of sensitivity) were obtained with heat-fixed than with formaldehyde-fixed embryos. Moreover, heat-fixed embryos showed much better preservation of the apical periplasm.
Data acquisition
Confocal images (1024×1024 pixels, 8 bits) were taken with a Leica TCS SP or a Zeiss LSM Pascal microscope. Image stacks were acquired as sagittal sections through entire embryos, and fluorescence intensities were taken from midsagittal planes. For collection of fluorescent signals, the parameters of the confocal microscope were kept the same in all experiments. Intensity graphs were obtained from a circular area moved along the dorsal cortex of midsagittal sections (Alexandrov et al., 2008) (a detailed description of the algorithms, scripts and tools used is available on request).
RESULTS
Revisiting the bcd mRNA gradient
The first molecular characterization of the bcd gene included the description of a bcd mRNA gradient along the anteroposterior axis of the embryo, visualized by in situ hybridization of a 3H-labeled bcd cDNA probe to frozen sections of pre-blastoderm embryos [see Fig. 5 in Frigerio et al. (Frigerio et al., 1986)]. The most striking of these pictures, illustrating a midsagittal section of an embryo at nuclear cycle 14, is shown again here (Fig. 1A,B). Though not presented at the time, we had further analyzed the shape and extent of this bcd mRNA gradient by counting the number of silver grains in the apical periplasm of each dorsal nucleus (Fig. 1B) and plotting this as a function of the position of the nucleus along the anteroposterior axis. The resulting curve of bcd mRNA concentration (Fig. 1C) decreases monotonically from a maximum at the anterior pole to background levels at ∼35% egg length (EL; anterior pole is 0%), as pointed out previously (Frigerio et al., 1986). Although more sensitive methods permit detection of bcd mRNA at more posterior positions (see below), Fig. 1A-C demonstrates unequivocally that (1) bcd mRNA is not strictly localized to the anterior pole of the embryo but forms a concentration gradient that decreases with distance from the anterior pole, and (2) bcd mRNA is localized to the apical periplasm at this stage of nuclear cycle 14. Both results have been emphasized previously (Frigerio et al., 1986).
Fig. 5.
Transport of bcd mRNA from basal to apical nuclear periplasm, and its degradation during early nuclear cycle 14. Anterior regions of Drosophila embryos during interphase (A) and mitosis (B) of nuclear cycle 13, at the onset of nuclear cycle 14 (C), and 4 (D), 10 (E) or 16 (F) minutes after onset of nuclear cycle 14, are shown in midsagittal planes with dorsal to the left. Embryos were collected from the same FISH experiment and analyzed in the same confocal session. Fluorescence intensities, reflecting bcd mRNA concentrations, were converted to a color scale, as shown to the right. For color conversion and interpretation of signal intensities, the OsiriX DICOM program was used. Insets are differential interference contrast (DIC) images of enlarged views of the cortical region with nuclei that reveal the stage (A,B) and the position of the leading edge (arrow) of the progressing cellular membranes (C-F), thereby permitting precise determination of the time (in minutes) after onset of nuclear cycle 14 (Foe and Alberts, 1983; Gutjahr et al., 1993). Confocal images were taken with a Leica TCS SP microscope.
Several years after our original publication, a non-radioactive and hence faster method of detecting mRNA became available, using probes labeled with DIG and alkaline phosphatase coupled to an anti-DIG antibody (Tautz and Pfeifle, 1989). This method, previously used to analyze bcd mRNA distributions (St Johnston et al., 1989), was tested by exposing the embryos for 30 minutes to the color substrate of alkaline phosphatase in order to detect the full extent of the gradient. This approach is indeed capable of detecting the bcd mRNA gradient, as evident from an embryo at nuclear cycle 10 (Fig. 1D) and its ImageJ scan (Fig. 1G). Considerably shorter exposures, however, generated the false impression that transcripts are trapped at the anterior pole of the embryo, in apparent agreement with the SDD model. Nevertheless, even in these faintly stained embryos, the bcd mRNA gradient was readily detectable by use of optical measurements combined with ImageJ, which are more sensitive than the eye.
Other chromogenic detection methods, such as using horseradish peroxidase (HRP), can also visualize the bcd mRNA gradient. Thus, a 2-minute exposure of an embryo at nuclear cycle 14 to diaminobenzidine (DAB), the substrate for HRP, easily detected the gradient (Fig. 1E,H). Moreover, the resolution of this method is superior to that using alkaline phosphatase and readily detected bcd transcripts in the apical periplasm (Fig. 1E and inset). To exclude the possibility that the bcd mRNA gradient arises as an artifact by diffusion of the color substrate, we stained embryos hybridized simultaneously with DIG-labeled bcd and nanos (nos) probes. nos is one of the posterior group genes, which are exclusively expressed at the posterior tip of the embryo (Wang and Lehmann, 1991). As is evident from Fig. 1F and its ImageJ scan (Fig. 1I), nos mRNA was clearly restricted to the posterior tip, excluding artifacts due to substrate diffusion.
These results raise the question of why others have not observed a bcd mRNA gradient as we do. As argued, the color reaction might have been too short or the stained images were not subjected to a quantitative analysis. Such an analysis of bcd mRNA concentrations has been shown in a graph representing the average of five embryos at nuclear cycle 13 (St Johnston et al., 1989). Although this graph was corrected for non-linearity of the staining method, it is not consistent with our analysis using modern experimental and analytical tools (see below). In addition, in some cases a bcd mRNA gradient can indeed be observed but was missed or ignored (T. Berleth, PhD thesis, University of Tübingen, 1988) (Berleth et al., 1988; St Johnston et al., 1989; Schnorrer et al., 2002; Song et al., 2007).
Analysis by FISH and confocal microscopy of bcd mRNA gradient formation
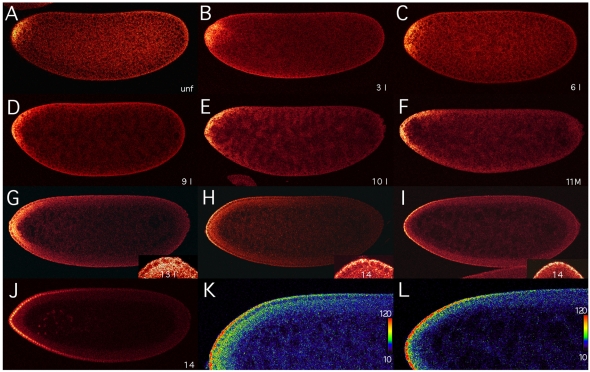
To visualize the bcd mRNA patterns, we modified a well-established in situ hybridization protocol (Tautz and Pfeifle, 1989) to raise its sensitivity. Moreover, we used FISH and confocal microscopy, which combine high sensitivity and high spatial resolution (Lécuyer et al., 2007). In unfertilized eggs, bcd transcripts are localized at the anterior tip of the embryo (Fig. 2A), where they are tightly associated with the cortex, as is evident from more-peripheral confocal sections (Weil et al., 2008) (data not shown). Their localization is very similar to that observed in mature (Frigerio et al., 1986) and live-imaged oocytes (Weil et al., 2006). A confocal stack (not shown) revealed a small `cap' of bcd mRNA with a posterior limit at 7-9% EL and a skew to the dorsal side (Fig. 2A). In 4-nuclei embryos, the cap extended posteriorly (Fig. 2B). In 32-nuclei embryos, bcd transcripts continued to move posteriorly along the periphery of the embryo (Fig. 2C). It is important to emphasize that transcripts do not diffuse as their movement is restricted to the cortex of the embryo. This movement continued during subsequent nuclear divisions (Fig. 2D) until the nuclei reached the periphery at nuclear cycle 10 (Fig. 2E). Little change in the transcript pattern was observed between nuclear cycles 10 and 13, although by this time two bcd mRNA gradients were evident: one along the basal, the other along the apical, periplasm of nuclei. Mitosis, as exemplified by an embryo at the end of nuclear cycle 11, did not affect the distribution of bcd mRNA (Fig. 2F). During nuclear cycle 13, basal and apical bcd gradients were prominent, though basal bcd transcripts covered a much wider layer of the cortex than did apical transcripts (Fig. 2G and inset). During early nuclear cycle 14, a striking change in pattern was observed: bcd transcripts began to disappear from the basal periplasm, while their apical concentration appeared unchanged (Fig. 2H and inset). The extended gradient was obvious from the monotonically decreasing bcd mRNA concentration (Fig. 2K). Subsequently, basal bcd transcripts disappeared within minutes, but apical transcripts remained (Fig. 2I,L). A few minutes later, the apical transcripts had also disappeared (data not shown).
Fig. 2.
Formation of bcd mRNA gradient analyzed by confocal microscopy. Confocal images (taken with a Leica TCS SP microscope) at midsagittal planes of Drosophila embryos oriented with their dorsal side up and anterior to the left. (A-I) bcd transcripts are detected by FISH with a DIG-labeled bcd cDNA in an unfertilized (unf) egg (A), and in embryos during interphase of nuclear cycle 3 (B), 6 (C), 9 (D), 10 (E), mitosis of nuclear cycle 11 (F), interphase of nuclear cycle 13 (G), and 4 (H) and 10 (I) minutes after onset of nuclear cycle 14. During cleavage stage, nuclear cycles were determined by counting the nuclei stained with DAPI (B-D). Insets in G-I show magnifications of the anterior region of the embryos. (J) Bcd protein gradient in an embryo 10 minutes after onset of nuclear cycle 14, visualized by an anti-Bcd antiserum and fluorescent immunostaining. (K,L) Magnified views of the dorsal anterior region of the embryos shown in H and I, respectively, visualizing the posterior extent of the bcd mRNA gradients using a color scale from 10 to 120, as shown to the right. For color conversion and interpretation of signal intensities, the OsiriX DICOM program was used (Rosset et al., 2004).
Quantitative analysis of bcd mRNA gradient formation
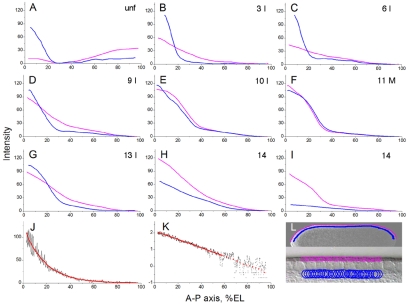
To measure bcd mRNA levels, we analyzed fluorescence intensities by a method similar to that used to determine the nuclear Bcd protein gradient (Houchmandzadeh et al., 2002). In unfertilized eggs and cleavage-stage embryos, where nuclei have not yet migrated to the periphery (Fig. 2A-C), basal and apical bcd transcript levels were measured along lines located at the same distance from the surface of the embryo as those employed at later stages (Fig. 3L). In unfertilized eggs and early cleavage-stage embryos, basal transcripts formed a steep gradient with a bend at ∼20% EL (Fig. 3A-C, blue) and thus might appear to be “strictly localized to the anterior cytoplasm” (Ephrussi and St Johnston, 2004). By contrast, apical transcripts began to form a shallow gradient (Fig. 3B,C, pink). Clearly, basal as well as apical bcd mRNAs already extended to posterior regions, as was apparent from a progressive increase in their concentration posterior to the bend (Fig. 3B,C).
Fig. 3.
Quantitative analysis of bcd mRNA gradient formation. (A-I) Distribution of basal and apical bcd transcripts along the dorsal midline of the anteroposterior axis in unfertilized eggs (A) and during early embryonic development (B-I) of Drosophila. The graphs, which were derived from raw data obtained by confocal microscopy of embryos shown in Fig. 2A-I (in the manner illustrated in L), show the intensities (arbitrary units) of basal (blue) and apical (pink) bcd transcripts. (J,K) Nuclear Bcd protein intensities, similarly measured along the anteroposterior axis of the embryo at nuclear cycle 14 shown in Fig. 2J, plotted against a linear (J) or base-10 logarithmic (K) scale to illustrate the exponential decay of the gradient between ∼10% and ∼65% EL. In both graphs, a background level of 58.9, reached at ∼65% EL and producing the widest range of exponentiality, was subtracted. (L) Overview of an embryo (above) with enlarged view of its surface (beneath), illustrating the positions at which basal (blue lines) and apical (pink lines) bcd transcript levels were measured in overlapping circular discs along the dorsal cortex of the embryos shown in Fig. 2A-I.
During nuclear cycles 7-9, the slope of the basal gradient decreased, moving the bend to ∼30% EL, whereas the slope of the apical gradient increased (Fig. 3D). By the time the nuclei reached the periphery, similarly shaped gradients of basal and apical bcd mRNAs were obvious (Fig. 3E). During the subsequent nuclear cycles, the appearance of these gradients did not change substantially (Fig. 3F,G). During early nuclear cycle 14, basal transcripts were first reduced, forming a shallow gradient, and then disappeared, whereas apical transcript levels were still high but also began to decrease (Fig. 3H,I). Before nuclear cycle 14, bcd mRNA must be stable because little, if any, degradation was apparent, as previously observed in activated unfertilized eggs (Surdej and Jacobs-Lorena, 1998).
The close similarity between the bcd mRNA (Fig. 3E-I) and protein gradients during syncytial blastoderm (Driever and Nüsslein-Volhard, 1988; Houchmandzadeh et al., 2002; Gregor et al., 2007) raised the question of whether the Bcd protein gradient, as analyzed by our method (Fig. 3L), displayed the same profile. Indeed, we found that the Bcd protein gradient (Fig. 3J) was in excellent agreement with published results (Houchmandzadeh et al., 2002): it decreased exponentially with increasing distance from the anterior pole to ∼65% EL (Fig. 3K), beyond which Bcd intensities no longer differed significantly from background.
Batch analysis of bcd mRNA gradients in embryos at nuclear cycles 13 and 14
To corroborate the bcd mRNA gradient during nuclear cycles 13 and 14 (Fig. 3G-I) and to ensure optimal comparability of results, 70 embryos hybridized together were analyzed in a single confocal session. Cycle-14 embryos were further subdivided into embryos at 0-4 minutes and at 8-12 minutes after onset of cycle 14, whereby the time after onset of nuclear cycle 14 was determined by the position of the leading edge of the progressing cellular membranes (Foe and Alberts, 1983; Gutjahr et al., 1993). At all stages, considerable variability in the bcd mRNA gradient profiles was observed, which was largest at the most anterior positions, in excellent agreement with previously published variability in Bcd protein profiles among embryos at similar stages (Houchmandzadeh et al., 2002; Holloway et al., 2006).
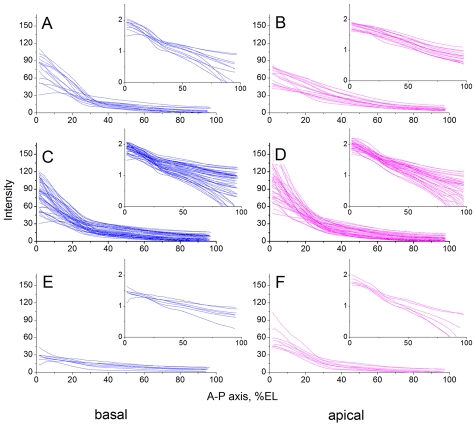
Basal bcd transcript profiles of 15 embryos at cycle 13 displayed a monotonic decrease with increasing distance from the anterior pole, with a minor deviation from exponentiality that was visible as a small bend at 30% EL when plotted on a logarithmic scale (Fig. 4A and inset). This deviation was not apparent in apical transcript profiles (Fig. 4B and inset). At this stage, gradients of apical bcd transcripts were shallower and anterior intensities lower than those of basal transcripts. Basal transcript profiles of 46 embryos at 0-4 minutes of nuclear cycle 14 (Fig. 4C) showed no significant difference to those of embryos at nuclear cycle 13 (Fig. 4A). By contrast, apical transcript profiles exhibited a marked increase in intensity (Fig. 4D) compared with those of cycle 13 embryos (Fig. 4B), which suggests a movement of bcd transcripts from basal to apical periplasm. At this time, basal and apical transcripts showed very similar profiles (Fig. 4C,D and insets). Finally, bcd transcript profiles of nine embryos at 8-12 minutes after onset of nuclear cycle 14 revealed the nearly complete absence of basal transcripts (Fig. 4E), while apical transcripts had decreased considerably (Fig. 4F). This drastic decrease in both basal and apical bcd transcripts reflects a rapid degradation of bcd mRNA during early cycle 14. Both the apical and basal profiles remained close to exponential (Fig. 4E,F, insets).
Fig. 4.
Batch analysis of the bcd mRNA gradient of embryos at nuclear cycles 13 and 14 and the exponentiality of the gradient. (A-F) Basal (A,C,E) and apical (B,D,F) bcd transcript profiles of 15 Drosophila embryos at nuclear cycle 13 (A,B), 46 embryos at 0-4 minutes of nuclear cycle 14 (C,D), and of nine embryos at 8-12 minutes of nuclear cycle 14 (E,F), were measured as illustrated in Fig. 3L. Insets show the same profiles with intensities of transcripts plotted on a base-10 logarithmic scale.
Transport of bcd mRNA from basal to apical periplasm and degradation of bcd mRNA during early nuclear cycle 14
An intriguing feature of the bcd mRNA patterns is the increase in apical, at the expense of basal, bcd mRNA levels during early nuclear cycle 14. Most likely, this results from an active transport of basal bcd mRNA to the apical periplasm (Fig. 3G,H) (Bullock and Ish-Horowicz, 2001; Wilkie and Davis, 2001). We analyzed this apical migration in detail in embryos from the same batch as in Fig. 4. To facilitate the analysis, fluorescence intensities, reflecting bcd mRNA concentrations, were converted to a color scale (Fig. 5). During nuclear cycle 13, bcd transcripts were still uniformly distributed between basal and apical periplasm (Fig. 5A), and no net apical migration of bcd mRNA was apparent up to this stage (Fig. 3E-G). During the following nuclear division, no changes in the patterns or ratio of basal to apical transcripts were apparent (Fig. 5B,C), a theme that continued up to ∼4 minutes after onset of nuclear cycle 14 at 25°C, although there was a slight decrease in the relative amount of basal versus apical bcd transcripts, while their relative maximum concentrations appeared unaltered (Fig. 5C,D). However, by ∼10 minutes after onset of nuclear cycle 14, most basal bcd transcripts had disappeared, while the concentrations of apical transcripts were slightly diminished (Fig. 5E). This drastic loss of basal transcripts results from the onset of bcd mRNA degradation. Apical transcripts are only mildly reduced because they have been replenished by an efficient apical transport of basal transcripts. By ∼16 minutes after onset of nuclear cycle 14, nearly all of the apical transcripts have disappeared as well (Fig. 5F). Thus, all bcd mRNAs are degraded within 15-20 minutes.
Virtually identical patterns of bcd mRNA and protein refute the SDD model
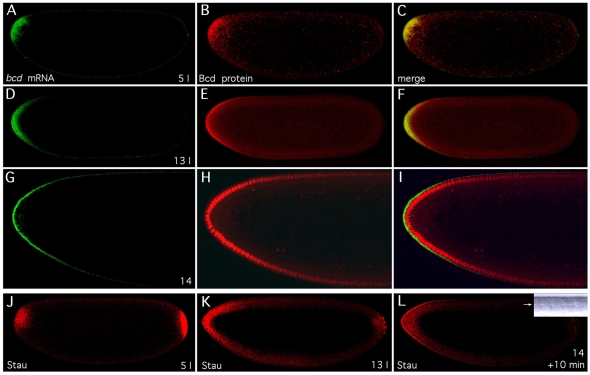
To assess whether diffusion of Bcd protein is instrumental in setting up its gradient, we compared bcd mRNA and protein gradients simultaneously. Translation of bcd mRNA takes only 2 minutes at a translation rate of 4-5 amino acids per second (Dintzis, 1961; Braakman et al., 1991). Congruence of protein and mRNA gradients at all times therefore implies that the Bcd protein gradient does not depend on protein diffusion but is dictated by the bcd mRNA gradient. As is evident from Fig. 6A-I, the bcd mRNA (green) and protein (red) patterns were virtually identical at all times, except that the protein was localized in the nucleus and the mRNA in the nuclear periplasm. Particularly striking is the similarity of the cleavage-stage patterns (Fig. 6A-C). During nuclear cycle 13, the Bcd protein is presumably translated from both basal and apical bcd mRNAs (Fig. 6D-F), whereas Bcd protein is imported into the nucleus only from the apical periplasm when the basal bcd mRNA has disappeared (Fig. 6G-I). Thus, these results demonstrate, in clear contradiction to the SDD model (Driever and Nüsslein-Volhard, 1988), that the Bcd protein gradient arises by translation of its mRNA, which forms the gradient.
Fig. 6.
Similarity of Bcd protein, bcd mRNA and Stau gradients. (A-I) Similarity of bcd mRNA and protein gradients. Patterns of bcd mRNA and Bcd protein are visualized by double-staining of bcd mRNA with Alexa 647 (A,D,G, green) and Bcd protein with Alexa 555 (B,E,H, red) in Drosophila embryos during interphase of nuclear cycle 5 (A-C) and 13 (D-F), and 10 minutes after onset of nuclear cycle 14 (G-I; enlarged views of the anterior of the embryo), with the merge shown to the right (C,F,I). (J-L) Colocalization of bcd mRNA and Stau protein. Drosophila embryos were stained with an anti-Stau antiserum during interphase of nuclear cycle 5 (J) and 13 (K), and 10 minutes after onset of nuclear cycle 14 (L). Inset in L is a DIC image (see Fig. 5). All embryos are oriented with anterior to the left and dorsal side up. Confocal images were taken with a Zeiss LSM Pascal (A-I) or a Leica TCS SP (J-L) microscope.
Similar localization and gradient of Staufen protein and bcd mRNA
Localization of bcd mRNA during late oogenesis and early embryogenesis has been shown to depend on the product of the staufen (stau) gene (St Johnston et al., 1989; Ferrandon et al., 1994; Weil et al., 2006; Weil et al., 2008). Stau protein binds to the 3′ UTR of bcd mRNA and is thought to be required to localize bcd mRNA to the anterior pole of the early embryo (Ferrandon et al., 1994). We tested whether Stau might also be associated with bcd mRNA in embryos by visualizing its distribution. In early cleavage embryos, Stau was localized to the posterior pole, but was also present in an anterior cap (Fig. 6J), similar to that of bcd mRNA (Fig. 6A). The similarity between the bcd mRNA and anterior Stau patterns persisted during subsequent stages (data not shown) through nuclear cycle 13 (Fig. 6D,K). In particular, Stau also formed an apical gradient (Fig. 6L) when basal bcd mRNA had disappeared (Fig. 6G). In contrast to bcd mRNA, however, some Stau remained in the basal periplasm, which indicates a function of Stau that is not associated with bcd mRNA. These results strongly suggest that bcd mRNA is bound to Stau protein in the embryo.
DISCUSSION
A model of bcd mRNA gradient formation
Revisiting the formation of the morphogenetic bcd gradient, we have corroborated and extended published results (Frigerio et al., 1986) by demonstrating a gradient of bcd mRNA that is very similar to that of the Bcd protein. Therefore, our results contradict the SDD model (Driever and Nüsslein-Volhard, 1988). Although the SDD model correctly predicts an exponential Bcd protein gradient, the diffusion coefficient of Bcd, as measured in syncytial-blastoderm embryos, is two orders of magnitude too low to account for the fact that the steady state of its gradient is reached at syncytial blastoderm (Gregor et al., 2007), a finding that is in serious conflict with this model. Our results strongly suggest that the bcd mRNA gradient forms the protein gradient and is established by transport of the mRNA along the cortex of the embryo.
To explain the formation of the bcd mRNA gradient, we arrived at the following model. The establishment of the bcd RNA gradient by nuclear cycle 10 and its disappearance during early nuclear cycle 14 occur in five phases. (1) The bcd RNA, which is associated with Stau and presumably many other proteins and anchored to the actin cytoskeleton at the anterior cortex of the mature oocyte, is released upon fertilization by calcium signaling. (2) This Stau-bcd mRNA complex is transported posteriorly along microtubules emanating from numerous microtubule-organizing centers (MTOCs) that are closely spaced and distributed throughout the cortex of the embryo (Karr and Alberts, 1986). The posterior transport of bcd mRNA is driven by its concentration gradient. (3) This transport is arrested by the breakdown of the cortical microtubular network when the nuclei reach the cortex (Foe and Alberts, 1983). At this time, a new microtubular network of astral microtubules forms. These extend from the centrosomes, located between the plasma membrane and each nucleus, and surround each nucleus with apical-basal polarity (Karr and Alberts, 1986). Thus, the bcd mRNA gradient is established by nuclear cycle 10 and does not change until the end of nuclear cycle 13. (4) During syncytial blastoderm, the bcd mRNP particles are transported apically on astral microtubules by the minus end-directed dynein/dynactin motor complex (Wilkie and Davis, 2001), a process that depends on the maternal proteins Bicaudal D (BicD) and Egalitarian (Egl) (Bullock and Ish-Horowicz, 2001). (5) Whereas bcd mRNA is stable until nuclear cycle 13, it is rapidly degraded during early nuclear cycle 14. We assume that this degradation is mediated by Stau and is triggered by apical factors and signals.
This model combines our results with those reported by others. Below, we discuss in detail the evidence that led us to this ARTS (active RNA transport and protein synthesis) model. Despite its speculative aspects, it should serve as a useful hypothesis for future experiments that test its predictions. In addition, the model postulates a new principle to explain the formation of the bcd mRNA gradient: a quasi-random transport through a cortical microtubular network that is driven by a high initial concentration of bcd mRNA at the anterior pole.
Release of bcd mRNA from the anterior cortex upon fertilization
Stau protein binds to the 3′UTR of bcd mRNA in oocytes and colocalizes with bcd mRNA at the anterior pole of freshly laid eggs (Ferrandon et al., 1994) (Fig. 6A,B). Additional proteins probably stabilize the interaction of Stau with bcd mRNA in the embryo, as in oocytes (Irion and St Johnston, 2007). Localization of bcd mRNA to the anterior pole is established by continual active transport of the Stau-bcd mRNA complex on microtubules, mediated by the minus end-directed motor dynein, when nurse cells empty their content into stage 10B-13 oocytes (Pokrywka and Stephenson, 1991; Weil et al., 2006). Subsequent anchoring of the Stau-bcd mRNA complex to the actin cytoskeleton stabilizes its anterior localization in mature oocytes (Weil et al., 2006; Weil et al., 2008). This anchoring step depends on swallow (swa), the product of which interacts with the dynein light chain and γTub37C, which is part of the MTOC at the anterior end of oocytes (Schnorrer et al., 2000; Schnorrer et al., 2002). Upon fertilization, calcium signaling releases the Stau-bcd mRNA complex from the actin cytoskeleton, which depends on the product of the sarah (sra) gene, an inhibitor of the calcium-dependent phosphatase calcineurin (Weil et al., 2008). Swa protein is no longer required and is degraded (Schnorrer et al., 2000).
A network of microtubules, in which the MTOCs are closely spaced (separated by a few microns), occupies the cortical region of embryos during nuclear cycles 1-9 (Karr and Alberts, 1986; Callaini and Riparbelli, 1997). Consistent with this observation is the pattern of cortical staining of early embryos for several Dgrips (S.B., unpublished), proteins of the γ-tubulin ring complex that caps the minus ends of microtubules at MTOCs (Gunawardane et al., 2000). Evidently, these microtubules nucleate from MTOCs that are established in late oocytes (Schnorrer et al., 2002; Vogt et al., 2006). To our knowledge, no function has been reported for this cortical microtubular network, which breaks down at the end of nuclear cycle 9. We propose that its existence is crucial for the formation of the bcd mRNA gradient.
Posterior cortical transport of bcd mRNA mediated by a nonpolar microtubular network
A mechanism based on diffusion of the bcd mRNA cannot explain the gradient observed because bcd mRNA is restricted to the cortex of the embryo. Diffusion of bcd mRNA to the interior would dramatically reduce its concentration along the cortex, where its function is required, because unlike for Bcd protein in the SDD model, there is no source replenishing the lost bcd mRNA. Active transport of a Stau-bcd mRNA complex on microtubules, similar to that observed in late-stage oocytes (Weil et al., 2006), is suggested by the striking colocalization of Stau and bcd mRNA until the latter disappears (Fig. 6). However, the microtubules with MTOCs located at the anterior pole are disassembled in late oocytes (Theurkauf et al., 1992; Weil et al., 2008). Indeed, the cortical microtubular network in embryos at nuclear cycles 1-9 shows no sign of an overall polarity, but appears to be nonpolar, with its plus ends growing in all directions from MTOCs closely spaced throughout the cortex (Karr and Alberts, 1986; Callaini and Riparbelli, 1997).
How can such a nonpolar microtubular network establish a bcd mRNA gradient by active transport of the Stau-bcd mRNP particles? Because the network exhibits no polarity, it supports only random transport as would occur by diffusion. The only restriction to the random transport is its confinement to the cortex of the embryo. Like diffusion, it is driven by the concentration gradient of the transported molecules, here by the high initial concentration of bcd mRNA at the anterior pole (Fig. 2B). Average transport velocities of Stau-bcd mRNA complexes on microtubules, as measured in stage 10B-13 oocytes, range from 0.36 to 2.15 μm/second (Weil et al., 2008). Such a non-random transport in the embryo would move bcd mRNA molecules within minutes from the anterior to the posterior pole and thus destroy its function as an anterior morphogen. Therefore, it seems crucial that bcd mRNA transport in the embryo occurs through a nonpolar microtubular network. It is additionally important that the time of 90 minutes that is required to establish the bcd mRNA gradient at 25°C is tuned finely with the time required for the first nine nuclear divisions, after which the nuclei reach the cortex.
The efficiency of a system employing random transport can be estimated from the average posterior drift velocity of bcd mRNAs along the cortex. When, 90 minutes after fertilization, syncytial blastoderm is reached, bcd mRNA has moved posteriorly on average by ∼50 μm (from 5% EL at fertilization to 15% EL), which corresponds to an average drift velocity of ∼0.01 μm/second. This is 100 times slower than the average transport velocity on a microtubule in the oocyte (Weil et al., 2008) and is thus rather inefficient. Since this transport of bcd mRNA occurs on a microtubular network with randomly oriented microtubules, it is irrelevant whether transport is mediated by the minus end-directed dynein/dynactin or the plus end-directed kinesin motors. In the oocyte, Stau-bcd mRNP particles are transported exclusively by dynein (Weil et al., 2006) in a process that depends on the presence of Exuperantia (Exu) in nurse cells (Cha et al., 2001). Since Exu disappears from late oocytes (Macdonald et al., 1991), this might permit Stau-bcd mRNPs to interact with dynein or kinesin upon their release from the actin cytoskeleton.
Just before the present study was submitted, transport in oocytes through a microtubular network exhibiting only a slight directional bias (57% of plus ends oriented posteriorly) was shown to localize Stau-oskar (osk) mRNA particles to the posterior pole (Zimyanin et al., 2008). Although it is conceivable that the bcd mRNA gradient is established through such a biased microtubular network, the net average posterior velocity in oocytes of 0.03 μm/second (Zimyanin et al., 2008) would displace the bcd mRNA on average by 162 μm towards the posterior pole of the embryo by the time the bcd RNA gradient is established. This is more than twice the observed average posterior displacement of bcd mRNA (Fig. 3E). Nevertheless, the bcd mRNA gradient might be established through such a biased microtubular network if transport is mediated by both minus- and plus-end motors. In such a case, however, the average posterior drift velocity would also depend on the availability of both motors. If the probability of Stau-bcd mRNP interacting with either motor is the same, transport by the microtubular network becomes independent of its directional bias, and the network behaves like the nonpolar microtubular network. However, we favor a nonpolar microtubular network in the embryo because it seems more robust to disturbances.
Apical transport of bcd mRNA during syncytial blastoderm
An intriguing feature of the bcd mRNA gradient during nuclear cycles 10-13 is the maintenance of a constant apical gradient similar to the basal gradient (Figs 3 and 5). It has been noted previously that bcd transcripts are localized to the narrow apical periplasm at late syncytial blastoderm (Frigerio et al., 1986) (Fig. 1A,B), and that this localization depends on a signal in their 3′UTR (Davis and Ish-Horowicz, 1991). Apical transport of bcd mRNA becomes evident during nuclear cycle 14, when the excess of basal bcd mRNA disappears more rapidly than its apical counterpart (Fig. 5C-F).
Although no net apical transport of bcd mRNA is apparent before its degradation during nuclear cycle 14, the establishment of an astral microtubular network during nuclear cycle 9 (Karr and Alberts, 1986) suggests that it might occur as early as nuclear cycle 10. Such a system, capable of transporting Stau-bcd mRNA particles to the apical periplasm, might be important to stabilize the bcd mRNA gradient against disturbances by the strong periplasmic flow that is observed in the cortex during nuclear cycles 10-13 (Foe and Alberts, 1983). Nevertheless, if Stau-bcd mRNA complexes detach when they reach the minus ends at the apical MTOCs, some apically localized bcd mRNAs might be subject to the periplasmic flow. Such a disturbance would be minor, as it would be corrected immediately by rapid apical transport of Stau-bcd mRNA particles, which occurs at a velocity of 0.5 μm/second (Bullock and Ish-Horowicz, 2001; Wilkie and Davis, 2001).
Why is it important to localize bcd mRNA not only to the basal, but also to the apical, nuclear periplasm? An answer is probably provided by elegant studies that have demonstrated that the nuclear concentration of Bcd protein remains approximately constant at a certain position along the anteroposterior axis during syncytial blastoderm (Gregor et al., 2007). This finding was surprising in view of the fact that the number of nuclei double after each nuclear division, their volume increases by 30% during interphase of each nuclear cycle, and the Bcd concentration drops fourfold when nuclear membranes disappear during mitosis. It was explained by measurements revealing that nuclear import of Bcd is sufficiently rapid to maintain a high and constant nuclear Bcd concentration. Hence, it might be crucial that Bcd can be imported through the entire nuclear surface (Gregor et al., 2007). Consistent with an accelerated nuclear import of Bcd by the product of the lesswright (lwr) gene (Epps and Tanda, 1998), we found Lwr in cleavage-stage and syncytial-blastoderm nuclei (K.F. and S.B., unpublished).
Is degradation of bcd mRNA also mediated by Stau?
Whereas bcd mRNA is stable before nuclear cycle 14, basal bcd mRNA disappears owing to its degradation and transport to the apical periplasm within ∼10 minutes of early nuclear cycle 14 (Fig. 5C-F). Thus, the estimated half-life of basal bcd mRNA is ∼2 minutes. Apical bcd mRNA decreases only when basal bcd mRNA becomes limiting (Fig. 5E,F). At this time, the estimated half-life of apical bcd mRNA is also ∼2 minutes (Fig. 5E,F). Therefore, bcd mRNA is degraded in the basal and apical periplasm, or only in the latter. This degradation is presumably mediated by a bcd instability element (BIE) located within a 43-nucleotide sequence following the stop codon (Surdej and Jacobs-Lorena, 1998). In mammals, Stau may trigger the degradation of an mRNA by binding to its 3′UTR and to the nonsense-mediated decay (NMD) factor Upf1, in a process that is different from NMD and is called Staufen-mediated mRNA decay (SMD) (Kim et al., 2005). As the Drosophila genome encodes a Upf1 homolog, Stau might well function not only in the transport of bcd mRNA but also in its degradation.
Since Bcd protein disappears ∼25 minutes after bcd mRNA, a lag during which its level decreases at least tenfold (S.B., unpublished), its half-life is less than 8 minutes at this time. The presence of a conserved PEST sequence in Bcd (Berleth et al., 1988; Gregor et al., 2008) might be responsible for its short half-life, presumably also during earlier stages, a hypothesis that is consistent with the similarity between the slopes of the bcd mRNA and protein gradients.
A fundamental difference between the ARTS and SDD models
There are many ways to generate a morphogenetic gradient. The original proposal of how the Bcd protein gradient forms (Driever and Nüsslein-Volhard, 1988) closely followed Wolpert's model of generating a morphogenetic gradient by a localized source synthesizing the morphogenetic molecules that are subject to diffusion and spatially uniform degradation (Wolpert, 1969). This model predicts a steady state at which the Bcd concentration decays exponentially along the anteroposterior axis (Gregor et al., 2007). We now see that the Bcd protein gradient is generated by an entirely different mechanism. Since there is no source of bcd mRNA, its posterior transport from the anterior pole must be arrested when the optimal gradient is reached. This arrest is triggered by the breakdown of the cortical microtubular network and is well timed with the arrival of the nuclei at the cortex, when gap genes are activated by the Bcd protein (Bergmann et al., 2007). At this time, the gradient is established and remains constant until nuclear cycle 14, when bcd mRNA is rapidly degraded. Thus, the bcd mRNA gradient is not established as a steady state, but by a process that is terminated by the breakdown of the microtubular network required for its formation.
Compared with the diffusion-based mechanism of the SDD model, a random active transport system has several advantages for the formation of the bcd mRNA gradient. The microtubular network is able to confine the movement of the bcd mRNA to the space where its function is required. The final shape of the gradient depends on several parameters: the initial concentration of bcd mRNA at the anterior pole, the transport velocity along microtubules, the average travel time per microtubule, the time between discharge from one and reloading onto another microtubule, and the time when transport is arrested by the breakdown of the microtubular network that supports the random transport. In addition, the availability of minus end- and plus end-directed motors might further influence the generation of the bcd mRNA gradient by random transport. Therefore, perhaps the greatest advantage of random active transport is that variations in these parameters during evolution permit the adaptation of the gradient to its optimal shape at the time when its function is required during development (Gregor et al., 2005). For these reasons, we suspect that random active transport represents a general mechanism that might have found wide application during evolution.
We are indebted to Doris Brentrup for the preparation and purification of anti-Bcd antiserum. We thank Sol Da Rocha and Michael Daube for excellent technical assistance; Daniel St Johnston for anti-Staufen antibodies; Yixian Zheng, Cayetano González and Brigitte Raynaud-Messina for antisera against Dgrips and γ-tubulin; David Holloway and Nina Golyandina for discussions; and Hans Noll and a reviewer for comments on the manuscript. S.B. thanks the Wenner-Gren Foundations for support during a sabbatical in Zürich. This work was supported by the Swedish `Vetenskapsrådet', the Swedish Cancer Foundation, the Medical Faculty of Lund (S.B.), by the Joint NSF/NIGMS BioMath Program Grant R01-GM072022 (A.S.), and by the Swiss National Science Foundation and the Kanton Zürich (M.N.). A.S. is supported by NIH NIGMS BioMath Program Grant R01-GM072022. Deposited in PMC for release after 12 months.
References
- Alexandrov, T., Golyandina, N. and Spirov, A. (2008). Singular spectrum analysis of gene expression profiles of early Drosophila embryo: exponential-in-distance patterns. Res. Lett. Signal Processing doi: 10.1155/2008/825758. [DOI] [PMC free article] [PubMed]
- Bergmann, S., Sandler, O., Sberro, H., Shnider, S., Schejter, E., Shilo, B.-Z. and Barkai, N. (2007). Pre-steady-state decoding of the Bicoid morphogen gradient. PLoS Biol. 5, 232-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berleth, T., Burri, M., Thoma, G., Bopp, D., Richstein, S., Frigerio, G., Noll, M. and Nüsslein-Volhard, C. (1988). The role of localization of bicoid RNA in organizing the anterior pattern of the Drosophila embryo. EMBO J. 7, 1749-1756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braakman, I., Hoover-Litty, H., Wagner, K. R. and Helenius, A. (1991). Folding of influenza hemagglutinin in the endoplasmic reticulum. J. Cell Biol. 114, 401-411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bullock, S. L. and Ish-Horowicz, D. (2001). Conserved signals and machinery for RNA transport in Drosophila oogenesis and embryogenesis. Nature 414, 611-616. [DOI] [PubMed] [Google Scholar]
- Callaini, G. and Riparbelli, M. G. (1997). Patterns of microtubule assembly in taxol-treated early Drosophila embryo. Cell Motil. Cytoskeleton 37, 300-307. [DOI] [PubMed] [Google Scholar]
- Cha, B.-J., Koppetsch, B. S. and Theurkauf, W. E. (2001). In vivo analysis of Drosophila bicoid mRNA localization reveals a novel microtubule-dependent axis specification pathway. Cell 106, 35-46. [DOI] [PubMed] [Google Scholar]
- Davis, I. and Ish-Horowicz, D. (1991). Apical localization of pair-rule transcripts requires 3′ sequences and limits protein diffusion in the Drosophila blastoderm embryo. Cell 67, 927-940. [DOI] [PubMed] [Google Scholar]
- Dintzis, H. M. (1961). Assembly of the peptide chains of hemoglobin. Proc. Natl. Acad. Sci. USA 47, 247-261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driever, W. (2004). The Bicoid morphogen papers (II): account from Wolfgang Driever. Cell S116, S7-S9. [DOI] [PubMed] [Google Scholar]
- Driever, W. and Nüsslein-Volhard, C. (1988). A gradient of bicoid protein in Drosophila embryos. Cell 54, 83-93. [DOI] [PubMed] [Google Scholar]
- Ephrussi, A. and St Johnston, D. (2004). Seeing is believing: the Bicoid morphogen gradient matures. Cell 116, 143-152. [DOI] [PubMed] [Google Scholar]
- Epps, J. L. and Tanda, S. (1998). The Drosophila semushi mutation blocks nuclear import of Bicoid during embryogenesis. Curr. Biol. 8, 1277-1280. [DOI] [PubMed] [Google Scholar]
- Ferrandon, D., Elphick, L., Nüsslein-Volhard, C. and St Johnston, D. (1994). Staufen protein associates with the 3′UTR of bicoid mRNA to form particles that move in a microtubule-dependent manner. Cell 79, 1221-1232. [DOI] [PubMed] [Google Scholar]
- Foe, V. E. and Alberts, B. M. (1983). Studies of nuclear and cytoplasmic behaviour during the five mitotic cycles that precede gastrulation in Drosophila embryogenesis. J. Cell Sci. 61, 31-70. [DOI] [PubMed] [Google Scholar]
- Frigerio, G., Burri, M., Bopp, D., Baumgartner, S. and Noll, M. (1986). Structure of the segmentation gene paired and the Drosophila PRD gene set as part of a gene network. Cell 47, 735-746. [DOI] [PubMed] [Google Scholar]
- Frohnhöfer, H. G. and Nüsslein-Volhard, C. (1986). Organization of anterior pattern in the Drosophila embryo by the maternal gene bicoid. Nature 324, 120-125. [Google Scholar]
- Gregor, T., Bialek, W., de Ruyter van Steveninck, R. R., Tank, D. W. and Wieschaus, E. F. (2005). Diffusion and scaling during early embryonic pattern formation. Proc. Natl. Acad. Sci. USA 102, 18403-18407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor, T., Wieschaus, E. F., McGregor, A. P., Bialek, W. and Tank, D. W. (2007). Stability and nuclear dynamics of the Bicoid morphogen gradient. Cell 130, 141-152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gregor, T., McGregor, A. P. and Wieschaus, E. F. (2008). Shape and function of the Bicoid morphogen gradient in dipteran species with different sized embryos. Dev. Biol. 316, 350-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunawardane, R. N., Martin, O. C., Cao, K., Zhang, L., Dej, K., Iwamatsu, A. and Zheng, Y. (2000). Characterization and reconstitution of Drosophila γ-tubulin ring complex subunits. J. Cell Biol. 151, 1513-1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutjahr, T., Frei, E. and Noll, M. (1993). Complex regulation of early paired expression: initial activation by gap genes and pattern modulation by pair-rule genes. Development 117, 609-623. [DOI] [PubMed] [Google Scholar]
- Holloway, D. M., Harrison, L. G., Kosman, D., Vanario-Alonso, C. E. and Spirov, A. V. (2006). Analysis of pattern precision shows that Drosophila segmentation develops substantial independence from gradients of maternal gene products. Dev. Dyn. 235, 2949-2960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Houchmandzadeh, B., Wieschaus, E. and Leibler, S. (2002). Establishment of developmental precision and proportions in the early Drosophila embryo. Nature 415, 798-802. [DOI] [PubMed] [Google Scholar]
- Irion, U. and St Johnston, D. (2007). bicoid RNA localization requires specific binding of an endosomal sorting complex. Nature 445, 554-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karr, T. L. and Alberts, B. M. (1986). Organization of the cytoskeleton in early Drosophila embryos. J. Cell Biol. 102, 1494-1509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim, Y. K., Furic, L., DesGroseillers, L. and Maquat, L. E. (2005). Mammalian Staufen1 recruits Upf1 to specific mRNA 3′UTRs so as to elicit mRNA decay. Cell 120, 195-208. [DOI] [PubMed] [Google Scholar]
- Kosman, D., Small, S. and Reinitz, J. (1998). Rapid preparation of a panel of polyclonal antibodies to Drosophila segmentation proteins. Dev. Genes Evol. 208, 290-294. [DOI] [PubMed] [Google Scholar]
- Lécuyer, E., Parthasarathy, N. and Krause, H. M. (2007). Fluorescent in situ hybridization protocols in Drosophila embryos and tissues. In Methods in Molecular Biology (ed. C. Dahmann) pp. 289-302. Totowa, NJ: Humana Press. [DOI] [PubMed]
- Macdonald, P. M., Luk, S. K.-S. and Kilpatrick, M. (1991). Protein encoded by the exuperantia gene is concentrated at sites of bicoid mRNA accumulation in Drosophila nurse cells but not in oocytes or embryos. Genes Dev. 5, 2455-2466. [DOI] [PubMed] [Google Scholar]
- Noll, M. (1993). Evolution and role of Pax genes. Curr. Opin. Genet. Dev. 3, 595-605. [DOI] [PubMed] [Google Scholar]
- Nüsslein-Volhard, C. N. (2004). The Bicoid morphogen papers (I): account from CNV. Cell S116, S1-S5. [DOI] [PubMed] [Google Scholar]
- Pokrywka, N. J. and Stephenson, E. C. (1991). Microtubules mediate the localization of bicoid RNA during Drosophila oogenesis. Development 113, 55-66. [DOI] [PubMed] [Google Scholar]
- Rosset, A., Spadola, L. and Ratib, O. (2004). OsiriX: an open-source software for navigating in multidimensional DICOM images. J. Digit. Imaging 17, 205-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnorrer, F., Bohmann, K. and Nüsslein-Volhard, C. (2000). The molecular motor dynein is involved in targeting Swallow and bicoid RNA to the anterior pole of Drosophila oocytes. Nat. Cell Biol. 2, 185-190. [DOI] [PubMed] [Google Scholar]
- Schnorrer, F., Luschnig, S., Koch, I. and Nüsslein-Volhard, C. (2002). γ -Tubulin37C and γ-tubulin ring complex protein 75 are essential for bicoid RNA localization during Drosophila oogenesis. Dev. Cell 3, 685-696. [DOI] [PubMed] [Google Scholar]
- Song, Y., Fee, L., Lee, T. H. and Wharton, R. P. (2007). The molecular chaperone Hsp90 is required for mRNA localization in Drosophila melanogaster embryos. Genetics 176, 2213-2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- St Johnston, D., Driever, W., Berleth, T., Richstein, S. and Nüsslein-Volhard, C. (1989). Multiple steps in the localization of bicoid RNA to the anterior pole of the Drosophila oocyte. Development 107Suppl., 13-19. [DOI] [PubMed] [Google Scholar]
- Surdej, P. and Jacobs-Lorena, M. (1998). Developmental regulation of bicoid mRNA stability is mediated by the first 43 nucleotides of the 3′ untranslated region. Mol. Cell. Biol. 18, 2892-2900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tautz, D. and Pfeifle, C. (1989). A non-radioactive in situ hybridization method for the localization of specific RNAs in Drosophila embryos reveals translational control of the segmentation gene hunchback. Chromosoma 98, 81-85. [DOI] [PubMed] [Google Scholar]
- Theurkauf, W. E., Smiley, S., Wong, M. L. and Alberts, B. M. (1992). Reorganization of the cytoskeleton during Drosophila oogenesis: implications for axis specification and intercellular transport. Development 115, 923-936. [DOI] [PubMed] [Google Scholar]
- Vogt, N., Koch, I., Schwarz, H., Schnorrer, F. and Nüsslein-Volhard, C. (2006). The γTuRC components Grip75 and Grip128 have an essential microtubule-anchoring function in the Drosophila germline. Development 133, 3963-3972. [DOI] [PubMed] [Google Scholar]
- Wang, C. and Lehmann, R. (1991). Nanos is the localized posterior determinant in Drosophila. Cell 66, 637-647. [DOI] [PubMed] [Google Scholar]
- Weil, T. T., Forrest, K. M. and Gavis, E. R. (2006). Localization of bicoid mRNA in late oocytes is maintained by continual active transport. Dev. Cell 11, 251-262. [DOI] [PubMed] [Google Scholar]
- Weil, T. T., Parton, R., Davis, I. and Gavis, E. R. (2008). Changes in bicoid mRNA anchoring highlight conserved mechanisms during the oocyte-to-embryo transition. Curr. Biol. 18, 1055-1061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkie, G. S. and Davis, I. (2001). Drosophila wingless and pair-rule transcripts localize apically by dynein-mediated transport of RNA particles. Cell 105, 209-219. [DOI] [PubMed] [Google Scholar]
- Wolpert, L. (1969). Positional information and the spatial pattern of cellular differentiation. J. Theoret. Biol. 25, 1-47. [DOI] [PubMed] [Google Scholar]
- Zimyanin, V. L., Belaya, K., Pecreaux, J., Gilchrist, M. J., Clark, A., Davis, I. and St Johnston, D. (2008). In vivo imaging of oskar mRNA transport reveals the mechanism of posterior localization. Cell 134, 843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]