Summary
Mind bomb1 (Mib1)-mediated endocytosis of the Notch ligand DeltaD is essential for activation of Notch in a neighboring cell. Although most DeltaD is localized in cytoplasmic puncta in zebrafish neural tissue, it is on the plasma membrane in mib1 mutants because Mib1-mediated endocytosis determines the normal subcellular localization of DeltaD. Knockdown of Notch increases cell surface DeltaA and DeltaD, but not DeltaC, suggesting that, like Mib1, Notch regulates endocytosis of specific ligands. Transplant experiments show that the interaction with Notch, both in the same cell (in cis) and in neighboring cells (in trans), regulates DeltaD endocytosis. Whereas DeltaD endocytosis following interaction in trans activates Notch in a neighboring cell, endocytosis of DeltaD and Notch following an interaction in cis is likely to inhibit Notch signaling by making both unavailable at the cell surface. The transplantation experiments reveal a heterogeneous population of progenitors: in some, cis interactions are more important; in others, trans interactions are more important; and in others, both cis and trans interactions are likely to contribute to DeltaD endocytosis. We suggest that this heterogeneity represents the process by which effective lateral inhibition leads to diversification of progenitors into cells that become specialized to deliver or receive Delta signals, where trans and cis interactions with Notch play differential roles in DeltaD endocytosis.
Keywords: Notch signaling, Endocytosis, DeltaD, DeltaA, DeltaC, Neurogenesis, Zebrafish, Lateral inhibition, Cis inhibition
INTRODUCTION
Notch signaling has an evolutionarily conserved role in determining distinct fates for adjacent cells in diverse contexts during metazoan development. Activation of Notch prevents cells from differentiating and plays a crucial role in maintaining progenitor populations during growth and development of an animal (reviewed by Artavanis-Tsakonas et al., 1999; Schweisguth, 2004).
The mature Notch receptor has an extracellular fragment, NotchEC, bound to the extracellular stub of a membrane-spanning fragment, Notch™, which is generated by S1 cleavage of the full-length receptor during synthesis. Cleavage in the extracellular stub (S2) followed by cleavage in the Notch™ transmembrane domain (S3) releases an intracellular fragment of Notch, NotchIC, into the cell. NotchIC contains a trans-activation domain and additional interaction domains that allow it to form a transcriptional activator complex with Mastermind and a member of the CSL family [CBF1/RBPjkappa, Su(H), Lag1]. Together, they drive expression of target genes recognized by the DNA-binding CSL protein (reviewed by Mumm and Kopan, 2000; Fiuza and Arias, 2007). The NotchEC fragment regulates release of NotchIC into the cell because its association with the Notch™ extracellular stub prevents access to the cleavage sites required for release of NotchIC. Removal of NotchEC is a key step in the `activation' of Notch as it leaves the S2 and S3 sites on Notch™ accessible to sequential cleavage by an ADAM protease and a γ-secretase complex (Gordon et al., 2007).
Separation of the NotchEC and Notch™ fragments is determined by interaction of Notch with DSL (Delta Serrate Lag2) ligands on the surface of a neighboring cell. The NotchEC fragment contains EGF repeats that bind DSL ligands, which are also trans-membrane proteins that have EGF repeats in their extracellular domain. It is thought that ubiquitin-dependent endocytosis of a Notch DSL ligand, like Delta, bound to NotchEC on the surface of a neighboring cell, promotes Notch activation because endocytosis of the DSL ligand-NotchEC complex facilitates separation of NotchEC from the extracellular stub of membrane-tethered Notch™ (Klueg and Muskavitch, 1999; Parks et al., 2000; Nichols et al., 2007a; Nichols et al., 2007b).
In zebrafish, the ubiquitin-mediated endocytosis of Delta homologues is regulated by the RING ubiquitin ligase Mind bomb (Mib). Previously, we have shown that a Mib homologue, now called Mib1, ubiquitylates DeltaD, promotes its endocytosis, and is essential for effective Notch activation in zebrafish during early neurogenesis (Itoh et al., 2003). It remained unclear, however, whether, in this context, Mib-mediated Delta endocytosis is triggered by interaction of Delta with Notch in a neighboring cell, or whether Mib-mediated endocytosis of Delta is a process that occurs independently of an interaction of Delta with Notch.
This study was initiated to examine the subcellular distribution of endogenous Delta in the central nervous system and to determine whether the previously described accumulation of DeltaD on the cell surface of hair cells in otic vesicles in Mib mutants (Itoh et al., 2003) is representative of changes in the rest of the nervous system. We show that Mib-mediated endocytosis is a crucial determinant of the subcellular distribution of DeltaD throughout the nervous system. We also show that Notch is a determinant of Delta endocytosis and that interaction with Notch, both within the same cell (in cis) and in a neighboring cell (in trans), is likely to regulate endocytosis of specific Delta homologues. We discuss the potential significance of these interactions during lateral inhibition.
MATERIALS AND METHODS
Fish maintenance and mutant strain
Zebrafish were maintained under standard conditions and staged according to Kimmel et al. (Kimmel et al., 1995). The mind bomb, mibta52b and mibm178 (Itoh et al., 2003; Jiang et al., 1996), and deltaD, aeiag49 (Holley et al., 2000) alleles have been described previously. Treatment with 50 μM DAPT (Calbiochem) was started at 80% epiboly to block Notch signaling.
Plasmid construction and embryo injections
Membrane-tethered mRFP-myc gene [modified from Moriyoshi et al. (Moriyoshi et al., 1996)] and HA-tagged zebrafish deltaD were cloned into pCS2 vector. Capped mRNAs were synthesized using Ambion mMessage mMachine kits. Morpholino and mRNA injections were performed at the one- or two-cell stage. 1.5 ng of each morpholino was injected for double morpholino experiments. Morpholinos were purchased from Gene Tools (Philomath, OR). mRNAs were injected at the following concentrations: 50 pg for HA-deltaD, 25 pg for memb-myc and 100 pg for DN-Su(H). Morpholinos used in this study are:
Notch1a-MO (5′-GAAACGGTTCATAACTCCGCCTCGG-3′) (Yeo et al., 2007);
Notch3-MO (5′-ATATCCAAAGGCTGTAATTCCCCAT-3′) (Yeo et al., 2007);
DeltaA-MO (5′-CTTCTCTTTTCGCCGACTGATTCAT-3′); and
DeltaD-MO (5′-AAACAGCTATCATTAGTCGTCCCAT-3′).
Whole-mount in situ hybridization
DIG-labeled anti-sense riboprobes were synthesized using the DIG labeling kit (Roche, Indianapolis, IN). Embryos were fixed in 4% paraformaldehyde overnight at 4°C. Signals were detected with alkaline phosphatase-labeled anti-DIG antibody and BM Purple substrate (Roche). Whole-mount embryos were imaged with a ProgRes C14 camera mounted on a Leica MZ12 stereomicroscope.
Immunohistochemistry
Embryos were fixed with ice-cold 10% TCA (Sigma) for 30 minutes and permeabilized with 0.2% Triton X-100 in PBS for 10 minutes. Primary antibodies used were: mouse anti-zebrafish deltaD, zdD2, anti-zebrafish deltaC, zdC1 (kindly provided by Dr Julian Lewis, Cancer Research UK London Research Institute), rabbit anti-β-catenin (Sigma) (dilution 1:500) and rabbit anti-myc (abCAM) (dilution 1:500), mouse anti-HA (Covance) (dilution 1:400). Photos were taken with a confocal microscope (LSM 510META, Carl Zeiss).
Cell transplantation and analysis
EK fish embryos were injected with either standard MO provided by Gene Tools or MOs against notch1a and notch3 with mRNA encoding memb-mRFP-myc or with mRNA encoding HA-DeltaD and memb-mRFP-myc, MycPM. Twenty to 30 cells from donor embryos were transplanted into host embryos at the 1000 cell stage, and embryos were allowed to develop until the appropriate stage as described in the Results section. Transplanted cells were visualized with rabbit anti-myc antibody, and endogenous DeltaD and exogenous DeltaD-HA proteins were detected with zdD2 or anti-HA antibodies, respectively. Z-series sections of the transplanted cells were taken on an LSMmeta510 confocal microscope.
RESULTS
Subcellular localization of DeltaD in neural ectoderm
DeltaD is expressed in `neurogenic' domains of the neural plate, where cells express a bHLH transcription factor neurogenin 1 (Ngn1) (Blader et al., 1997; Kim et al., 1997). Ngn1 expression gives cells the potential to become neurons and drives its own expression. If cells acquire high enough levels of Ngn1, they initiate expression of additional genes that allow them to differentiate as neurons. However, Ngn1 also drives the expression of Delta, which activates Notch in neighboring cells. Notch activation drives expression of transcriptional repressors in the Hairy E(Spl) Related family, which in turn inhibit expression of Ngn1 (Takke et al., 1999). So, by driving Delta expression, each Ngn1-expressing cell acquires the ability to inhibit Ngn1 and Delta expression in neighboring cells. As a consequence of this process of `lateral inhibition', only a subset of cells in the neurogenic domain acquire high enough levels of Ngn1 expression to differentiate as neurons. In the absence of effective Notch signaling, most cells within the neurogenic domain express high levels of Ngn1 to become neurons (reviewed by Chitnis, 1999; Louvi and Artavanis-Tsakonas, 2006).
We examined the distribution of endogenous Delta protein in neurogenic domains using a monoclonal antibody for DeltaD, zdD2 (Itoh et al., 2003). Fixation of zebrafish embryos with trichloracetic acid (TCA) improved the ability to visualize endogenous DeltaD distribution. zdD2 staining correlates with the distribution of deltaD transcripts both at the taibud stage, when deltaD is expressed in a subset of cells within longitudinal neurogenic domains in the prospective hindbrain (Fig. 1A,B) (Haddon et al., 1998; Appel and Eisen, 1998), and at 30 hours post fertilization (hpf), when deltaD is expressed adjacent to rhombomere boundaries (Fig. 1C,D) (Cheng et al., 2004; Riley et al., 2004; Amoyel et al., 2005). To examine the subcellular distribution of DeltaD protein, embryos were double-stained with zdD2 mAb and anti-β-catenin pAb, an adherens junction protein, which reveals the shape of individual cells. DeltaD protein was mainly detected as cytoplasmic puncta without overlap with β-catenin associated with the plasma membrane, both at the tail bud stage (Fig. 1E,E′) and later in the hindbrain at 30 hpf (Fig. 1F,F′).
Fig. 1.
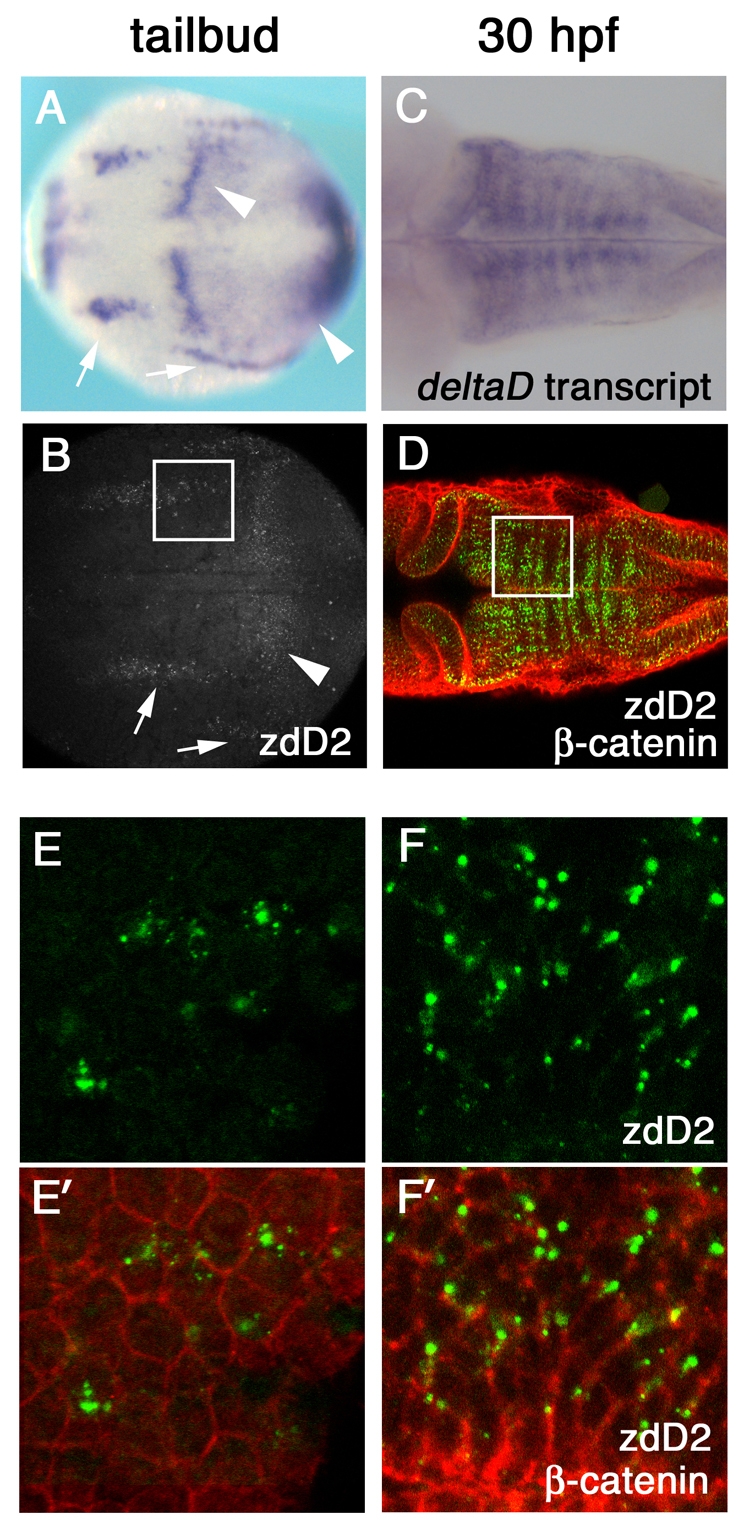
DeltaD protein is primarily localized in cytoplasmic puncta. (A-D) deltaD transcript distribution (A,C) and anti-DeltaD mAb zdD2 staining (B,D) are similar. (A,B) Arrows indicate neural expression and arrowheads indicate the mesodermal expression in tail bud stage zebrafish embryos. (C,D) DeltaD expression in the hindbrain at 30 hpf. zdD2 in green and β-catenin in red. (E-F′) Magnified images correspond to squares in B and D. (E,F) zdD2 staining. (E′,F′) Merged images with zdD2 (green) and β-catenin (red). DeltaD protein is mainly detected as cytoplasmic puncta and does not colocalize with β-catenin.
Mib1 requirement for DeltaD endocytosis
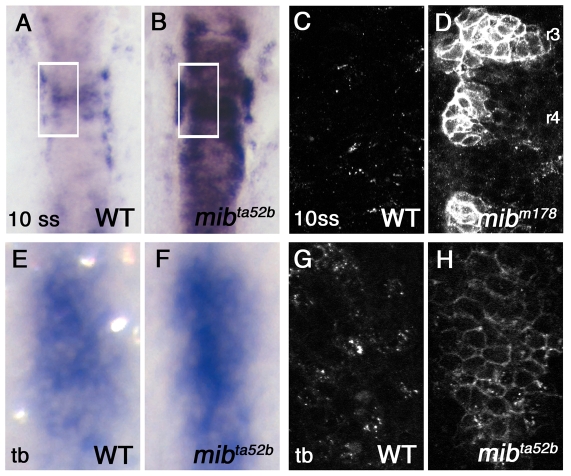
At the 10-somite stage, most of the DeltaD protein accumulated on cell surface in mib1 mutants (Fig. 2D), whereas DeltaD was in cytoplasmic puncta in wild-type siblings (Fig. 2C), suggesting that Mib1-mediated endocytosis is a crucial determinant of endogenous DeltaD distribution. However, at this stage, the deltaD transcript and DeltaD protein in mib1 mutants is exaggerated (compare Fig. 2A,B with Fig. 2C,D) as absence of Notch signaling allows cells in the neurogenic domain to express much higher levels of ngn1 and deltaD. The exaggerated deltaD expression in mib1 mutants raised the possibility that surface accumulation of DeltaD could be due to higher production of DeltaD rather than to reduced internalization from the cell surface (Lai et al., 2005; Le Borgne et al., 2005; Pitsouli and Delidakis, 2005; Wang and Struhl, 2005).
Fig. 2.
Accumulation of DeltaD protein on the plasma membrane in the mib mutant. (A,B) Hindbrain expression of deltaD transcript in wild-type (WT) and mibta52b 10-somite stage (10 ss) zebrafish embryos. (C,D) DeltaD protein in areas corresponding to the rectangles in A and B. Embryos are stained with zdD2. The patches of DeltaD expression in D are in rhombomeres 3, 4 and 5. (E,F) deltaD transcript in wild type and mibta52b at the tail bud stage (tb) in hindbrain neurogenic domains. Area shown here corresponds approximately to rectangular areas in Fig. S1A,B (see supplementary material). mibta52b embryos do not have significantly exaggerated deltaD expression. (G,H) zdD2 staining in corresponding wild-type and mibta52b neurogenic domains. In mibta52b mutants, most DeltaD is at the cell surface, even when expression is not so exaggerated.
To distinguish between these possibilities, we compared DeltaD protein distribution at the tail bud stage when the overall level of deltaD transcript does not look as significantly exaggerated (compare Fig. 2A,B with Fig. 2E,F). In wild-type siblings, DeltaD was distributed in small puncta in a small subset of cells within neurogenic domains. In mib1 mutants, failure of Notch signaling allowed most of the cells in the neurogenic domains to express DeltaD protein. Though DeltaD expression was significantly lower at this stage, most of it was still on the cell surface (Fig. 2G,H). Two different mib1 alleles, mibta52b and mibm178, had similar changes in DeltaD distribution (Fig. 2H; Fig. 4C). These results suggest Mib1-mediated endocytosis has a significant role in determining endogenous DeltaD distribution in the neural tissue.
Fig. 4.
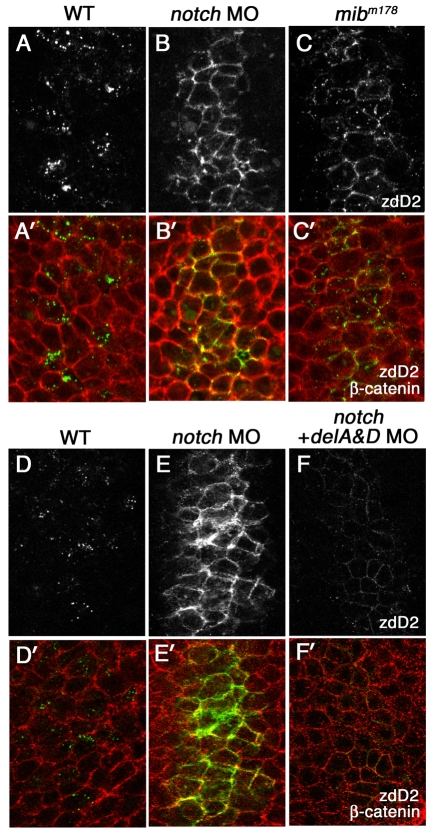
The effect of loss of Notch receptors on surface accumulation of DeltaD. (A-C′) A comparison of DeltaD distribution in wild type, notch1a and notch3 morphants, and mibm178 mutant zebrafish embryos at the tail bud stage. Areas shown here approximately correspond to rectangular areas in Fig. S1A-D (see supplementary material). Embryos are double-stained with zdD2 (grey in A-C, green in A′-C′) and β-catenin (red in A′-C′). DeltaD-expressing cells are increased and DeltaD is mainly localized at cell surface in both Notch morphants (B,B′) and mibm178 mutants (C,C′). (D-F′) DeltaD distribution in Notch morphants with (F,F′) or without (E,E′) co-injection of deltaA and deltaD (deltaA&D) MOs. Though co-injection of deltaA&D MO dramatically reduces zdD2 staining (compare E with F), DeltaD is still localized at the cell surface.
DeltaD distribution in DAPT and DN-Su(H) embryos
To further evaluate how increased deltaD production following loss of Notch signaling contributes to surface expression of DeltaD, we examined the changes in DeltaD distribution following other manipulations that inhibit Notch signaling: DAPT treatment, which inhibits the γ-secretase dependent S3 cleavage of Notch (Geling et al., 2002); injection of mRNA encoding a dominant negative form of Su(H), DN-Su(H), which lacks its DNA binding domain (Wettstein et al., 1997); and injection of morpholinos (MO) against two homologues of Notch, notch1a and notch3 (previously called notch5) expressed in the neural plate during early neurogenesis (Hsiao et al., 2007; Yeo et al., 2007).
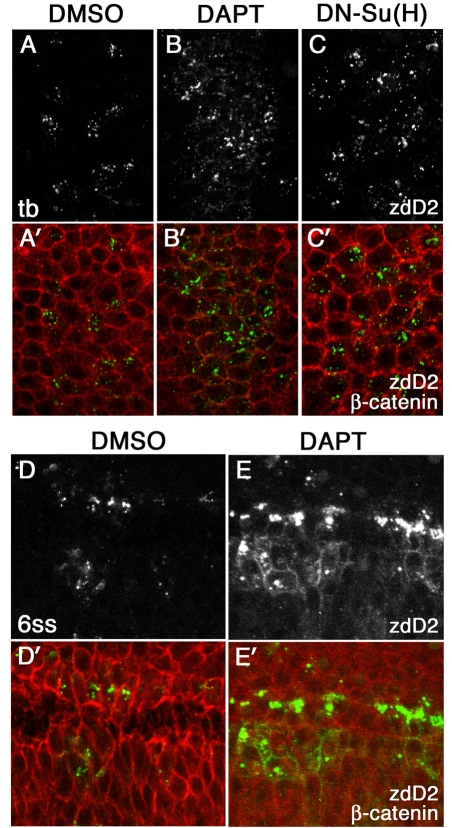
As in mib1 mutants, these manipulations induced a neurogenic phenotype with more cells in the neurogenic domain expressing DeltaD. However, at the tail bud stage, when cells did not show significant exaggeration in deltaD transcript (see Fig. S1 in the supplementary material), DAPT-treated or DN-Su(H)-expressing embryos had zdD2 in cytoplasmic puncta (Fig. 3B,B′ and Fig. 3C,C′, respectively), not at the cell surface as seen in mib1 mutants (Fig. 2H). Later, at the six-somite stage, when there was exaggerated expression of DeltaD (compare Fig. 3D,D′ and Fig. 3E,E′), zdD2 did accumulate at the cell surface in DAPT-treated (Fig. 3E,E′) and DN-Su(H)-injected (data not shown) embryos. However, unlike the situation in mib1 mutants, the surface accumulation was now accompanied by intense cytoplasmic puncta of DeltaD. These observations suggest that surface accumulation seen in mib1 mutants at an early stage is specifically due to failure of Mib1-mediated endocytosis. Later, as failure of Notch signaling results in exaggerated deltaD expression, some of the DeltaD does accumulate at the cell surface because it exceeds the cells capacity for DeltaD endocytosis.
Fig. 3.
The effect of loss of Notch signaling on surface accumulation of DeltaD. DeltaD distribution in DMSO-treated (A,A′), DAPT-treated (B,B′) or DN-Su(H)-expressing zebrafish embryos (C,C′) at the tail bud stage, and in DMSO-treated (D,D′) and DAPT-treated (E,E′)embryos at the six-somite stage (6 ss). Areas shown in A-C′ correspond approximately to rectangular areas in Fig. S1E,F (see supplementary material). Embryos were stained with zdD2 (A-E) and double-stained (A′-E′) with β-catenin (zdD2 in green, β-catenin in red). DAPT and DN-Su(H) treatment increased the numbers of cells expressing DeltaD but did not result in DeltaD accumulation at the cell surface at the tail bud stage. Some cell surface DeltaD is detected in DAPT-treated embryos by the six-somite stage; however, it is accompanied by intense cytoplasmic puncta.
Interactions with Notch are required for DeltaD endocytosis
In contrast to changes seen following DAPT treatment or injection of DN-Su(H), embryos with notch1a and notch3 simultaneously knocked down had an increase in surface DeltaD similar to that seen in mib mutants: DeltaD was at the cell surface at the tail bud stage (Fig. 4B,B′) before there was a significant increase in the level of deltaD (supplementary material Fig. S1C-E′) and the surface accumulation was not accompanied by prominent cytoplasmic distribution of DeltaD. In fact, the surface accumulation was more prominent than in mib1m178 mutants (compare Fig. 4B,B′ with Fig. 4C,C′). To demonstrate that excess DeltaD production is not responsible for its accumulation at the cell surface, DeltaD protein was reduced in Notch MO embryos by co-injection of deltaA and deltaD MO (compare Fig. 4E with Fig. 4F). The resulting reduction of DeltaD protein was still accompanied by surface accumulation of DeltaD (Fig. 4F,F′). This suggested that DeltaD interactions with Notch are essential for effective DeltaD endocytosis.
Interactions with Notch in trans and cis are required for DeltaD endocytosis
Cell transplantation experiments were performed to determine whether interaction of DeltaD with Notch in the same cell (in cis) or in a neighboring cell (in trans) is required for triggering endocytosis. Donor embryos were injected with mRNA encoding a plasma membrane-tethered mRFP and Myc-tag (MycPM), so that donor cells could be identified in host embryos by surface expression of RFP or a red fluorescent secondary antibody to visualize MycPM. DeltaD was labeled with zdD2 and visualized with a green fluorescent secondary antibody. Embryos were co-injected with notch1a and notch3 MO (Notch MO) or control MO (WT) to evaluate how the reduction of Notch affects Delta distribution. When WT cells were transplanted into WT embryos, DeltaD protein was localized in cytoplasmic puncta both in the donor cells and host embryos (28/28 of the transplanted cells) (Fig. 5G,G′). When Notch MO cells were transplanted into Notch MO embryos, DeltaD protein remained at the cell surface (24/24 cells) (Fig. 5I,I′). This showed that the transplantation process does not significantly alter the distribution of DeltaD.
Fig. 5.
Delta-Notch interactions in trans and cis contribute to DeltaD endocytosis in zebrafish. (A-F) Schematic of changes in DeltaD distribution expected following transplantation if endocytosis was determined primarily by interactions in trans (C,D) or in cis (E,F). If Notch interactions in `trans' were essential for Delta endocytosis: (C) DeltaD would be in cytoplasmic puncta in Notch MO donor cells in a WT host, (D) DeltaD would accumulate on the cell surface of WT donor cells in Notch MO host, and some Delta would now also be internalized in host Notch MO cells following interaction with Notch in adjacent WT donor cells. If `cis' interactions with Notch were essential for Delta endocytosis: (E) DeltaD would remain on the surface of donor Notch MO cells in a WT host and (F) DeltaD would remain in cytoplasmic puncta in a donor WT cell in Notch MO hosts. (G-J′) Transplantation results. (G,G′) In WT to WT transplants, DeltaD remains in intracellular puncta in WT donor and host cells. (H,H′) In Notch MO to WT transplants, some DeltaD in Notch MO donor cells becomes localized in cytoplasmic puncta (arrows in H′), suggesting that interactions with in trans promote endocytosis. However, some DeltaD remains on the cell surface (arrowheads in H) in Notch MO donor cells, suggesting an additional requirement for interactions in cis. (I,I′) In Notch MO to Notch MO transplants, DeltaD remains on the cell surface in Notch MO donor cells in Notch MO hosts. (J,J′) In WT to Notch MO transplants, some adjacent Notch MO host cells now show DeltaD in cytoplasmic puncta (arrows in J), suggesting DeltaD endocytosis following interactions in trans. Observation of cytoplasmic DeltaD puncta in WT donor cells in Notch MO hosts (black arrowheads in J′) suggest Notch interactions in cis contribute to DeltaD endocytosis. (K,L) Quantitative analysis. Cumulative pixel intensity within the cell or at the cell surface, representing the fluorescence of a secondary antibody recognizing zdD2, was measured in individual cells to quantify the distribution of DeltaD protein. Each spot represents an individual cell. (K) Scatter plot of relative intracellular (x-axis) versus cell surface (y-axis) distribution in transplanted cells from WT to WT (blue), Notch MO to Notch MO (green) and Notch MO to WT (red). The broken line indicates 10 K units and the area below 5 K units is shadowed. (L) Cumulative intracellular intensity in Notch MO host cells adjacent to WT transplants and Notch MO host adjacent to Notch MO transplants.
Transplantation of Notch MO cells into WT embryos, and WT cells into a Notch MO embryo was expected to have distinct effects on the distribution of DeltaD if interactions with Notch either in cis or trans were primarily responsible for triggering endocytosis (schematized in Fig. 5A-F). If interactions in trans were essential for endocytosis, DeltaD would be effectively internalized in a Notch MO donor cell transplanted to a WT embryo, as DeltaD in the Notch MO donor cell would interact with Notch in surrounding WT host cells (Fig. 5C). Similarly, Notch MO host cells adjacent to a transplanted WT cell would show some internalized DeltaD following successful interaction with Notch on the cell surface of transplanted WT cells (Fig. 5D). At the same time, the transplanted WT cell itself would now be expected to accumulate DeltaD on the cell surface due to absence of Notch in neighboring Notch MO host cells (Fig. 5D). However, if interactions with Notch in cis were essential to trigger DeltaD endocytosis, DeltaD would remain on the surface of a Notch MO donor cell when placed adjacent to WT host cells (Fig. 5E), and DeltaD would continue to be internalized in a WT donor cell when placed in a Notch MO host embryo (Fig. 5F).
Our results suggest that neither cis nor trans interactions are exclusively responsible for DeltaD endocytosis, but rather both types of interactions are likely to contribute. When Notch MO cells were transplanted into WT host embryos (Fig. 5H,H′), some Notch MO donor cells still had some DeltaD on the cell surface (Fig. 5H, arrowheads), suggesting that interactions with Notch in cis are required for normal levels of DeltaD endocytosis. However, some of the Notch MO cells (Fig. 5H′, arrows) now had significant intracellular DeltaD puncta, suggesting that interactions in trans with surrounding Notch-expressing WT cells had triggered DeltaD endocytosis. When WT donor cells were transplanted into a Notch MO host (Fig. 5J,J′), some donor WT cells had prominent intracellular DeltaD puncta (Fig. 5J′, black arrowheads), suggesting that cis interactions with Notch within the transplanted cell were adequate for some internalization of DeltaD. In addition, some host Notch MO cells adjacent to donor WT cells now had intracellular DeltaD (Fig. 5J, arrows), suggesting that trans interactions with Notch in transplanted WT cells had promoted DeltaD endocytosis in Notch MO host cells.
To quantify how much interactions in cis versus trans contribute to endocytosis of DeltaD, we compared surface DeltaD to cytoplasmic DeltaD in individual transplanted cells, where DeltaD had the ability to interact with Notch both in trans or in cis (WT cells into WT hosts), neither in trans nor in cis (Notch MO cells into Notch MO hosts), and only in trans (Notch MO cells into WT hosts). We did not evaluate WT cells in Notch MO hosts, where interactions only take place in cis, because DeltaD on the surface of the transplanted cells could not be distinguished from DeltaD on the surface of surrounding host cells. The cell surface or cytoplasmic boundary was outlined in a representative confocal image of each cell and surface versus intracellular DeltaD was quantified by measuring the cumulative pixel intensity within the defined boundaries using Image J software. The relative distribution of DeltaD for each cell, within each experimental group, was represented in a scatter plot with cumulative intracellular pixel intensity on the x-axis and cumulative cell surface pixel intensity on the y-axis (Fig. 5K).
The scatter plot revealed that, although WT cells transplanted into WT host embryos (Fig. 5K, blue spots) had variable amounts of intracellular DeltaD, none had a surface intensity higher than 10,000 (10 K) units (Fig. 5K, broken line). By contrast, all but one of the Notch MO cells transplanted into Notch MO host embryos (Fig. 5K, green spots) had surface intensity higher than 10 K and intracellular intensity less than 10 K units. These changes in distribution illustrate ineffective DeltaD endocytosis in the absence of both trans and cis interactions. The scatter diagram also illustrates that although all the WT cells transplanted into WT hosts have low cell surface DeltaD, 10/28 (36%) of the cells had lower than 5 K units of both surface and intracellular intensity (Fig. 5K, shadowed area). By contrast, 23/24 (96%) of the Notch MO cells transplanted into Notch MO embryos had cumulative intensities greater than 5 K. This difference is likely to be related to the role of lateral inhibition in restricting Delta expression to a subset of WT cells and failure of lateral inhibition to allow a larger fraction of transplanted Notch MO cells to express DeltaD.
DeltaD in Notch MO cells transplanted into WT hosts (Fig. 5K, red spots) was neither primarily restricted to the cell surface as in Notch MO to Notch MO transplants, nor primarily intracellular as in WT to WT transplants. In 11/32 (34%) of the cells, more than 10 K units intensity remained on the cell surface and fewer than 10 K units was intracellular, suggesting that in these cells trans interactions with Notch in surrounding WT cells were not capable of restoring high intracellular DeltaD distribution as seen in WT cells. In another 5/32 (16%) of the cells, however, fewer than 10 K units pixel intensity was on the cell surface and more than 10 K units was intracellular, suggesting that interactions in trans had restored sufficient endocytosis to promote significant intracellular distribution in these cells. In another 5/32 (16%) of cells, there was greater than 10 K units of intensity both on the cell surface and in an intracellular compartment, suggesting that although interactions in trans had permitted significant amounts of Delta endocytosis, the absence of interactions in cis allowed significant amounts of DeltaD to remain on the cell surface. These observations suggest that interactions with Notch in cis and trans have differential roles in the endocytosis of DeltaD in different cells.
As seen in Notch MO cells transplanted into Notch MO embryos, a large fraction of Notch MO cells transplanted into WT embryos (21/32 or 66%) had levels of DeltaD intensity higher than 10 K units and only a small fraction had lower than 5K units of pixel intensity. As discussed earlier, it is likely that reduced Notch signaling in Notch MO cells allowed a large fraction of these cells to express high levels of DeltaD when transplanted into WT embryos.
Intracellular DeltaD levels in Notch MO host cells adjacent to either transplanted Notch MO or transplanted WT cells were also quantified (Fig. 5L). Most host Notch MO cells (21/22 or 95%) had lower than 10 K units intracellular intensity when they were adjacent to transplanted Notch MO cells. However, when the host Notch MO cells were adjacent to transplanted WT cells, 10/39 (26%) of the cells had higher than 10 K units intensity, suggesting that interaction of DeltaD in trans with Notch in adjacent transplanted WT cells triggered endocytosis of DeltaD in host Notch MO cells.
Stabilization of DeltaD protein in Notch morphant
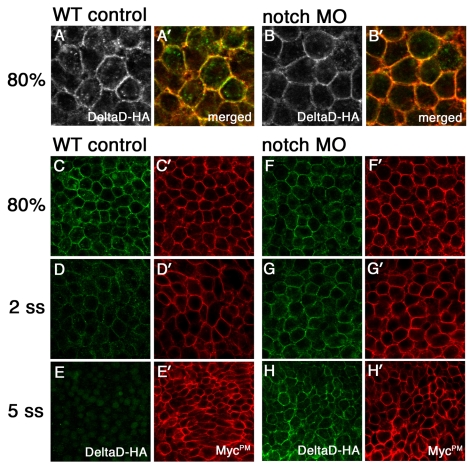
Next, we asked whether Notch-dependent DeltaD endocytosis determines DeltaD degradation. As loss of Notch signaling allows increased deltaD transcription, it is hard to evaluate if the change in the amount of endogenous DeltaD protein is determined by increased synthesis or decreased degradation. To avoid complexities introduced by endogenous transcriptional regulation, mRNAs for deltaD-HA and mycPM were co-injected into one-cell stage embryos and expression levels of DeltaD-HA and MycPM were compared in the presence and absence of Notch (Fig. 6). It should be noted that for reasons that remain unclear, significantly higher amounts of exogenously provided DeltaD-HA protein localize at the cell surface compared with endogenous DeltaD (Fig. 6A,A′). Nevertheless, intracellular DeltaD-HA puncta seen in WT embryos are no longer seen in Notch MO embryos (compare Fig. 6A,A′ with Fig. 6B,B′), confirming that endocytosis of DeltaD-HA is also regulated by Notch.
Fig. 6.
Notch-mediated DeltaD endocytosis results in DeltaD degradation. (A-B′) Distribution of DeltaD-HA in WT and Notch MO zebrafish embryos. In vitro synthesized mRNA encoding DeltaD-HA and MycPM was co-injected with control (WT in A,A′) or notch1a and notch3 MOs (Notch MO in B,B′). Embryos are labeled with anti-HA mAb (green) and anti-myc pAb (red) at 80% epiboly stage. More DeltaD-HA is localized in cytoplasmic puncta in WT than in Notch MO cells. (C-H′) Comparison of DeltaD-HA (green) and MycPM stability with (F-H) and without Notch morpholinos (C-E). DeltaD-HA expression reduces from 80% epiboly (C) to the two-somite stage (2 ss) (D) and is barely detectable at the five-somite stage (5 ss) (E) in WT. By comparison, MycPM expression levels are progressively increased from 80% epiboly to 5 ss (C′-E′). In Notch MO embryos, Delta-HA (F-H) behaves like MycPM (F′-H′) and its expression is not decreased.
Following co-injection of deltaD-HA and mycPM mRNA, MycPM expression levels progressively increased when examined at 80% epiboly the two-somite and the five-somite stage (compare Fig. 6C′-E′). This suggested that MycPM is relatively stable and it progressively accumulates on the cell surface as more is translated. By contrast, progressively lower levels of DeltaD-HA were detected at the cell surface between 80% epiboly (Fig. 6C) (n=6 embryos) and the two-somite stage (Fig. 6D) (n=10), and it was hard to see by the five-somite stage (Fig. 6E) (n=9). As Delta-HA and MycPM were transcribed from identical vectors with the same 5′- and 3′-UTR, this difference suggests that DeltaD-HA is degraded more rapidly than it is translated during this period in control embryos. When notch1a and notch3 MOs were co-injected, there was no longer a progressive reduction in DeltaD-HA expression. Instead, similar levels of DeltaD-HA were seen at 80% epiboly (Fig. 6F) and at the two-somite stage (Fig. 6G), while slightly increased DeltaD-HA was seen at the five-somite stage (Fig. 6H). These observations suggest that Notch knockdown reduces DeltaD-HA protein degradation. At this stage, we have not been able evaluate whether interactions with Notch in cis or in trans are more effective promoting degradation of DeltaD-HA.
Interactions with Notch are also required for DeltaA endocytosis
Although we used zdD2 as a specific antibody for zebrafish DeltaD protein, we noticed that deltaD/aeiAG49 mutants, with an early termination signal in the deltaD coding sequence (Holley et al., 2000), have faint zdD2 labeling when the sensitivity of fluorescence detection is increased (see Fig. S2C,E in the supplementary material). As the distribution of the faint signal resembled the deltaA expression pattern, we surmised that the staining was due to cross reactivity of zdD2 with DeltaA. Injection of deltaA MO into deltaD/aeiAG49 mutant eliminated this faint signal (data not shown), confirming that zdD2 faintly labels DeltaA in deltaD/aeiAG49 mutants. We took advantage of this cross-reactivity and examined the role of Notch interactions in DeltaA endocytosis. We found that whereas zdD2 is primarily in intracellular puncta (see Fig. S2E,E′ in the supplementary material), its distribution becomes restricted to the plasma membrane when Notch MOs are injected into deltaD/aeiAG49 mutants (see Fig. S2F,F′ in the supplementary material). As zdD2 labels DeltaA in deltaD/aeiAG49 mutants, this suggests that interactions with Notch are also crucial for DeltaA endocytosis.
DeltaC endocytosis is regulated differently from DeltaD and DeltaA
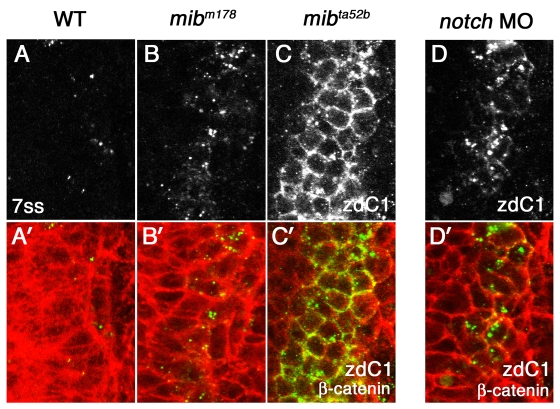
Previous studies have suggested that whereas DeltaD is a substrate for Mib1, DeltaC, expressed in a small subset of neuronal progenitors (Smithers et al., 2000), is a substrate for both Mib1 and its paralog, Mib2 (Zhang et al., 2007a; Zhang et al., 2007b). Consistent with the specific requirement of Mib1 for DeltaD ubiquitylation, DeltaD is on the cell surface in both mib1ta52b and mib1m178 embryos (Fig. 2H; Fig. 4C,C′). By contrast, DeltaC cellular distribution is not dramatically affected in mib1m178 embryos (compare Fig. 7A,A′ with Fig. 7B,B′), even though its expression is exaggerated as a consequence of reduced Notch signaling. This is consistent with Mib2 function being sufficient to mediate DeltaC endocytosis in mib1m178 embryos. However, DeltaC is primarily on the cell surface in mib1ta52b mutants (Fig. 7C,C′). This is consistent with the reported dominant inhibitory effect of the Mib1ta52b protein reducing both Mib1 and Mib2 function (Fig. 7C,C′) and therefore, DeltaC endocytosis (Zhang et al., 2007a).
Fig. 7.
Mib1, Notch1a and Notch3 are not essential for DeltaC endocytosis. (A-C′) Distribution of DeltaC in wild-type zebrafish embryos (A,A′) and in two mib alleles, mibm178 (B,B′) and mibta5ab (C,C′). (D,D′) Distribution of DeltaC in Notch MO embryos. Eight-somite stage embryos were double stained with zebrafish DeltaC antibody zdC1 (grey in A-D, green in A′-D′) and β-catenin (red in A′-D′). A dorsal view of a lateral neurogenic domain in the caudal neural plate is shown. In WT embryos, only subset of cells express DeltaC in cytoplasmic puncta (A,A′). A neurogenic phenotype allows more cells to express DeltaC in both mib mutants and Notch MO embryos as Notch signaling is reduced (B-D). However, DeltaC distribution remains cytoplasmic in mibm178 (B,B′) and Notch MO embryos (D,D′), and it only accumulates on the cell surface in mibta52b mutant embryos (C,C′).
We also examined the requirement for Notch interactions in DeltaC endocytosis. Surprisingly, DeltaC endocytosis was not dramatically affected by notch1a and notch3 knockdown (Fig. 7D,D′), though DeltaC expression itself was exaggerated as a consequence of failed Notch signaling (compare Fig. 7A,D). These results suggest that DeltaC and DeltaD not only have distinct requirements for Mib1 and Mib2, their endocytosis is differentially regulated by interactions with Notch.
DISCUSSION
In this study, we examined the subcellular localization of endogenous Delta protein in zebrafish neural tissue and we showed that DeltaA, DeltaC and DeltaD is primarily located in cytoplasmic puncta. Our analysis indicates that whereas Mib1 is crucial for endocytosis of DeltaA and DeltaD, Mib1 and Mib2 have redundant roles in mediating DeltaC endocytosis. Furthermore, while Mib1-mediated endocytosis is likely to have a crucial role in determining the cytoplasmic distribution of DeltaD, overproduction of deltaD in the neural tissue following loss of Notch signaling can also eventually contribute to some DeltaD accumulation on the cell surface.
Surface accumulation of DeltaD induced by dominant-negative Su(H) expression or DAPT treatment is accompanied by accumulation of intense cytoplasmic puncta of DeltaD. By contrast, DeltaA and DeltaD accumulation on the cell surface is not accompanied by cytoplasmic puncta when notch1a and notch3 function is knocked down. This observation revealed that interactions with Notch1a and Notch3 have a crucial role in regulating DeltaA and DeltaD endocytosis. Loss of Notch1a and Notch3 does not, however, affect the cellular distribution of DeltaC. This suggests that although interactions with Notch1a and Notch3 are likely to regulate endocytosis of DeltaA and DeltaD, they are not crucial for endocytosis of DeltaC.
Surface localization of DeltaA and DeltaD in mib1m178 mutant embryos revealed a crucial role for Mib1 in determining cytoplasmic distribution of DeltaA and DeltaD. The mib1m178 allele encodes a truncated Mib1m178 protein that lacks the third RING domain essential for its ubiquitin ligase function (Itoh et al., 2003). Nonsense-mediated degradation of mib1m178 transcript significantly reduces the amounts of the mutant protein and its phenotype is like that of a null mutant (Zhang et al., 2007b). By contrast, the mib1ta52b transcript, which is relatively stable, encodes a Met to Arg substitution in the third RING domain. This interferes with its ubiquitin ligase function and Mib1ta52b has a dominant-negative effect on both Mib1 and Mib2. Consistent with these distinctions and with the reported ability of both Mib1 and Mib2 to promote DeltaC endocytosis in cultured cells, DeltaC endocytosis does not appear to be significantly reduced in mib1m178 mutants where only Mib1 function is lost, whereas endocytosis is compromised in mib1ta52b mutants where the function of both Mib1 and Mib2 is compromised.
Though our study suggests that interactions with Notch1a and Notch3 regulate DeltaA and DeltaD endocytosis in zebrafish embryos, it remains unclear whether Delta interactions with Notch are specifically required for Mib-mediated endocytosis. Previously, we have shown that Mib1 promotes DeltaD ubiquitylation and its endocytosis in Cos-7 cells (Itoh et al., 2003). In that context, however, where both DeltaD and Mib1 were provided in excess, an interaction with Notch did not appear to be essential for Mib1-mediated endocytosis. Nevertheless, it is likely that in vivo, under physiological conditions, interaction with Notch in trans triggers Mib1-mediated DeltaD endocytosis as Mib-mediated Delta endocytosis is required for Notch activation. However, it is not clear whether Mib is required for Delta endocytosis when Delta and Notch interact in cis. Determining when and how Delta-Notch interactions trigger Mib1-mediated Delta endocytosis remains a question for future studies.
Interactions with Notch both in cis and trans regulate DeltaD endocytosis
One particularly interesting role for Notch revealed in this study is that Delta-Notch interactions both `in trans' and `in cis' regulate Delta endocytosis. Endocytosis of Delta following its interaction with Notch in trans is expected to `activate' Notch, as it helps to separate the NotchEC domain from the Notch™ domain, and this in turn facilitates sequential S2 and S3 cleavage of Notch™ and release of the NotchIC fragment into the cytoplasm. By contrast, Delta endocytosis following Delta-Notch interactions in cis, on the surface of the same cell, is not expected to activate Notch. Instead, if Delta and Notch remain bound in this context, endocytosis is likely to help to clear Delta and Notch from the cell surface, making both unavailable for interactions with partners in a neighboring cell.
Internalization of Delta-Notch complexes following interactions in cis would leave either a net excess of Delta or Notch at the cell surface, making cells specialized to deliver or receive Notch activation, respectively. As a cell with an excess of Delta becomes specialized in activating Notch in its neighbor, endocytosis of Delta might become progressively more dependent on interactions with Notch in trans. By contrast, Delta endocytosis in a cell with an excess of Notch might become progressively less dependent on interactions in trans.
We found that when Notch MO cells were transplanted into wild-type embryos, three types of cells were identified: (1) cells with relatively low surface DeltaD and high intracellular DeltaD where interactions in trans might have been adequate for effective endocytosis; (2) a population with high surface Delta and low intracellular DeltaD where interactions in cis might have been more important; and (3) a third population with high levels of both surface and intracellular DeltaD where both interactions in cis and in trans were likely to have been contributing to DeltaD endocytosis. The distinctions in the requirement for interactions in trans or cis with Notch for DeltaD endocytosis seen in transplanted cells may reflect progressive differentiation into distinct populations specialized to deliver or receive Delta signals. It remains to be seen whether interactions in trans predominantly determine Delta endocytosis in cells specialized to deliver Delta signals, whether Delta interactions with Notch in cis determine endocytosis in cells that are not as yet specialized or whether they dominate in cells with an excess of Notch specialized in receiving activation.
Interactions in cis may help tune a system for more effective lateral inhibition
Lateral inhibition is expected to amplify differences in neurogenic potential between adjacent cells and allow the progenitors with greatest neurogenic potential to differentiate, while they inhibit their neighbors from doing the same. However, when cells have high levels of both Delta and Notch on their surface, relative differences in the amounts of Delta and Notch may not be enough to prevent significant cross activation of Notch by neighbors and mutual inhibition of neurogenesis. In this context, internalization of Delta-Notch complexes following interactions in cis may help to tune the system for more effective lateral inhibition as it leaves a net excess of Delta or Notch on the surface of cells. In this context, cells with an excess of Delta would activate Notch in neighbors left with a net excess of Notch, without receiving effective activation from their neighbors. However, this model also suggests that if all the cells expressed an excess of Delta, internalization of Delta-Notch complexes following interactions in cis could deplete surface Notch and lead to failure of Notch signaling. But we know that ectopic expression of delta mRNA results in inhibition of neurogenesis (Chitnis et al., 1995; Dornseifer et al., 1997; Haddon et al., 1998; Appel and Eisen, 1998). This suggests that when all cells express an excess of Delta, Notch signaling does not fail. Instead, interactions with Notch in trans dominate and mutual Notch activation inhibits neurogenesis. This implies that when cells express both Delta and Notch, and in principle both interactions in cis and trans are possible, interactions in trans are likely to dominate and are more likely to be productive compared with cis interactions. Future studies will determine whether this is true.
The models described above assume that both Delta and Notch are internalized following an interaction in cis. Our data at present show only that Delta endocytosis is regulated by Notch. We do not have evidence for Delta regulating Notch trafficking in zebrafish. Tests of these models await future experiments using tools for visualizing the trafficking of Notch receptors in zebrafish. In Drosophila, it has been shown that cis interactions of another DSL ligand, Serrate, with Notch inhibit Notch signaling (Jacobsen et al., 1998; Li and Baker, 2004) by promoting endocytosis of Notch (Glittenberg et al., 2006). Similarly in C. elegans it has been suggested that interactions of a Notch homologue, Lin12, with a DSL ligand in cis inhibit the ability of a prospective vulval cell to deliver a signal and activate Lin12 in its neighboring cells (Chen and Greenwald, 2004).
In conclusion, our study confirms the role of Mib proteins in regulating Delta endocytosis and it reveals a crucial role for interactions with Notch in regulating endocytosis of specific Notch ligands. Future studies will reveal how differential trafficking of different Notch ligands contributes in unique ways to regulation of cell fate and behavior in distinct developmental contexts.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/2/197/DC1
Supplementary Material
We thank Julian Lewis (Cancer Research UK, London) for providing zdC1 and zdD2 antibodies; Motoyuki Itoh (Nagoya University, Japan) for valuable discussions and preliminary exploration of these questions; and all the members of Chitnis laboratory for their comments. This work was supported by the Intramural Research Program of the NIH/NICHD. This work was also supported by a JSPS research fellowship (M.M.). Deposited in PMC for release after 12 months.
References
- Amoyel, M., Cheng, Y. C., Jiang, Y. J. and Wilkinson, D. G. (2005). Wnt1 regulates neurogenesis and mediates lateral inhibition of boundary cell specification in the zebrafish hindbrain. Development 132, 775-785. [DOI] [PubMed] [Google Scholar]
- Appel, B. and Eisen, J. S. (1998). Regulation of neuronal specification in the zebrafish spinal cord by Delta function. Development 125, 371-380. [DOI] [PubMed] [Google Scholar]
- Artavanis-Tsakonas, S., Rand, M. D. and Lake, R. J. (1999). Notch signaling: cell fate control and signal integration in development. Science 284, 770-776. [DOI] [PubMed] [Google Scholar]
- Blader, P., Fischer, N., Gradwohl, G., Guillemot, F. and Strahle, U. (1997). The activity of neurogenin1 is controlled by local cues in the zebrafish embryo. Development 124, 4557-4569. [DOI] [PubMed] [Google Scholar]
- Chen, N. and Greenwald, I. (2004). The lateral signal for LIN-12/Notch in C. elegans vulval development comprises redundant secreted and transmembrane DSL proteins. Dev. Cell 6, 183-192. [DOI] [PubMed] [Google Scholar]
- Cheng, Y. C., Amoyel, M., Qiu, X., Jiang, Y. J., Xu, Q. and Wilkinson, D. G. (2004). Notch activation regulates the segregation and differentiation of rhombomere boundary cells in the zebrafish hindbrain. Dev. Cell 6, 539-550. [DOI] [PubMed] [Google Scholar]
- Chitnis, A. B. (1999). Control of neurogenesis-lessons from frogs, fish and flies. Curr. Opin. Neurobiol. 9, 18-25. [DOI] [PubMed] [Google Scholar]
- Chitnis, A., Henrique, D., Lewis, J., Ish-Horowicz, D. and Kintner, C. (1995). Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature 375, 761-766. [DOI] [PubMed] [Google Scholar]
- Dornseifer, P., Takke, C. and Campos-Ortega, J. A. (1997). Overexpression of a zebrafish homologue of the Drosophila neurogenic gene Delta perturbs differentiation of primary neurons and somite development. Mech. Dev. 63, 159-171. [DOI] [PubMed] [Google Scholar]
- Fiuza, U. M. and Arias, A. M. (2007). Cell and molecular biology of Notch. J. Endocrinol. 194, 459-474. [DOI] [PubMed] [Google Scholar]
- Geling, A., Steiner, H., Willem, M., Bally-Cuif, L. and Haass, C. (2002). A gamma-secretase inhibitor blocks Notch signaling in vivo and causes a severe neurogenic phenotype in zebrafish. EMBO Rep. 3, 688-694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glittenberg, M., Pitsouli, C., Garvey, C., Delidakis, C. and Bray, S. (2006). Role of conserved intracellular motifs in Serrate signalling, cis-inhibition and endocytosis. EMBO J. 25, 4697-4706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon, W. R., Vardar-Ulu, D., Histen, G., Sanchez-Irizarry, C., Aster, J. C. and Blacklow, S. C. (2007). Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14, 295-300. [DOI] [PubMed] [Google Scholar]
- Haddon, C., Smithers, L., Schneider-Maunoury, S., Coche, T., Henrique, D. and Lewis, J. (1998). Multiple delta genes and lateral inhibition in zebrafish primary neurogenesis. Development 125, 359-370. [DOI] [PubMed] [Google Scholar]
- Holley, S. A., Geisler, R. and Nusslein-Volhard, C. (2000). Control of her1 expression during zebrafish somitogenesis by a delta-dependent oscillator and an independent wave-front activity. Genes Dev. 14, 1678-1690. [PMC free article] [PubMed] [Google Scholar]
- Hsiao, C. D., You, M. S., Guh, Y. J., Ma, M., Jiang, Y. J. and Hwang, P. P. (2007). A positive regulatory loop between foxi3a and foxi3b is essential for specification and differentiation of zebrafish epidermal ionocytes. PLoS ONE 2, e302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Itoh, M., Kim, C. H., Palardy, G., Oda, T., Jiang, Y. J., Maust, D., Yeo, S. Y., Lorick, K., Wright, G. J., Ariza-McNaughton, L. et al. (2003). Mind bomb is a ubiquitin ligase that is essential for efficient activation of Notch signaling by Delta. Dev. Cell 4, 67-82. [DOI] [PubMed] [Google Scholar]
- Jacobsen, T. L., Brennan, K., Arias, A. M. and Muskavitch, M. A. (1998). Cis-interactions between Delta and Notch modulate neurogenic signalling in Drosophila. Development 125, 4531-4540. [DOI] [PubMed] [Google Scholar]
- Jiang, Y. J., Brand, M., Heisenberg, C. P., Beuchle, D., Furutani-Seiki, M., Kelsh, R. N., Warga, R. M., Granato, M., Haffter, P., Hammerschmidt, M. et al. (1996). Mutations affecting neurogenesis and brain morphology in the zebrafish, Danio rerio. Development 123, 205-216. [DOI] [PubMed] [Google Scholar]
- Kim, C. H., Bae, Y. K., Yamanaka, Y., Yamashita, S., Shimizu, T., Fujii, R., Park, H. C., Yeo, S. Y., Huh, T. L., Hibi, M. et al. (1997). Overexpression of neurogenin induces ectopic expression of HuC in zebrafish. Neuroscience Lett. 239, 113-116. [DOI] [PubMed] [Google Scholar]
- Kimmel, C. B., Ballard, W. W., Kimmel, S. R., Ullmann, B. and Schilling, T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310. [DOI] [PubMed] [Google Scholar]
- Klueg, K. M. and Muskavitch, M. A. (1999). Ligand-receptor interactions and trans-endocytosis of Delta, Serrate and Notch: members of the Notch signalling pathway in Drosophila. J. Cell Sci. 112, 3289-3297. [DOI] [PubMed] [Google Scholar]
- Lai, E. C., Roegiers, F., Qin, X., Jan, Y. N. and Rubin, G. M. (2005). The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 132, 2319-2332. [DOI] [PubMed] [Google Scholar]
- Le Borgne, R., Remaud, S., Hamel, S. and Schweisguth, F. (2005). Two distinct E3 ubiquitin ligases have complementary functions in the regulation of delta and serrate signaling in Drosophila. PLoS Biol. 3, e96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, Y. and Baker, N. E. (2004). The roles of cis-inactivation by Notch ligands and of neuralized during eye and bristle patterning in Drosophila. BMC Dev. Biol. 4, 5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi, A. and Artavanis-Tsakonas, S. (2006). Notch signalling in vertebrate neural development. Nat. Rev. Neurosci. 7, 93-102. [DOI] [PubMed] [Google Scholar]
- Moriyoshi, K., Richards, L. J., Akazawa, C., O'Leary, D. D. and Nakanishi, S. (1996). Labeling neural cells using adenoviral gene transfer of membrane-targeted GFP. Neuron 16, 255-260. [DOI] [PubMed] [Google Scholar]
- Mumm, J. S. and Kopan, R. (2000). Notch signaling: from the outside in. Dev. Biol. 228, 151-165. [DOI] [PubMed] [Google Scholar]
- Nichols, J. T., Miyamoto, A., Olsen, S. L., D'Souza, B., Yao, C. and Weinmaster, G. (2007a). DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol. 176, 445-458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols, J. T., Miyamoto, A. and Weinmaster, G. (2007b). Notch signaling-constantly on the move. Traffic 8, 959-969. [DOI] [PubMed] [Google Scholar]
- Parks, A. L., Klueg, K. M., Stout, J. R. and Muskavitch, M. A. (2000). Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 127, 1373-1385. [DOI] [PubMed] [Google Scholar]
- Pitsouli, C. and Delidakis, C. (2005). The interplay between DSL proteins and ubiquitin ligases in Notch signaling. Development 132, 4041-4050. [DOI] [PubMed] [Google Scholar]
- Riley, B. B., Chiang, M. Y., Storch, E. M., Heck, R., Buckles, G. R. and Lekven, A. C. (2004). Rhombomere boundaries are Wnt signaling centers that regulate metameric patterning in the zebrafish hindbrain. Dev. Dyn. 231, 278-291. [DOI] [PubMed] [Google Scholar]
- Schweisguth, F. (2004). Regulation of notch signaling activity. Curr. Biol. 14, R129-R138. [PubMed] [Google Scholar]
- Smithers, L., Haddon, C., Jiang, Y. J. and Lewis, J. (2000). Sequence and embryonic expression of deltaC in the zebrafish. Mech. Dev. 90, 119-123. [DOI] [PubMed] [Google Scholar]
- Takke, C., Dornseifer, P. v., Weizsacker, E. and Campos-Ortega, J. A. (1999). her4, a zebrafish homologue of the Drosophila neurogenic gene E(spl), is a target of NOTCH signalling. Development 126, 1811-1821. [DOI] [PubMed] [Google Scholar]
- Wang, W. and Struhl, G. (2005). Distinct roles for Mind bomb, Neuralized and Epsin in mediating DSL endocytosis and signaling in Drosophila. Development 132, 2883-2894. [DOI] [PubMed] [Google Scholar]
- Wettstein, D. A., Turner, D. L. and Kintner, C. (1997). The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development 124, 693-702. [DOI] [PubMed] [Google Scholar]
- Yeo, S. Y., Kim, M., Kim, H. S., Huh, T. L. and Chitnis, A. B. (2007). Fluorescent protein expression driven by her4 regulatory elements reveals the spatiotemporal pattern of Notch signaling in the nervous system of zebrafish embryos. Dev. Biol. 301, 555-567. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Li, Q. and Jiang, Y. J. (2007a). Zebrafish Mib and Mib2 are mutual E3 ubiquitin ligases with common and specific delta substrates. J. Mol. Biol. 366, 1115-1128. [DOI] [PubMed] [Google Scholar]
- Zhang, C., Li, Q., Lim, C. H., Qiu, X. and Jiang, Y. J. (2007b). The characterization of zebrafish antimorphic mib alleles reveals that Mib and Mind bomb-2 (Mib2) function redundantly. Dev. Biol. 305, 14-27. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.