Summary
In mouse, the LIM-homeodomain transcription factor Islet1 (Isl1) has been shown to demarcate a separate cardiac cell population that is essential for the formation of the right ventricle and the outflow tract of the heart. Whether Isl1 plays a crucial role in the early regulatory network of transcription factors that establishes a cardiac fate in mesodermal cells has not been fully resolved. We have analyzed the role of the Drosophila homolog of Isl1, tailup (tup), in cardiac specification and formation of the dorsal vessel. The early expression of Tup in the cardiac mesoderm suggests that Tup functions in cardiac specification. Indeed, tup mutants are characterized by a reduction of the essential early cardiac transcription factors Tin, Pnr and Dorsocross1-3 (Doc). Conversely, Tup expression depends on each of these cardiac factors, as well as on the early inductive signals Dpp and Wg. Genetic interactions show that tup cooperates with tin, pnr and Doc in heart cell specification. Germ layer-specific loss-of-function and rescue experiments reveal that Tup also functions in the ectoderm to regulate cardiogenesis and implicate the involvement of different LIM-domain-interacting proteins in the mesoderm and ectoderm. Gain-of-function analyses for tup and pnr suggest that a proper balance of these factors is also required for the specification of Eve-expressing pericardial cells. Since tup is required for proper cardiogenesis in an invertebrate organism, we believe it is appropriate to include tup/Isl1 in the core set of ancestral cardiac transcription factors that govern a cardiac fate.
Keywords: Drosophila, Cardiogenesis, Islet1, Tailup, Second heart field
INTRODUCTION
In our effort to decipher the molecular network that determines a cardiac fate, we attempt to identify all the key players in this process. Some time ago, the LIM-homeodomain transcription factor Islet1 (Isl1), known for its role in neural development, was introduced as a novel gene with a function in mouse heart development (Korzh et al., 1993; Pfaff et al., 1996; Thor and Thomas, 1997; Cai et al., 2003). Initial analyses of the murine Isl1 expression pattern, combined with the missing right ventricle and outflow tract of the heart in Isl1 knockout mouse embryos, suggested that Isl1 demarcates a separate cardiac lineage, also called the second heart field (Cai et al., 2003) (reviewed by Buckingham et al., 2005; Abu-Issa and Kirby, 2007). However, in vitro studies in cell culture systems and analysis of Xenopus Isl1 implicate that Isl1 is part of the early transcriptional network that establishes a cardiac fate in mesodermal cells (reviewed by Anton et al., 2007; Brade et al., 2007). Here, we took advantage of the Drosophila model to genetically determine whether tailup (tup), the fly homolog of Isl1, is required for the specification of heart precursor cells.
The Drosophila heart, although a simple tube, has become a paradigm of studying complex genetic interactions that determine cell fate. Two major cell types comprise the fly heart, which forms at the dorsal midline of the embryo. The contractile myocardial cells form the lumen of the dorsal vessel. The six myocardial cells per hemisegment are flanked by a group of pericardial cells that are needed for normal heart function. During embryogenesis, two of the six myocardial cells further differentiate into specialized myocardial cells, the ostia, which serve as inflow tracts in the posterior heart portion of the fly. Heart development in Drosophila is initiated in the dorsal mesoderm when a particular group of cells in each hemisegment receives input from the ectodermal growth factors Wingless (Wg) and Decapentaplegic (Dpp) (Wu et al., 1995; Frasch, 1995; Park et al., 1996). These signaling pathways induce in Tinman (Tin)-positive mesodermal cells a complex network of transcription factors that distinguishes the cardiac mesoderm from the adjacent visceral mesoderm and dorsal somatic muscles (Frasch, 1995; Riechmann et al., 1997; Lee and Frasch, 2000; Lockwood and Bodmer, 2002; Jagla et al., 2002). In addition to the homeobox transcription factor tin (Nkx2.5), early specification requires the function of the T-box factors Dorsocross1-3 (herein referred to as Doc) and of the GATA factor pannier (pnr) (Gajewski et al., 1999; Alvarez et al., 2003; Klinedinst and Bodmer, 2003; Reim and Frasch, 2005). Once cardiac specification has taken place, tin, Doc and pnr cross-regulate each other to maintain their expression and to initiate the differentiation of the cardiac cells (reviewed by Zaffran and Frasch, 2002; Qian et al., 2008). The latter requires additional transcription factors, including the Tbx20-related gene neuromancer (nmr1 and nmr2; also known as H15 and mid, respectively), the COUP-TFII-related gene seven up (svp), and the homeobox gene ladybird (lb), which are involved in regulating the diversity of myocardial and pericardial cell fates (Miskolczi-McCallum et al., 2005; Qian et al., 2005; Reim et al., 2005; Lo and Frasch, 2001; Jagla et al., 1997; Jagla et al., 2002). Most of these transcription factors have a fairly dynamic expression pattern during heart development, which suggests that their specific function in cardiogenesis can vary depending on the cellular context. For example, tin and Doc initially cooperate to properly specify cardiac progenitors (Reim and Frasch, 2005). However, during the differentiation of myocardial cells, tin represses Doc genes in four out of the six myocardial cells in each hemisegment, thereby restricting Doc expression to the two Tin-negative cells that form the ostia in segments A2 to A7 (Zaffran et al., 2006).
A phenotypic characterization of tup mutants has shown that tup plays an important role in Drosophila heart and hematopoietic organ formation (Tao et al., 2007). However, these analyses, which included a description of the cardiac expression pattern of Tup, were restricted to stages well past the time when cardiac specification occurs. Hence, the question has remained whether tup is required for the proper specification of heart cells.
Here, we present a detailed study of Tup expression and function during cardiogenesis and show that tup is indeed required for the specification of a cardiac fate. Analyses of genetic interactions establish tup as a crucial factor that cooperates with tin, pnr and Doc during cardiogenesis. Germ layer-specific inhibition of Tup function shows that ectodermal Tup is also required for normal Tin expression at early stages. Rescue experiments suggest that there might be a different set of mesodermal and ectodermal factors with which Tup can interact through its LIM domains. Cell-specific inhibition of Tup function shows that Tup is required to maintain expression of Odd in pericardial cells. Overexpression experiments show that a balance of Tup and Pnr is required for the correct specification of Eve-expressing cell clusters. Taken together, these findings place tup as a crucial factor in the early cardiac transcriptional network.
MATERIALS AND METHODS
Drosophila stocks and crosses
The following mutant fly stocks were used: tupisl-1 [isl37Aa (Thor and Thomas, 1997], pnrVX6, wgCX4, dppd6, Df(2L)OD15 (all from The Bloomington Stock Center), tin346 (Azpiazu and Frasch, 1993) and Df(3L)DocA (Reim et al., 2003). The tupisl-1, wgCX4 and the Df(2L)OD15 stocks were rebalanced with CyO, wg-lacZ, and the pnrVX6 stock was rebalanced with TM3, ftz-lacZ to identify homozygous mutant embryos. CantonS served as a wild-type stock. Analysis of cuticles of tupisl-1/CyO embryos (n=415) showed that 19% of the homozygous tup mutants (n=104) had an obvious germ band retraction phenotype. These embryos were not included in our analyses. For the genetic interactions, embryos that were single or double heterozygous for the investigated allele(s) were selected based on the lack of staining for β-galactosidase activity present on the corresponding balancer chromosomes. Statistical computing was performed using R (www.r-project.org). The following Gal4 and UAS lines were used: twi-Gal4 (Greig and Akam, 1993), 69B-Gal4 (Brand and Perrimon, 1993), Dot-Gal4 (Kimbrell et al., 2002), tinCΔ4-Gal4 (Lo and Frasch, 2001), UAS-tup, UAS-tupΔHD (Thor and Thomas, 1997; O'Keefe et al., 1998), UAS-tupΔLIM (Biryukova and Heitzler, 2005), UAS-pnrD4 (Haenlin et al., 1997) and UAS-tin (Ranganayakulu et al., 1998). For co-overexpression of UAS-tup and UAS-pnrD4, the individual UAS constructs were recombined on the third chromosome. The UAS-tin;UAS-tup stock was generated by standard genetic crossings.
Immunohistochemistry and in situ hybridization
Antibody staining (single and double labeling) was performed essentially as described (Qian et al., 2005; Liu et al., 2006). Primary antibodies were detected with a Cy3-conjugated AffiniPure donkey anti-rabbit IgG (H+L) (1:200) (Dianova, Hamburg, Germany). If amplification of the signal was necessary, biotinylated secondary antibodies were used (1:200) in combination with the Tyramide Signal Amplification System (Perkin Elmer) and dichlorotriazinylamino fluorescein (1:200) (Dianova). Embryos were mounted in Vectashield (Vector Laboratories). Embryos from single immunostainings were analyzed using Olympus BX60 (Olympus, Hamburg, Germany) or Keyence BZ-8000K epifluorescence microscopes, with the image-analyzing software BZ-Analyzer (Keyence, Neu-Isenburg, Germany). Embryos from double immunostainings were analyzed using a Leica TCS SP confocal microscope. Primary antibodies were used at the following dilutions: mouse anti-chicken Isl1 (Tup), 1:50 with TSA [Developmental Studies Hybridoma Bank (DSHB)]; rabbit anti-Dmef2, 1:2000 (Lilly et al., 1995); rabbit anti-Tin, 1:50 (Venkatesh et al., 2000); mouse anti-Pericardin (EC11), 1:10 with TSA (DSHB); rabbit anti-Eve, 1:3000 (Frasch et al., 1987); rabbit anti-Odd, 1:100 (Ward and Skeath, 2000); and mouse anti-Pnr, 1:400 with TSA (Herranz and Morata, 2001). Fluorescent in situ hybridization for dpp was performed essentially as described (Klinedinst and Bodmer, 2003). Double fluorescent in situ hybridization and immunostaining was adapted from Knirr et al. (Knirr et al., 1999). The digoxigenin-labeled dpp and pnr in situ probes were generated using the DIG RNA Labeling Mix from Roche (Mannheim, Germany).
RESULTS
Expression pattern of Tup during dorsal vessel development
Tup protein, as detected by a monoclonal mouse antibody against chicken Isl1, was observed in a broad domain along the dorsal side of the Drosophila embryo at stage 10, within the ectodermal layer (Fig. 1A). This expression pattern is reminiscent of the expression domain of ectodermal Dpp, as well as of those of Tin (mesoderm) and Pnr (initially only in the ectoderm), two transcription factors that are crucial for proper cardiac specification. Double labeling for tup transcripts and Wg protein demonstrated more clearly the ectodermal expression of tup (Fig. 1B). Double immunostainings for Tup and Tin were performed to identify Tup-positive cardiac cells within this broad domain. This analysis revealed that Tup expression in the cardiac mesoderm initiates at mid-stage 11, when Tin becomes restricted to the dorsal-most mesoderm, in ∼10 clusters, each consisting of ∼2 cells (Fig. 1C). Cells of the Tup clusters co-expressed Eve and therefore belong to the pericardial cell lineage (Fig. 1D). By late stage 11, Tup expression expanded within the Tin-expressing cardiac mesoderm (Fig. 1E) and continued in all myocardial cells during embryogenesis (Fig. 1F-I) (Tao et al., 2007). We also detected Tup in at least two of the Tin-positive pericardial cells and in all four Odd-expressing pericardial cells in each hemisegment (Fig. 1G-I). In the lymph glands, Tup was only expressed in some of the Odd-positive cells (Fig. 1I). tup transcripts were also present in the cytoplasm of the Dmef2 (Mef2)-positive myocardial cells, demonstrating that the expression patterns of tup RNA and Tup protein were identical (Fig. 1J). Consistent with its function in amnioserosa development, Tup was also detected in this tissue. Here, we focus on the role of tup in heart development and our analysis demonstrates that the observed heart phenotype is not primarily an effect of a defect in germ band retraction.
Fig. 1.
Tup expression during cardiogenesis in wild-type Drosophila embryos. (A) At stage 10, Tup is expressed in a broad domain in the dorsal ectoderm (arrowhead). The expression in the amnioserosa (as) persists throughout embryogenesis. (B) Double labeling for Wg protein and tup RNA confirms the ectodermal expression of tup. (C) At mid-stage 11, Tup starts to be expressed in the cardiac mesoderm in ∼10 small clusters of cells (arrowheads). (D) These clusters are also positive for Eve (arrowheads). (E) By late stage 11, Tup is co-expressed with Tin throughout the cardiac mesoderm (arrowheads). (F-H) Tup is expressed in all six myocardial cells (arrowheads) and in the Tin-positive pericardial cells (arrows in H). Arrowheads in H point to the two Tin-negative, Tup-positive myocardial cells. (I) Tup is expressed in all Odd-positive pericardial cells (arrows) and in a subset of Odd-expressing cells of the lymph glands (lg). (J) tup RNA expression in myocardial Dmef2-expressing cells matches Tup protein localization (arrowheads), as seen in G. Except for H and I, which are dorsal views of stage 15 embryos, all images are lateral views. Anterior is to the left. WT, wild type.
tup is required for the formation of the dorsal vessel
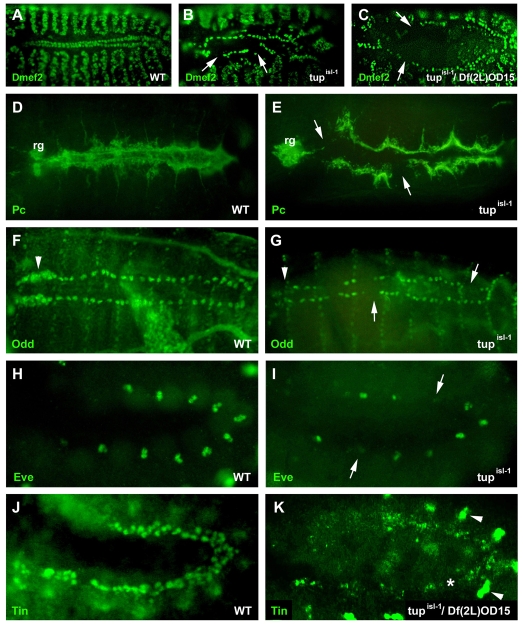
All analyses were performed using embryos harboring the tupisl-1 allele. Molecular analysis of the tupisl-1 allele suggests that it has a mutation in the transcriptional regulatory region (Tao et al., 2007). The extremely low expression level of Tup protein or tup RNA in mutant embryos indicates that the tupisl-1 allele is a strong hypomorph (see Fig. S1A-F in the supplementary material). Consistent with its cardiac expression pattern, formation of the dorsal vessel is severely affected in tupisl-1 embryos, as can be seen by the loss of Dmef2-expressing myocardial cells, as well as by the disrupted Pericardin (Pc) expression (Fig. 2A,B,D,E). Pericardin normally demarcates pericardial cells and accumulates at the basal membrane of the myocardial cells (Chartier et al., 2002). The loss of pericardial cells in tupisl-1 embryos is shown by gaps in Odd and Eve expression (Fig. 2F-I). The late cardiac phenotype at stage 15/16 has essentially been described before (Tao et al., 2007). Our study aimed to determine the position of tup in the early cardiac transcriptional network, and whether the cause of the cardiac phenotype was distinct from secondary effects of problems in germ band retraction.
Fig. 2.
Heart phenotypes in tupisl-1 mutants. (A,B,D-J) Compared with wild-type Drosophila embryos, tupisl-1 mutants are characterized by gaps in expression of all examined myocardial (Dmef2 and Tin) and all pericardial (Pc, Odd and Eve) cell markers. (C,K) Embryos that are transheterozygotic for tupisl-1 and a deficiency that includes the tup locus, Df(2L)OD15, also show gaps in Dmef2 expression at stage 14 (arrows in C) and show a strong reduction of Tin-expressing cardiac cells at late stage 11 (asterisk in K). Arrowheads in K point to Tin-positive visceral mesodermal cells. as, amnioserosa; rg, ring glands.
The loss of some Eve-expressing cell clusters is already seen by late stage 11 and indicates that the defects in heart development are not restricted to later stages when the two rows of myocardial cells come together at the dorsal midline. Reduced Tin and Dmef2 expression was also observed in tupisl-1/Df(2L)OD15 transheterozygotes (Fig. 2C,J,K). Since tup is expressed in the dorsal mesoderm around the time when cardiac progenitor cells become specified, it is likely that tup plays a role in this process. Proper cardiac specification requires interactions between Tin, Pnr and Doc. Therefore, we examined whether the expression of these factors is affected in tupisl-1 embryos. Indeed, tup mutants were characterized by a strong reduction in Tin-, Pnr- and Doc2-expressing cells at stage 11 (Fig. 3A-D,I,J). Since Tin and Doc are already expressed in the cardiac mesoderm at stage 10, before the onset of mesodermal Tup expression, these findings indicate that tup is required for their maintenance rather than their induction. The onset of cardiac expression of Pnr and Tup seems to coincide at stage 11 (Klinedinst and Bodmer, 2003; Reim and Frasch, 2005). Moreover, like Tup, Pnr is also expressed in the ectodermal layer and double staining for pnr RNA and Dmef2 or Wg protein demonstrated that pnr expression is reduced in the mesoderm and ectoderm in tup mutants (Fig. 3E-H). This suggests that Tup function is also required in the ectoderm to maintain Pnr expression.
Fig. 3.
tup is required for the normal expression of early cardiac transcription factors. (A,B) Drosophila stage 11 tupisl-1 mutants are characterized by a reduction in Tin-expressing cells (arrows). (C,D) The Pnr expression domain is strongly reduced in tupisl-1 mutants (arrows). (E,F) Double fluorescence labeling for Dmef2 protein and pnr RNA shows the mesodermal reduction of pnr expression in tupisl-1 mutants (arrows). (G,H) Reduced pnr expression (arrows) in the ectoderm is demonstrated by co-staining for Wg protein. (I,J) Stage 11 tupisl-1 mutants lack cardiac Doc2-positive cells (arrows). Arrowheads indicate missing Eve-expressing cells.
To further evaluate the functional relationship between tup, tin, pnr and Doc, we analyzed the expression of Tup in tin346, pnrVX6 and Df(3L)DocA embryos. Staining for Tup protein in tin346 embryos showed that the early Tup clusters are present, suggesting that they are initially independent of tin (Fig. 4A,B). However, Tup expression was not maintained (Fig. 4C,D). Df(3L)DocA and pnrVX6 mutants also showed a strong reduction in, or lack of, Tup-expressing cells (Fig. 4E,F). Together with the data above, these results point to an interdependency of all four factors: tup, tin, pnr and Doc.
Fig. 4.
Tup expression requires the presence of early cardiac transcription factors and depends on wg and dpp signaling. (A-D) Tup expression is initiated in the cell clusters in Drosophila tin346 mutants (arrowheads in B) but is not maintained at later stages (compare C with D). (E) Myocardial Tup and Dmef2 expression is absent in Df(3L)DocA mutants. Since Doc mutants have been shown to also lack pericardial cells, the remaining Tup-expressing cells (green) are unlikely to be cardiac-related cells. (F) pnrVX6 mutants also show a dramatic reduction in myocardial Tup- and Dmef2-expressing cells. (G,H) Tup expression at stage 13/14 depends on Wg (G) and Dpp (H) signaling. Arrowheads in all images point to Dmef2/Tup co-expressing cells, which appear yellow in the merged optical sections. Asterisks are placed in the region of the myocardial cell row, which has defects to various degrees in all mutants shown.
Since Wg and Dpp are crucial growth factors in heart development, we also analyzed Tup expression in wgCX4 and dppd6 embryos. Both mutants were characterized by a strong reduction in, or loss of, Tup expression (Fig. 4G,H). Similarly, as observed in tin346 mutants, Tup was initially present in the early cell clusters in wgCX4 embryos at stage 11 (data not shown). Early steps in visceral mesoderm formation seemed to be unaffected in tupisl-1 mutants, whereas tup might play a role in the specification of the Kr-expressing dorsal somatic muscle cells (see Fig. S2A-D in the supplementary material). In summary, these data demonstrate that tup is required for the proper specification of cardiac progenitor cells and for the formation of the dorsal vessel.
tup cooperates with tin, pnr and Doc during cardiogenesis
Next we tested whether tup interacts genetically with tin, pnr and Doc. For this purpose, we analyzed embryos that are transheterozygous for tupisl-1 and Df(3L)DocA, tupisl-1 and tin346, or tupisl-1 and pnrVX6. The phenotypes of these embryos were compared with the phenotypes of single heterozygotes for each of the investigated alleles. Each double transheterozygous combination resulted in obvious gaps within the Dmef2-expressing myocardial cell rows in ∼30% of the embryos analyzed (Fig. 5A-D and Tables 1 and 2). Also, tup and tin cooperated to maintain normal Pnr expression, as tupisl-1/+;tin346/+ transheterozygotes showed reduced staining for Pnr (72%, n=102) (Fig. 5E,F). Tin expression was reduced in tupisl-1/+;Df(3L)DocA/+ (51%, n=98) and tupisl-1/+;pnrVX6/+ (45%, n=111) embryos, demonstrating that the combined action of these factors is required for proper cardiac specification (Fig. 5G-I). Only ∼8-13% of the single heterozygous embryos (∼100 embryos for each combination were counted) had phenotypes comparable to the double transheterozygotes. Taken together, these results further indicate that tup is required in combination with tin, pnr and Doc to properly specify and maintain a cardiac fate.
Fig. 5.
Genetic interactions between tup, tin, pnr and Doc. The cardiac phenotypes in transheterozygotic Drosophila embryos demonstrate that tup interacts genetically with all three factors. The phenotypes were compared with those of the cardiac markers in single heterozygotes, and were evaluated statistically for Dmef2 (see Tables 1 and 2). (A-D) Dmef2 expression in the wild type (A) and in embryos transheterozygotic for tupisl-1 and pnrVX6 (B), tupisl-1 and tin346 (C), tupisl-1 and Df(3L)DocA (D). Dorsal views of embryos at stage 15/16 are shown. Arrows point to gaps in the myocardial rows of the dorsal vessel. (E,F) Pnr is reduced in tup/tin transheterozygotic embryos (arrows in F). A lateral view of a stage 11 embryo is shown. (G-I) Tin expression in the wild type (G), and in embryos transheterozygotic for tup and pnr (H), and tup and DocA (I). Reduced Tin expression is seen in both cases (arrows in H,I). Dorsal views of stage 14 embryos are shown.
Table 1.
The number of double transheterozygous embryos that have gaps in the myocardial cell rows is significantly higher than in embryos with only one mutant allele
| Genotype (group 1) | # Embryos w/o gap | # Embryos with gap | Genotype (group 2) | # Embryos w/o gap | # Embryos with gap | Genotype (group 3) | # Embryos w/o gap | # Embryos with gap |
|---|---|---|---|---|---|---|---|---|
| tupisl-1/+;Df(3L)DocA/+ | 50 | 20 | pnrVX6/tupisl-1 | 38 | 18 | tupisl-1/+;tin346/+ | 58 | 25 |
| Df(3L)DocA/+ | 38 | 4 | pnrVX6/+ | 48 | 6 | tin346/+ | 63 | 7 |
| tupisl-1/+ | 60 | 6 | tupisl-1/+ | 60 | 6 | tupisl-1/+ | 60 | 6 |
| Chi-square test (χ2), P | χ2=11.99, P1=0.0005 | χ2=11.72, P2=0.0006 | χ2=13.80, P3=0.0002 | |||||
The χ2 test revealed in all three groups that the proportion of embryos with and without gaps is statistically different for single heterozygous and double transheterozygous embryos. Adjustment for multiple comparisons was performed with the Bonferroni correction (P̃1=0.0016, P̃2=0.0019, P̃3=0.00061).
Table 2.
There is significant difference between the number of Dmef2-positive cells in embryos with and without gaps in the myocardial cell rows
| Genotype | # Dmef2+ cells in embryos w/o gap* | # Dmef2+ cells in embryos with gap* | ||
|---|---|---|---|---|
| Wild type | 105 | - | ||
| tupisl-1/+;Df(3L)DocA/+ | 96 | (91; 101) | 90 | (85.5; 95) |
| tupisl-1/+;pnrVX6/+ | 93 | (89; 95) | 84.5 | (73; 92) |
| tupisl-1/+;tin346/+ | 100 | (98; 104) | 96 | (86; 100.25) |
| Df(3L)DocA/+ | 100 | (97; 102) | 94.5 | (93; 96) |
| pnrVX6/+ | 100.5 | (98; 105) | 92.5 | (87; 97) |
| tin346/+ | 101 | (95; 107) | 93 | (78; 98.75) |
| tupisl-1/+ | 99 | (96; 102) | 94 | (92; 98) |
The data were statistically tested for significant differences using the Wilcoxon rank sum test (P=2.58−13).
Showing the median, with the interquartile range in parentheses.
Tissue- and cell-specific requirement for tup during cardiogenesis
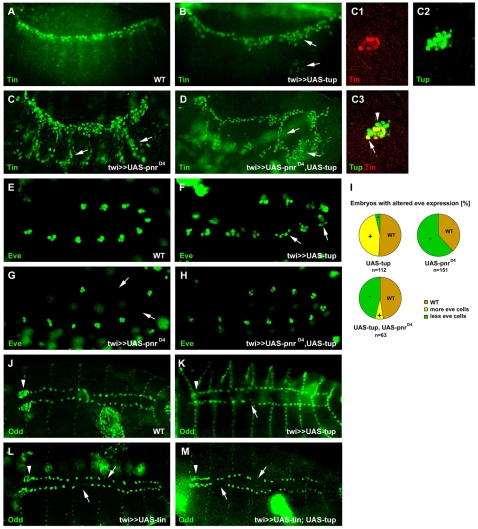
As shown above, Tup is expressed in the dorsal ectoderm as well as in the cardiac mesoderm, and in tup mutants expression of Tup protein is almost absent in both germ layers. Therefore, we wanted to distinguish between the mesodermal and a possible ectodermal contribution of tup function in cardiogenesis. We interfered with endogenous Tup function by expressing a deletion construct of tup, which lacks the homeodomain but contains both LIM domains and is likely to act as a dominant-negative (UAS-tupΔHD) (O'Keefe et al., 1998). When expressing UAS-tupΔHD in the mesoderm, we observed a reduction in Tin-expressing cells (Fig. 6A,B). A similar phenotype was induced after expressing UAS-tupΔHD in the ectoderm (Fig. 6D). To further investigate the mesodermal and ectodermal contribution of Tup for cardiogenesis, we aimed to rescue the Tin phenotype by co-expressing the full-length tup cDNA. When both constructs were expressed in the ectoderm, we observed a partial rescue in ∼53% of the embryos (n=74) (Fig. 6E). However, ∼82% of the embryos (n=48) in which UAS-tupΔHD and UAS-tup were co-expressed in the mesoderm, still exhibited a strong phenotype (Fig. 6C). Since the LIM domains are known to act as protein-interaction domains (Schmeichel and Beckerle, 1994; Kadrmas and Beckerle, 2004), this result implicates that the Tup LIM domains interact with, and thereby inhibit, other mesodermal factors, the functions of which appear to be required for normal cardiogenesis but cannot be rescued by simultaneous expression of tup. To test whether the LIM domains are required for Tup function in cardiogenesis, we expressed a UAS-tupΔLIM deletion construct in the mesoderm. This construct is expected to still bind to the DNA, but LIM domain-mediated interactions with other proteins are disrupted. Mesodermal expression of UAS-tupΔLIM also resulted in a reduction of Tin-expressing cells (Fig. 6F), demonstrating the requirement of the LIM domains to mediate proper interactions between Tup and other proteins in cardiogenesis.
Fig. 6.
Germ layer- and cell-specific requirements of Tup at various stages of cardiogenesis. Drosophila embryos at stage 12 (A-F) or between stages 10 and 11 (G-I), shown from the lateral side with anterior to the left; or at stage 15/16 (J-M) or 14 (N) shown from the dorsal side. (A,B,D) Mesodermal and ectodermal inhibition of Tup function by expressing UAS-tupΔHD results in a reduction of Tin-positive cells (arrows in B,D). (C,E) Full-length tup (UAS-tup) can partially restore Tin when co-expressed in the ectoderm but not in the mesoderm. (F) Mesodermal expression of a Tup construct lacking the LIM domains (UAS-tupΔLIM) also affects Tin expression (arrows). (G-I) Tup is required for normal dpp expression as shown by in situ hybridization. dpp is reduced after ectodermal inhibition of Tup function (arrows in H). dpp is strongly reduced in tupisl-1 mutants (I). (J-K′) Mesodermal expression of UAS-tupΔHD results in a reduced number of Dmef2-positive myocardial cells. (K′) An enlargement of the two segments (as delineated by the vertical lines) indicated by the brackets in K. (L-N) Inhibition of Tup function in the pericardial cell lineage results in loss of Odd-positive cells (arrow in M), including a subset of Odd-expressing lymph gland cells (arrowhead in M). Overexpression of Tup in this lineage induces additional Odd-positive pericardial cells (arrows in N). Odd expression in the lymph glands appears unaffected (arrowhead in N).
The reduction of mesodermal Tin-expressing cells after inhibiting Tup in the ectoderm can only be explained if the function of a secreted cardiogenic factor is impaired. Owing to their highly similar expression patterns, Dpp is a likely candidate. Although dpp expression was reduced in embryos expressing UAS-tupΔHD in the ectoderm (Fig. 6G,H), we observed a stronger phenotype in tupisl-1 mutants (Fig. 6I). Hence, ectodermal Tup can regulate dpp expression, either directly or indirectly through Pnr. In contrast to the relatively strong downregulation of early Tin expression, there was still a considerable number of Dmef2-expressing myocardial cells, with only some segments that had fewer than the normal six Dmef2-positive cells (Fig. 6J,K,K′). Therefore, we analyzed Tin expression throughout embryogenesis and observed that the initial, strong reduction of Tin in embryos expressing UAS-tupΔHD in the mesoderm appears to recover over time, and by stage 16 Tin expression was comparable to that of wild-type embryos (see Fig. S3B,E,H in the supplementary material).
This pattern of expression could either point to a temporal requirement of tup for Tin expression, or be due to the twi-Gal4 driver, the activity of which becomes weaker during embryogenesis. Because we have observed a similar phenomenon for Tin expression in tupisl-1 mutants (see Fig. S3C,F,I in the supplementary material), we favor the first possibility. Since Tup is expressed in pericardial cells throughout embryogenesis, we tested whether Tup function is required at later stages to maintain this pericardial fate. We expressed UAS-tupΔHD in the pericardial cell lineage using the Dot-Gal4 driver (Kimbrell et al., 2002). Inhibition of Tup function in pericardial cells resulted predominantly in the loss of Odd-positive cells in one or more hemisegments in 63% of the embryos (n=122), as well as in lymph glands of reduced size (Fig. 6L,M). Conversely, overexpression of Tup in the pericardial cell lineage yielded additional Odd-expressing cells in several hemisegments in 42% of the embryos (n=119) (Fig. 6N).
Our data show that Tup functions in the mesoderm, as well as in the ectoderm regulating dpp expression to guarantee normal heart development. The experimental approach of inhibiting and rescuing Tup function implicates that Tup interacts with different proteins in the ectoderm and in the mesoderm to ensure normal cardiogenesis.
Mesodermal overexpression of Tup
The requirement of tup in cardiogenesis prompted us to investigate whether Tup might be sufficient to induce additional cardiac and/or pericardial cells. Early pan-mesodermal overexpression of Tup induced only a slight increase in Tin-positive cells (Fig. 7A,B). Expression of UAS-pnrD4 results in a strong induction of ectopic Tin expression, as reported by Klinedinst and Bodmer (Klinedinst and Bodmer, 2003) (Fig. 7C). Co-expression of UAS-pnrD4 and UAS-tup resulted in a similar phenotype to that seen upon mesodermal overexpression of UAS-pnrD4 alone (Fig. 7D). Double immunostaining for Tin and Tup in embryos overexpressing UAS-pnrD4 revealed that the ectopic Tin cells co-express Tup, but the clusters are heterogeneous because they also contain cells that only express Tup (Fig. 7C1-C3). Mesodermal overexpression of Tup induced an enlargement of the Eve-positive cell clusters in 46% of the embryos (n=112) (Fig. 7E,F,I), whereas mesodermal overexpression of UAS-pnrD4 resulted in the opposite phenotype in 62% of the embryos (n=151) at stage 11 (Fig. 7G,I). Co-expression of UAS-tup and UAS-pnrD4 was able to rescue the effect induced by overexpression of each factor singly (Fig. 7H,I), but to different extents. It is important to note here that pnrD4 is a very active allele and therefore cannot be fully counteracted by tup. As a result, when both constructs are co-overexpressed, the phenotype of enlarged Eve-expressing clusters induced by UAS-tup was more efficiently `rescued' (from 46% to 6%) than the phenotype induced by UAS-pnrD4 (from 62% to 46%). Mesodermal overexpression of UAS-tup resulted in a moderate loss of Odd-positive pericardial cells and in the complete loss of Odd-positive cells in the lymph glands (Fig. 7J,K). The same phenotype was observed when UAS-tin was expressed early throughout the mesoderm (Fig. 7L). When both factors were overexpressed, the effect on Odd-expressing pericardial cells did not appear to be synergistic (Fig. 7M).
Fig. 7.
Mesodermal overexpression of Tup reveals different functional relationships with other cardiac transcription factors. (A-D) Overexpression of Tup leads to a moderate expansion of Tin and some ectopic Tin-expressing cells on the lateral side of the embryo (arrows in B). Overexpression the Pnr allele pnrD4 results in a strong ectopic induction of Tin across the whole lateral side of the embryo (arrows in C). Co-overexpression of Tup and PnrD4 mimics the phenotype of PnrD4 overexpression alone (arrows in D point to ectopic Tin-expressing cells). (C1-C3) The ectopic Tin-positive cell clusters induced by overexpression of PnrD4 alone are heterogenous. Some cells co-express Tin and Tup (arrow in C3), whereas others are only positive for Tup (arrowhead in C3). (E-I) Tup and Pnr counteract each other in Eve-positive pericardial cell specification. Overexpression of Tup results in additional Eve-positive cells within the clusters (arrows in F), whereas overexpression of PnrD4 leads to the complete loss of Eve-positive cell clusters (arrows in G). (H) Co-overexpression of Tup and PnrD4 can reduce the effects induced by each factor singly. (I) Pie charts showing the percentage of embryos with wild-type (WT, brown), expanded (+, yellow) or reduced (-, green) Eve-positive cell clusters. (J-M) Overexpression of Tup results in a moderate loss of Odd-positive pericardial cells (arrow in K) and to a strong reduction of Odd-positive lymph gland cells (arrowheads in J,K). Overexpression of Tin has a slightly stronger negative effect on the Odd-positive pericardial cells (arrows in L); however, the reduction of Odd-positive cells in the lymph glands appears to be less strong (arrow in L) than that caused by Tup overexpression (arrowhead in K). (M) Co-overexpression of Tup and Tin results in a similar phenotype to that seen for overexpression of Tin alone. Arrows point to the absence of Odd-positive pericardial cells; the arrowhead points to Odd-positive lymph gland cells.
In summary, an early pan-mesodermal overexpression of UAS-tup does not result in a dramatic overspecification of cardiac cells. Nonetheless, Tup can promote Tin expression, whereas it has a negative effect on Odd-positive cells. These results provide initial clues that Tup regulates heart and hematopoietic organ development on a transcriptional level by acting as both an activator and a repressor, depending on the context.
DISCUSSION
The specification of a subset of mesodermal cells towards a cardiac fate requires well-orchestrated interactions of a plethora of factors. Drosophila is the model system of choice to decipher the complex transcriptional network that initiates and sustains a cardiac lineage. Our data place the LIM-homeodomain transcription factor tup as an essential component in the early transcriptional network that specifies cardiac mesoderm.
After the initially broad expression domain of Tin has become restricted to the dorsal mesodermal margin, we first see Tup expression in the cardiac mesoderm in ∼10 small clusters, which co-express Eve. Slightly later, Tup is present throughout the Tin-positive cardiac mesoderm and gene expression analyses in tupisl-1, tin346, pnrVX6 and Df(3L)DocA embryos demonstrate that all four factors are required to maintain each other's expression (Fig. 8). Additionally, analyses of cardiac gene expression in embryos that are transheterozygotic for tup and tin, pnr or Doc, showed that these factors interact genetically to specify heart cells.
Fig. 8.
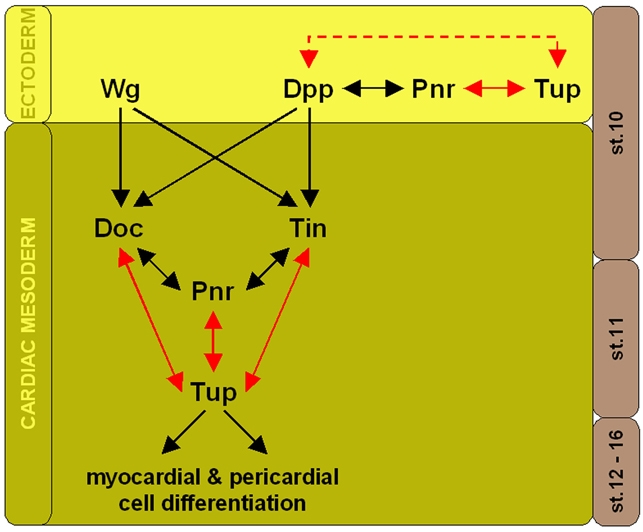
Tup as a new component of the Drosophila early cardiac transcriptional network. At stage 10, Tup is expressed in the ectoderm and is required for normal Pnr and dpp expression. Regulation of dpp expression through Tup may be direct or indirect (dashed line). Likewise, ectodermal Tup expression may be regulated by Dpp directly or indirectly through Pnr. After Wg and Dpp have induced a cardiac fate in the dorsal mesoderm by initiating and maintaining Doc and Tin expression, respectively, Pnr and Tup start to be expressed in the cardiac mesoderm by stage 11. All four factors are required to ensure proper cardiac specification of mesodermal cells. Black arrows indicate previously characterized interactions; red arrows indicate novel interactions with Tup as proposed in this study.
Although it might be expected that Tup expression is lost in tin mutants as these embryos are devoid of heart cells, it is interesting that Tup expression in the early cell clusters is still initiated. This finding is somewhat reminiscent of the observation that the initiation of Doc expression is also independent of tin (Reim and Frasch, 2005). According to the temporal appearance of Tup in the cardiac mesoderm with respect to Tin and Doc, tup is required for their maintenance rather than their initiation. By contrast, the onset of mesodermal Pnr and Tup expression appears to coincide (Klinedinst and Bodmer, 2003; Reim and Frasch, 2005). We did not resolve whether Tup is induced by Pnr or directly by Dpp. A direct regulation by Dpp was implicated by the reduced expression of Tup after mesodermal overexpression of UAS-brinker (data not shown), which is known to bind to dpp-response elements of dpp target genes (Kirkpatrick et al., 2001). Conversely, we show that dpp expression depends on tup and our present data suggest that this regulation requires pnr.
Germ layer-specific inhibition of Tup using a construct that lacks the homeodomain, but contains the two LIM domains, revealed that Tup can regulate cardiogenesis in the mesoderm as well as from the ectoderm. Since the 69B-Gal4 driver has been reported not to be strictly ectodermal (Klinedinst and Bodmer, 2003), it is possible that we also interfered with mesodermal Tup function. However, the mesodermal expression of 69B-Gal4 seems to be negligible (Baylies et al., 1995). The effect of ectodermal Tup inhibition on cardiogenesis in the mesoderm can only be explained if the function of a secreted growth factor is impaired. We have analyzed dpp expression and observed a slight downregulation of its transcripts in embryos expressing UAS-tupΔHD in the ectoderm. Since this effect might not be sufficient to account for the strong Tin phenotype, further experiments will be required to determine whether additional growth factors are affected.
To better determine the germ layer-specific contribution of Tup in cardiogenesis, we attempted to rescue the Tin phenotype by co-expressing the full-length tup cDNA. Somewhat unexpectedly, we obtained a better rescue when both constructs were expressed in the ectoderm rather than in the mesoderm. Since the LIM domains present in tupΔHD can sequester LIM-domain-binding proteins (O'Keefe et al., 1998), a simple explanation for this finding is that Tup interacts with proteins that are present in the mesoderm but not in the ectoderm. Based on the data published by O'Keefe et al. (O'Keefe et al., 1998), it is reasonable to hypothesize that in the mesoderm the LIM domains of tupΔHD not only act as a dominant-negative for Tup, but additionally for another, perhaps as yet unidentified, LIM-domain containing protein. Since it has been shown that Pnr can bind Tup through the LIM domains (Biryukova and Heitzler, 2005), we are likely to have interfered with Pnr function by overexpressing UAS-tupΔHD. The requirement of the LIM domains for proper cardiac specification is shown by the reduction of Tin-expressing cells after mesodermal expression of the UAS-tupΔLIM construct. Further experiments are under way to better resolve the molecular function of Tup in the different tissues.
Since the mesodermal expression of UAS-tupΔHD resulted in a strong reduction of Tin-expressing cells at early stages of cardiac mesoderm formation, it was surprising to observe a rather low reduction of Dmef2-positive myocardial cells at later stages (15/16). To exclude the possibility that the twi-Gal4 driver does not sufficiently express UAS-tupΔHD throughout embryogenesis, we repeated this experiment using the combined mesodermal driver twi-Gal4; 24B-Gal4. However, the phenotypes were not enhanced (data not shown). A time course for Tin expression in these crosses revealed that Tin appears to recover over time. A similar phenomenon can be seen in tupisl-1 mutants, although it might not be as obvious because the mutants also lack ectodermal tup expression. In any case, the data is suggestive of a different temporal requirement for tup with respect to tin expression. It is known that tin expression depends on different transcriptional activation events (Yin et al., 1997). Consistent with the onset of Tup expression in the cardiac mesoderm at mid-stage 11, the earlier phases of Tin expression are unlikely to depend on Tup. Hence, the initial Tin expression at stages 8-10 is sufficient to generate a considerable number of Dmef2-positive myocardial cells at later stages (Zaffran et al., 2006).
Our analyses further implicate that Tup might act as a transcriptional activator or repressor depending on the cellular context and on the factors with which it is co-expressed. This is most strikingly observed with respect to the Odd-expressing pericardial and lymph gland cells. In tup mutants, Odd-positive cells are missing in both organs (Tao et al., 2007) (this study). A similar phenotype is seen when Tup is overexpressed in the mesoderm using the twi-Gal4 driver. The loss of Odd-expressing cells in lymph glands is reminiscent of the phenotype observed in tup mutants, although it is less severe. This differential occurrence of the phenotype indicates that tup can differentially regulate factors involved in cardiogenesis versus lymph gland development. This is substantiated by the finding of Tao et al. (Tao et al., 2007), who showed that mesodermal overexpression of tup results in an increase in Hand expression in the lymph glands, while Hand expression throughout the dorsal vessel is only mildly affected. Despite the loss of Odd-positive cells after early mesodermal tup overexpression, Tup is required in the pericardial and lymph gland cells at later stages to maintain Odd expression. Moreover, overexpressing tup in the pericardial cell lineage yields additional Odd-expressing pericardial cells and rescues Odd expression in the lymph glands.
To obtain more insight into possible functional interactions with other cardiac transcription factors, we overexpressed tup in combination with pnrD4. The latter is a highly active variant of wild-type pnr that contains an amino acid substitution in the N-terminal zinc finger, which abolishes binding of Ush to Pnr (Haenlin et al., 1997). Mesodermal overexpression of pnrD4 results in robust ectopic activation of Tin (Klinedinst and Bodmer, 2003) and embryos co-overexpressing tup and pnrD4 exhibit the same phenotype. Most likely, a possible influence of Tup on Pnr activity, regardless of whether it is positive or negative, is concealed by the strong gain-of-function pnr allele. However, analysis of Eve expression does provide insight into possible regulatory interactions between Tup and Pnr. Mesodermal overexpression of each factor alone yields opposing phenotypes, and when both factors are co-overexpressed PnrD4 can efficiently counteract Tup activity and prevent the overspecification of Eve cells. Vice versa, Tup can, although only moderately, counteract the effect of PnrD4. It has been shown that during patterning of the thorax, Tup can antagonize the proneural activity of Pnr by forming a heterodimer, and that the physical interaction between Pnr and Tup is mediated by the two zinc fingers of Pnr (Biryukova and Heitzler, 2005). Hence, the somewhat weak, but possibly antagonistic, function of Tup towards PnrD4 in Eve-positive cell specification could be due to the amino acid substitution encoded in the pnrD4 allele, which might weaken the interaction between the two factors, as compared with wild-type Pnr. Overexpression of a Tup construct that lacks both LIM domains did not result in expanded Eve-positive clusters (data not shown), which strongly suggests that the effect of Pnr on Tup activity, as seen when both factors are co-expressed, requires the presence of the LIM domains.
In summary, our data demonstrate the crucial role of tup in the proper specification of cardiac mesoderm in an invertebrate organism. Therefore, tup/Isl1 should be added to the core set of ancestral cardiac transcription factors. Consequently, this implicates that the evolution of the vertebrate four-chambered heart does not necessarily require the acquisition of a novel network of cardiac transcription factors. At least, it is unlikely that tup/Isl1 is part of a regulatory network separate from that of tin/Nkx2.5, pnr/Gata4 and Doc/Tbx5/6 because it is an essential factor for the formation of the simple linear heart tube in the fly.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/cgi/content/full/136/2/317/DC1
Supplementary Material
We are grateful to M. Frasch, J. Skeath, G. Morata, B. Paterson, J. B. Thomas, S. Thor, I. Biryukova, P. Heitzler, the Bloomington Stock Center and the Developmental Studies Hybridoma Bank for providing fly stocks and antibodies. We also thank S. Liebau, H. Kestler and I. O. Sirbu for confocal microscopy, statistics and comments, respectively? This research was financed by a grant of the SFB 497, A8, to P.P. R.B. is supported by grants from NHLBI at NIH. Deposited in PMC for release after 12 months.
References
- Abu-Issa, R. and Kirby, M. L. (2007). Heart field: from mesoderm to heart tube. Annu. Rev. Cell Dev. Biol. 23, 45-68. [DOI] [PubMed] [Google Scholar]
- Alvarez, A. D., Shi, W., Wilson, B. A. and Skeath, J. B. (2003). pannier and pointedP2 act sequentially to regulate Drosophila heart development. Development 130, 3015-3026. [DOI] [PubMed] [Google Scholar]
- Anton, R., Kuhl, M. and Pandur, P. (2007). A molecular signature for the `master' heart cell. BioEssays 29, 422-426. [DOI] [PubMed] [Google Scholar]
- Azpiazu, N. and Frasch, M. (1993). tinman and bagpipe: two homeo box genes that determine cell fates in the dorsal mesoderm of Drosophila. Genes Dev. 7, 1325-1340. [DOI] [PubMed] [Google Scholar]
- Baylies, M. K., Martinez Arias, A. and Bate, M. (1995). wingless is required for the formation of a subset of muscle founder cells during Drosophila embryogenesis. Development 121, 3829-3837. [DOI] [PubMed] [Google Scholar]
- Biryukova, I. and Heitzler, P. (2005). The Drosophila LIM-homeo domain protein Islet antagonizes pro-neural cell specification in the peripheral nervous system. Dev. Biol. 288, 559-570. [DOI] [PubMed] [Google Scholar]
- Brade, T., Gessert, S., Kuhl, M. and Pandur, P. (2007). The amphibian second heart field: Xenopus islet-1 is required for cardiovascular development. Dev. Biol. 311, 297-310. [DOI] [PubMed] [Google Scholar]
- Brand, A. H. and Perrimon, N. (1993). Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development 118, 401-415. [DOI] [PubMed] [Google Scholar]
- Buckingham, M., Meilhac, S. and Zaffran, S. (2005). Building the mammalian heart from two sources of myocardial cells. Nat. Rev. Genet. 6, 826-835. [DOI] [PubMed] [Google Scholar]
- Cai, C. L., Liang, X., Shi, Y., Chu, P. H., Pfaff, S. L., Chen, J. and Evans, S. (2003). Isl1 identifies a cardiac progenitor population that proliferates prior to differentiation and contributes a majority of cells to the heart. Dev. Cell 5, 877-889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chartier, A., Zaffran, S., Astier, M., Semeriva, M. and Gratecos, D. (2002). Pericardin, a Drosophila type IV collagen-like protein is involved in the morphogenesis and maintenance of the heart epithelium during dorsal ectoderm closure. Development 129, 3241-3253. [DOI] [PubMed] [Google Scholar]
- Frasch, M. (1995). Induction of visceral and cardiac mesoderm by ectodermal Dpp in the early Drosophila embryo. Nature 374, 464-467. [DOI] [PubMed] [Google Scholar]
- Frasch, M., Hoey, T., Rushlow, C., Doyle, H. and Levine, M. (1987). Characterization and localization of the even-skipped protein of Drosophila. EMBO J. 6, 749-759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gajewski, K., Fossett, N., Molkentin, J. D. and Schulz, R. A. (1999). The zinc finger proteins Pannier and GATA4 function as cardiogenic factors in Drosophila. Development 126, 5679-5688. [DOI] [PubMed] [Google Scholar]
- Greig, S. and Akam, M. (1993). Homeotic genes autonomously specify one aspect of pattern in the Drosophila mesoderm. Nature 362, 630-632. [DOI] [PubMed] [Google Scholar]
- Haenlin, M., Cubadda, Y., Blondeau, F., Heitzler, P., Lutz, Y., Simpson, P. and Ramain, P. (1997). Transcriptional activity of pannier is regulated negatively by heterodimerization of the GATA DNA-binding domain with a cofactor encoded by the u-shaped gene of Drosophila. Genes Dev. 11, 3096-3108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herranz, H. and Morata, G. (2001). The functions of pannier during Drosophila embryogenesis. Development 128, 4837-4846. [DOI] [PubMed] [Google Scholar]
- Jagla, K., Frasch, M., Jagla, T., Dretzen, G., Bellard, F. and Bellard, M. (1997). ladybird, a new component of the cardiogenic pathway in Drosophila required for diversification of heart precursors. Development 124, 3471-3479. [DOI] [PubMed] [Google Scholar]
- Jagla, T., Bidet, Y., Da Ponte, J. P., Dastugue, B. and Jagla, K. (2002). Cross-repressive interactions of identity genes are essential for proper specification of cardiac and muscular fates in Drosophila. Development 129, 1037-1047. [DOI] [PubMed] [Google Scholar]
- Kadrmas, J. L. and Beckerle, M. C. (2004). The LIM domain: from the cytoskeleton to the nucleus. Nat. Rev. Mol. Cell Biol. 5, 920-931. [DOI] [PubMed] [Google Scholar]
- Kimbrell, D. A., Hice, C., Bolduc, C., Kleinhesselink, K. and Beckingham, K. (2002). The Dorothy enhancer has Tinman binding sites and drives hopscotch-induced tumor formation. Genesis 34, 23-28. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick, H., Johnson, K. and Laughon, A. (2001). Repression of dpp targets by binding of brinker to mad sites. J. Biol. Chem. 276, 18216-18222. [DOI] [PubMed] [Google Scholar]
- Klinedinst, S. L. and Bodmer, R. (2003). Gata factor Pannier is required to establish competence for heart progenitor formation. Development 130, 3027-3038. [DOI] [PubMed] [Google Scholar]
- Knirr, S., Azpiazu, N. and Frasch, M. (1999). The role of the NK-homeobox gene slouch (S59) in somatic muscle patterning. Development 126, 4525-4535. [DOI] [PubMed] [Google Scholar]
- Korzh, V., Edlund, T. and Thor, S. (1993). Zebrafish primary neurons initiate expression of the LIM homeodomain protein Isl-1 at the end of gastrulation. Development 118, 417-425. [DOI] [PubMed] [Google Scholar]
- Lee, H. H. and Frasch, M. (2000). Wingless effects mesoderm patterning and ectoderm segmentation events via induction of its downstream target sloppy paired. Development 127, 5497-5508. [DOI] [PubMed] [Google Scholar]
- Lilly, B., Zhao, B., Ranganayakulu, G., Paterson, B. M., Schulz, R. A. and Olson, E. N. (1995). Requirement of MADS domain transcription factor D-MEF2 for muscle formation in Drosophila. Science 267, 688-693. [DOI] [PubMed] [Google Scholar]
- Liu, J., Qian, L., Wessells, R. J., Bidet, Y., Jagla, K. and Bodmer, R. (2006). Hedgehog and RAS pathways cooperate in the anterior-posterior specification and positioning of cardiac progenitor cells. Dev. Biol. 290, 373-385. [DOI] [PubMed] [Google Scholar]
- Lo, P. C. and Frasch, M. (2001). A role for the COUP-TF-related gene seven-up in the diversification of cardioblast identities in the dorsal vessel of Drosophila. Mech. Dev. 104, 49-60. [DOI] [PubMed] [Google Scholar]
- Lockwood, W. K. and Bodmer, R. (2002). The patterns of wingless, decapentaplegic, and tinman position the Drosophila heart. Mech. Dev. 114, 13-26. [DOI] [PubMed] [Google Scholar]
- Miskolczi-McCallum, C. M., Scavetta, R. J., Svendsen, P. C., Soanes, K. H. and Brook, W. J. (2005). The Drosophila melanogaster T-box genes midline and H15 are conserved regulators of heart development. Dev. Biol. 278, 459-472. [DOI] [PubMed] [Google Scholar]
- O'Keefe, D. D., Thor, S. and Thomas, J. B. (1998). Function and specificity of LIM domains in Drosophila nervous system and wing development. Development 125, 3915-3923. [DOI] [PubMed] [Google Scholar]
- Park, M., Wu, X., Golden, K., Axelrod, J. D. and Bodmer, R. (1996). The wingless signaling pathway is directly involved in Drosophila heart development. Dev. Biol. 177, 104-116. [DOI] [PubMed] [Google Scholar]
- Pfaff, S. L., Mendelsohn, M., Stewart, C. L., Edlund, T. and Jessell, T. M. (1996). Requirement for LIM homeobox gene Isl1 in motor neuron generation reveals a motor neuron-dependent step in interneuron differentiation. Cell 84, 309-320. [DOI] [PubMed] [Google Scholar]
- Qian, L., Liu, J. and Bodmer, R. (2005). Neuromancer Tbx20-related genes (H15/midline) promote cell fate specification and morphogenesis of the Drosophila heart. Dev. Biol. 279, 509-524. [DOI] [PubMed] [Google Scholar]
- Qian, L., Liu, J. and Bodmer, R. (2008). Heart development in Drosophila. In Advances in Developmental Biology (ed. R. Bodmer), pp. 1-29. Amsterdam: Elsevier Publishing.
- Ranganayakulu, G., Elliott, D. A., Harvey, R. P. and Olson, E. N. (1998). Divergent roles for NK-2 class homeobox genes in cardiogenesis in flies and mice. Development 125, 3037-3048. [DOI] [PubMed] [Google Scholar]
- Reim, I. and Frasch, M. (2005). The Dorsocross T-box genes are key components of the regulatory network controlling early cardiogenesis in Drosophila. Development 132, 4911-4925. [DOI] [PubMed] [Google Scholar]
- Reim, I., Lee, H. H. and Frasch, M. (2003). The T-box-encoding Dorsocross genes function in amnioserosa development and the patterning of the dorsolateral germ band downstream of Dpp. Development 130, 3187-3204. [DOI] [PubMed] [Google Scholar]
- Reim, I., Mohler, J. P. and Frasch, M. (2005). Tbx20-related genes, mid and H15, are required for tinman expression, proper patterning, and normal differentiation of cardioblasts in Drosophila. Mech. Dev. 122, 1056-1069. [DOI] [PubMed] [Google Scholar]
- Riechmann, V., Irion, U., Wilson, R., Grosskortenhaus, R. and Leptin, M. (1997). Control of cell fates and segmentation in the Drosophila mesoderm. Development 124, 2915-2922. [DOI] [PubMed] [Google Scholar]
- Schmeichel, K. L. and Beckerle, M. C. (1994). The LIM domain is a modular protein-binding interface. Cell 79, 211-219. [DOI] [PubMed] [Google Scholar]
- Tao, Y., Wang, J., Tokusumi, T., Gajewski, K. and Schulz, R. A. (2007). Requirement of the LIM homeodomain transcription factor tailup for normal heart and hematopoietic organ formation in Drosophila melanogaster. Mol. Cell. Biol. 27, 3962-3969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thor, S. and Thomas, J. B. (1997). The Drosophila islet gene governs axon pathfinding and neurotransmitter identity. Neuron 18, 397-409. [DOI] [PubMed] [Google Scholar]
- Venkatesh, T. V., Park, M., Ocorr, K., Nemaceck, J., Golden, K., Wemple, M. and Bodmer, R. (2000). Cardiac enhancer activity of the homeobox gene tinman depends on CREB consensus binding sites in Drosophila. Genesis 26, 55-66. [PubMed] [Google Scholar]
- Ward, E. J. and Skeath, J. B. (2000). Characterization of a novel subset of cardiac cells and their progenitors in the Drosophila embryo. Development 127, 4959-4969. [DOI] [PubMed] [Google Scholar]
- Wu, X., Golden, K. and Bodmer, R. (1995). Heart development in Drosophila requires the segment polarity gene wingless. Dev. Biol. 169, 619-628. [DOI] [PubMed] [Google Scholar]
- Yin, Z., Xu, X. L. and Frasch, M. (1997). Regulation of the Twist target gene tinman by modular cis-regulatory elements during early mesoderm development. Development 124, 4971-4982. [DOI] [PubMed] [Google Scholar]
- Zaffran, S. and Frasch, M. (2002). Early signals in cardiac development. Circ. Res. 91, 457-469. [DOI] [PubMed] [Google Scholar]
- Zaffran, S., Reim, I., Qian, L., Lo, P. C., Bodmer, R. and Frasch, M. (2006). Cardioblast-intrinsic Tinman activity controls proper diversification and differentiation of myocardial cells in Drosophila. Development 133, 4073-4083. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.