Abstract
Marine photosynthetic microorganisms (phytoplankton) are the basis of marine foodwebs and are responsible for nearly 50% of the global annual carbon-based primary production.1 Phytoplankton can grow rapidly and form massive blooms that can be regulated by environmental factors such as nutrients and light availability and biotic interaction with grazers and viruses.2,3 Their crucial role in drawing down atmospheric CO2 and their potential use for future biofuel production4 raises the critical need for better understanding of fundamental features of their biology.5 Although traditionally phytoplankton were considered passive drifters with the currents (from Greek-“Planktos”), our recent reports demonstrate how cells employ a complex mechanism to sense changes in environmental cues and activate chemical-based defense strategies.
Key words: phytoplankton, diatoms, chemical signaling, infochemicals, nitric oxide, calcium, oxylipins, stress response
Diatoms are a highly diverse and abundant group of phytoplankton in the aquatic environment. As photosynthetic secondary endosymbionts, they are responsible for about 25% of global primary productivity,6 and play a central role in the biogeochemical cycling of important nutrients such as carbon, nitrogen and silica.
Their evolutionary and ecological success in contemporary oceans suggests they utilize sophisticated mechanisms to monitor and adapt appropriately to changes in environmental stress conditions.7 Only recently we have began to elucidate how chemical signals (e.g., infochemicals) derived from biotic interactions have the potential to shape complex community structures in aquatic systems.8,9
When diatom populations are subjected to grazing or nutrient stress, cells can rapidly induce the biosynthesis of diatom-derived unsaturated aldehydes (DD).10,11 These oxylipins, products of the fatty-acid oxidation pathway, can be derived both from plasma membrane-localized phospholipid and chloroplast membrane-localized glycolipid precursors.12,13 DD molecules have been implicated in diatom's chemical defense against competing species14 and as birth control of copepods, their principal grazers. Thus, these aldehydes may lead to compromised transfer of energy through the marine food web. They can also posses teratogenic and pro-apoptotic activities in human carcinoma cells.10
We recently demonstrated how diatoms can accurately sense a potent DD (2E,4E/Z-decadienal) and employ it as a signaling molecule to control diatom population sizes.15 This aldehyde triggered a dose-dependent calcium transient that has derived from intracellular store,15 in contrast to the external origin of cytosolic calcium induced in response to abiotic stress.7 Subsequently, Ca2+ increase led to nitric oxide (NO) generation by a calcium-dependent NO synthase-like activity,15 resulting in cell death in diatoms. Interestingly, cells pretreated with a sublethal dose of this aldehyde prior to application of lethal dose had far better survival and growth rates than cells treated with only a single high dose. These results suggest that signaling levels of DD may immunize cells, stimulating resistance to lethal DD concentrations. Perception of the immunized signal modulated calcium and NO signatures.
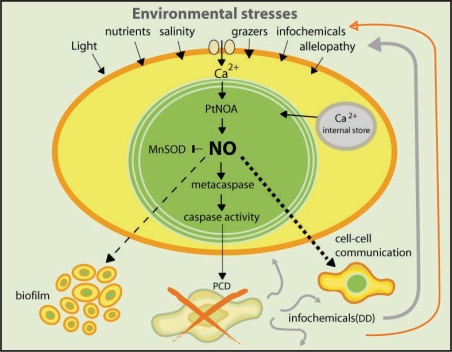
Furthermore, healthy diatom cells were able to utilize NO-based system to monitor stressed bystanders cells. Therefore, monitoring of this infochemical concentrations thereby provides a sophisticated stress surveillance system whereby subthreshold levels could serve as an early-warning protective mechanism, and lethal doses would initiate the cascade responsible for cell death and bloom termination (Fig. 1).15 Utilization of the gaseous NO molecule for regulating cell fate in marine phytoplankton provides a novel context for understanding cell signaling and diatom cell-cell communication during algal bloom succession.
Figure 1.
Chemical signaling and programmed cell death in diatoms. Diatom cells can perceive environmental stress conditions and chemical cues (infochemicals) by intracellular calcium transients. The source of calcium depends on nature of the environmental signal. The formation of infochemicals (e.g., diatom-derived aldehydes-DD) by stressed cells, regulate intercellular NO signalling, stress surveillance and defence against grazers. Accurate sensing of these aldehydes is mediated by Ca2+ and NO signaling. Integration of these signals in a spatio-temporal context would determine the fate of the cells and their adhesion to surfaces. Under low levels, (orange arrow) DD can act as a signal that immunizes cells against subsequent stress conditions while in high doses (gray arrow) it will induce PCD and initiate bloom termination. Nitric-oxide-associated protein (NOA) plays a key role in linking the initial perception of infochemical to cellular functions such as biofilm formation, cell-cell communication and cell death.
NO is widely used as a signaling molecule in vasodilation, neurotransmission and immune system responses in animals and in defense and hormonal signaling in plants and pathogenic bacteria.16,17 Nonetheless, the highly conserved arginine-dependent nitric oxide synthases (NOS)18 found in diverse animal and bacteria, can not be detected with significant homology in any genome from a photosynthetic organism reported to date.19
In order to dissect the signaling pathway involved in mediating sensing of this aldehyde to regulate diatom cell fate, we took a functional genomics approach to study the role of a DD-induced gene from the marine diatom Phaeodactylum tricornutum designated PtNOA1 (nitric oxide-associated protein). This protein belongs to the highly conserved YqeH subfamily of GTP-binding proteins, and is thought to play a role in ribosome biogenesis,20 sporulation and nitric oxide (NO) generation.21
Overexpressing this gene in P. tricornutum led to higher NO production, reduced growth and impaired photosynthetic efficiency compared to wild-type cells. Cells expressing PtNOA-YFP displayed localized signal within the plastid, differentiating it from a mitochondrial-localized plant orthologue.22,23 PtNOA transformants were hypersensitive to sublethal levels of this aldehyde, manifesting through altered expression of superoxide dismutase, metacaspases and caspase activity, key components of stress and cell death pathways (Fig. 1).23 The critical function of metacaspases was recently revealed in diatoms response to iron starvation and in the coccolithophore response to viral infection.24,25 Widespread documentation of programmed cell death (PCD) in diverse classes of phytoplankton in response to environmental stresses26 raises fundamental ecological and evolutionary questions for its origin and function in a population composed of unicellular cells. The occurrence of PCD in unicells can be explained when coupled to developmental changes (cyst/spore formation),27 when it acts as a selective mechanism in intra-species competition28 or as a viral exclusion strategy.25 Differential susceptibility to infochemicals may be a way to eliminate propagation of deleterious mutations in stressed-cells with high reactive oxygen species.27
Microbial mats and biofilm are important ecological niches where diatoms cells are likely to operate cell-cell chemical signaling. Changes in environmental conditions such as salinity and nutrients availability can shift diatom cells between pelagic and benthic forms to facilitate diatom biofilms. Adhesion of diatoms to surfaces, mediated by cell surface properties and secretion of extracellular polymeric substances (e.g., biofouling), is an important ecological strategy for growth and survival. Recent work in diatoms showed how cells can selectivity sense and adhere to surfaces via NO signaling.29 Consistent with this finding, PtNOA transformants were compromised in their ability to form biofilm by a reduced attachment strength compare to wild-type cells. This suggests NO plays an important role not only in diatom's perception of chemical cues but also in sensing optimal substrata and regulation of biofouling production.
We also detected NOA-like sequences from diverse oceanic samples, suggesting this novel NO-based system may be widespread among phytoplankton. These data also provide a biological context for NO production in the upper ocean, where it was suggested to originate solely from abiotic nitrite photolysis or bacterial (de) nitrification.30
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/6867
References
- 1.Field CB, Behrenfeld MJ, Randerson JT, Falkowski PG. Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science. 1998:237–240. doi: 10.1126/science.281.5374.237. [DOI] [PubMed] [Google Scholar]
- 2.Falkowski PG, Barber RT, Smetacek V. Biogeochemical Controls and Feedbacks on Ocean Primary Production. Science. 1998:200–206. doi: 10.1126/science.281.5374.200. [DOI] [PubMed] [Google Scholar]
- 3.Suttle CA. Marine viruses—major players in the global ecosystem. Nat Rev Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 4.Hu Q, Sommerfeld M, Jarvis E, Ghirardi M, Posewitz M, Seibert M, et al. Microalgal triacylglycerols as feedstocks for biofuel production: perspectives and advances. Plant J. 2008;54:621–639. doi: 10.1111/j.1365-313X.2008.03492.x. [DOI] [PubMed] [Google Scholar]
- 5.Armbrust EV, Berges JA, Bowler C, Green BR, Martinez D, Putnam NH, et al. The genome of the diatom Thalassiosira pseudonana: Ecology, evolution and metabolism. Science. 2004;306:79–86. doi: 10.1126/science.1101156. [DOI] [PubMed] [Google Scholar]
- 6.Nelson DM, Treguer P, Brzezinski MA, Leynaert A, Queguiner B. Production and Dissolution of Biogenic Silica in the Ocean—Revised Global Estimates, Comparison with Regional Data and Relationship to Biogenic Sedimentation. Global Biogeochem Cycles. 1995;9:359–372. [Google Scholar]
- 7.Falciatore A, d'Alcala MR, Croot P, Bowler C. Perception of environmental signal by a marine diatom. Science. 2000;288:2363–2366. doi: 10.1126/science.288.5475.2363. [DOI] [PubMed] [Google Scholar]
- 8.Long JD, Smalley GW, Barsby T, Anderson JT, Hay ME. Chemical cues induce consumer-specific defenses in a bloom-forming marine phytoplankton. Proc Natl Acad Sci USA. 2007;104:10512–10517. doi: 10.1073/pnas.0611600104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vardi A, Schatz D, Beeri K, Motro U, Sukenik A, Levine A, et al. Dinoflagellate-cyanobacterium communication may determine the composition of phytoplankton assemblage in a mesotrophic lake. Curr Biol. 2002;12:1767–1772. doi: 10.1016/s0960-9822(02)01217-4. [DOI] [PubMed] [Google Scholar]
- 10.Miralto A, Barone G, Romano G, Poulet SA, Ianora A, Russo GL, et al. The insidious effect of diatoms on copepod reproduction. Nature. 1999;402:173–176. [Google Scholar]
- 11.Ribalet F, Wichard T, Pohnert G, Ianora A, Miralto A, Casotti R. Age and nutrient limitation enhance polyunsaturated aldehyde production in marine diatoms. Phytochem. 2007;68:2059–2067. doi: 10.1016/j.phytochem.2007.05.012. [DOI] [PubMed] [Google Scholar]
- 12.Fontana A, d'Ippolito G, Cutignano A, Miralto A, Ianora A, Romano G, et al. Chemistry of oxylipin pathways in marine diatoms. Pure and Applied Chemistry. 2007;79:481–490. [Google Scholar]
- 13.Pohnert G. Phospholipase A(2) activity triggers the wound-activated chemical defense in the diatom Thalassiosira rotula. Plant Physiol. 2002;129:103–111. doi: 10.1104/pp.010974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Casotti R, Mazza S, Brunet C, Vantrepotte V, Ianora A, Miralto A. Growth inhibition and toxicity of the diatom aldehyde 2-trans, 4-trans-decadienal on Thalassiosira weissflogii (Bacillariophyceae) J Phycol. 2005;41:7–20. [Google Scholar]
- 15.Vardi A, Formiggini F, Casotti R, De Martino A, Ribalet F, Miralto A, et al. A stress surveillance system based on calcium and nitric oxide in marine diatoms. PLoS Biol. 2006;4:411–419. doi: 10.1371/journal.pbio.0040060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Singel DJ, Stamler JS. Chemical physiology of blood flow regulation by red blood cells: The role of nitric oxide and S-nitrosohemoglobin. Ann Rev Physiol. 2005;67:99–145. doi: 10.1146/annurev.physiol.67.060603.090918. [DOI] [PubMed] [Google Scholar]
- 17.Lamattina L, Garcia-Mata C, Graziano M, Pagnussat G. Nitric oxide: The versatility of an extensive signal molecule. Ann Rev Plant Biol. 2003;54:109–136. doi: 10.1146/annurev.arplant.54.031902.134752. [DOI] [PubMed] [Google Scholar]
- 18.Stuehr DJ. Mammalian nitric oxide synthases. Biochim Biophys Acta-Bioenerg. 1999;1411:217–230. doi: 10.1016/s0005-2728(99)00016-x. [DOI] [PubMed] [Google Scholar]
- 19.Zemojtel T, Frohlich A, Palmieri MC, Kolanczyk M, Mikula I, Wyrwicz LS, et al. Plant nitric oxide synthase: a never-ending story? Trends Plant Sci. 2006;11:524–525. doi: 10.1016/j.tplants.2006.09.008. [DOI] [PubMed] [Google Scholar]
- 20.Uicker WC, Schaefer L, Koenigsknecht M, Britton RA. The essential GTPase YqeH is required for proper ribosome assembly in Bacillus subtilis. J Bac. 2007;189:2926–2929. doi: 10.1128/JB.01654-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Guo FQ, Okamoto M, Crawford NM. Identification of a plant nitric oxide synthase gene involved in hormonal signaling. Science. 2003;302:100–103. doi: 10.1126/science.1086770. [DOI] [PubMed] [Google Scholar]
- 22.Guo FQ, Crawford NM. Arabidopsis nitric oxide synthase1 is targeted to mitochondria and protects against oxidative damage and dark-induced senescence. Plant Cell. 2005;17:3436–3450. doi: 10.1105/tpc.105.037770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vardi A, Bidle KD, Kwityn C, Hirsh DJ, Thompson SM, Callow JA, et al. A diatom gene regulating nitric-oxide signaling and susceptibility to diatom-derived aldehydes. Curr Biol. 2008;18:895–899. doi: 10.1016/j.cub.2008.05.037. [DOI] [PubMed] [Google Scholar]
- 24.Bidle KD, Bender SJ. Iron Starvation and Culture Age Activate Metacaspases and Programmed Cell Death in the Marine Diatom Thalassiosira pseudonana. Eukaryotic Cell. 2008;7:223–236. doi: 10.1128/EC.00296-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bidle KD, Haramaty L, Ramos JBE, Falkowski P. Viral activation and recruitment of metacaspases in the unicellular coccolithophore, Emiliania huxleyi. Proc Natl Acad Sci USA. 2007;104:6049–6054. doi: 10.1073/pnas.0701240104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bidle KD, Falkowski PG. Cell death in planktonic, photosynthetic microorganisms. Nat Rev Microbiol. 2004;2:643–655. doi: 10.1038/nrmicro956. [DOI] [PubMed] [Google Scholar]
- 27.Vardi A, Berman-Frank I, Rozenberg T, Hadas O, Kaplan A, Levine A. Programmed cell death of the dinoflagellate Peridinium gatunense is mediated by CO2 limitation and oxidative stress. Curr Biol. 1999;9:1061–1064. doi: 10.1016/s0960-9822(99)80459-x. [DOI] [PubMed] [Google Scholar]
- 28.Vardi A, Eisenstadt D, Murik O, Berman-Frank I, Zohary T, Levine A, et al. Synchronization of cell death in a dinoflagellate population is mediated by an excreted thiol protease. Environ Microbiol. 2007;9:360–369. doi: 10.1111/j.1462-2920.2006.01146.x. [DOI] [PubMed] [Google Scholar]
- 29.Thompson SM, Taylor AR, Brownlee C, Callow M E A CJ. The role of nitric oxide in diatom adhesion in relation to substratum properties. J Phycol. 2008;44:967–976. doi: 10.1111/j.1529-8817.2008.00531.x. [DOI] [PubMed] [Google Scholar]
- 30.Zafiriou OC, McFarland M, Bromund RH. Nitric-oxide in seawater. Science. 1980;207:637–639. doi: 10.1126/science.207.4431.637. [DOI] [PubMed] [Google Scholar]