Abstract
Cross-talk between salicylic acid (SA) and jasmonic acid (JA) defense signaling pathways allows a plant to finely tune its response to the attacker encountered. In Arabidopsis, pharmacological experiments revealed that SA exerts a strong antagonistic effect on JA-responsive genes, such as PDF1.2, indicating that the SA pathway can be prioritized over the JA pathway. We investigated the putative role of histone modifications in the regulation of SA-mediated suppression of PDF1.2 transcription. Chromatin immunoprecipitation analysis using an antibody directed against acetylated histone H3 revealed that SA does not affect the association of this histone modification at the PDF1.2 promoter, suggesting that chromatin remodeling does not play a major role in SA/JA cross-talk.
Key words: Arabidopsis, chromatin remodeling, cross-talk, jasmonic acid, plant immune response, salicylic acid
Plants can activate specific defense responses in order to resist attack by deleterious organisms. Apart from pre-existing chemical or structural barriers, inducible defenses are triggered, which often act systemically throughout the plant and confer broad-spectrum resistance. The plant hormones salicylic acid (SA), jasmonic acid (JA), and ethylene (ET) play important roles in the regulation of these induced defenses.1–7 The corresponding signal transduction pathways cross-communicate, providing the plant with a highly flexible defense signaling network. Cross-talk between signaling pathways is thought to optimize the defense reaction to a particular attacker by enhancing the appropriate response, while suppressing suboptimal reactions. Indeed, trade-offs between defense signaling pathways have been demonstrated in several plant species.8–10
Elucidation of the molecular mechanism underlying this pathway cross-talk will provide insight into how a plant copes with multiple stress inputs and relays these to an appropriate defense reaction.11,12 Several key regulatory proteins involved in SA-mediated suppression of JA-responsive genes have been identified in Arabidopsis thaliana.13 Previously, we demonstrated that the defense regulatory protein NPR1 is required for this SA/JA cross-talk.14 In mutant npr1-1 plants, the SA-mediated suppression of JA-responsive gene expression was completely abolished. The transcription factor WRKY70 was shown to act as an activator of SA-responsive genes and a repressor of JA-inducible genes, thereby functioning as a node of convergence between both pathways.15,16 Recently, overexpression of the SA-regulated glutaredoxin GRX480 was found to antagonize JA-responsive PDF1.2 transcription.17 Furthermore, a role for redox modulation was suggested in the SA/JA antagonism.12
To identify novel key players in SA/JA cross-talk we assessed whether chromatin remodeling plays a role in the SA-mediated down-regulation of JA-responsive genes. DNA is packaged with histone proteins into chromatin, which physically restricts the accessibility of the genome to regulatory proteins, such as transcription factors. The chromatin configuration can be altered to allow or prevent access of the transcription machinery by covalent modifications of the exposed N-terminal histone tails in the nucleosome. Histone acetylation is mediated by the activity of histone acetyltransferases (HATs) and is often associated with increased gene activity. Histone deacetylation, mediated by histone deacetylases (HDACs), and methylation are generally correlated with transcriptional repression.18,19 Thus, gene expression can be regulated at the level of histone modifications. Previously, a HDAC was found to interact with the JA regulatory protein COI1.20 The expression of this gene, as well as another HDAC, HDA19, was shown to be upregulated by JA and the ET precursor 1-aminocyclopropane-1-carboxylic acid (ACC).21 Overexpression of HDA19 enhanced expression of several JA- and ET-responsive genes and conferred increased resistance to the necrotrophic pathogen Alternaria brassicicola.21 Chromatin immunoprecipitation analysis demonstrated that induction of SA-responsive PR-1 gene expression is associated with an increase in histone acetylation at the PR-1 promoter in Arabidopsis and tobacco,22,23 suggesting involvement of histone modifications in the activation of SA-responsive genes too. Since both SA-responsive and JA-responsive defense signaling pathways involve chromatin remodeling,21–23 we investigated the potential influence of histone acetylation on the SA-mediated suppression of JA-responsive gene expression.
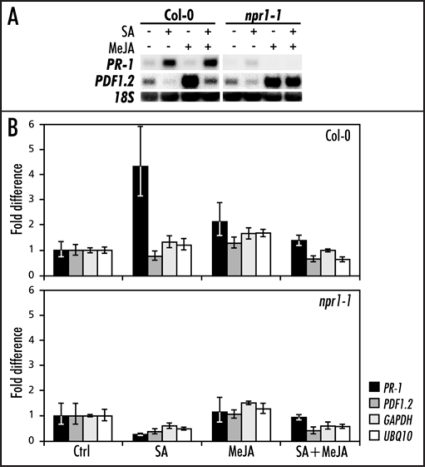
Five-week-old A. thaliana wild-type Col-0 and mutant npr1-1 plants were treated with an aqueous solution of 1 mM SA, 0.1 mM MeJA, or a combination of both chemicals as described previously.12 Leaf tissue was harvested 24 h after induction, and the expression of SA-responsive PR-1 (At2g14610) and MeJA-responsive PDF1.2 (At5g44420) marker genes was assessed. As expected, SA induced PR-1 expression and suppressed MeJA-induced expression of PDF1.2 in Col-0 plants (Fig. 1A). Both induction of PR-1 and suppression of PDF1.2 by SA was blocked in mutant npr1-1, confirming that the plant material displayed NPR1-dependent SA/JA cross-talk (Fig. 1A).14 This plant material was subjected to chromatin immunoprecipitation (ChIP) analysis,7 using an antibody directed against acetylated histone H3 (AcH3; 06-599; Upstate, Lake Placid, USA), which is a marker for a more transcriptionally permissive state.19,24 RT-Q-PCR analysis was performed on 1.5 µL immunoprecipitated and non-immunoprecipitated input DNA with primers designed to amplify fragments of the PR-1 promoter (PR-1 Fw; 5′ TCG GTC CCT AGA GTT TTT CAA 3′ and PR-1 Rv; 5′ CCG CCA CAT CTA TGA CGT AAG 3′) and the PDF1.2 promoter (PDF1.2 Fw; 5′ TTC AGT AAT AGG TGT GTC CCA GG 3′ and PDF1.2 Rv; 5′ GCG GCT GGT TAA TCT GAA TGG 3′). As a control, primer sets were included that amplify promoter fragments of two constitutively expressed marker genes, GAPDH (GAPDH Fw; 5′ GCA AAG CTC ATT GGC TGT CA 3′ and GAPDH Rv; 5′ GGA AAC TAA TGG CGC TTG GA 3′) and UBQ10 (UBQ10 Fw; 5′ TTG CCA ATT TTC AGC TCC AC 3′ and UBQ10 Rv; 5′ TGA CTC GTC GAC AAC CAC AA 3′).25 The amount of immunoprecipitated DNA was calculated relative to the input DNA using the 2-ΔΔCT method.26 The CT values of input and ChIP DNA were averaged before performing the ΔCT calculation, and the variance estimated from the replicate CT values was carried through to the final calculation of relative quantities using standard propagation of error methods. The error was estimated by calculating the 2-ΔΔCT term using ΔCT plus the standard deviation and ΔCT minus the standard deviation.26 Next, the amount of immunoprecipitated DNA was corrected for dilution factors, and control-treated samples of Col-0 and npr1-1 were set at 1.
Figure 1.
Chromatin immunoprecipitation (ChIP) analysis of histone H3 acetylation at the PR-1 and PDF1.2 promoters in Col-0 and npr1-1. (A) Northern blot analysis of PDF1.2 and PR-1 expression in Col-0 and npr1-1 leaves treated with 1 mM SA, 0.1 mM MeJA, or a combination of both chemicals. Leaf tissue was harvested 24 h after chemical treatment. Equal loading of RNA samples was checked using a probe for 18S rRNA. (B) ChIP analysis of Col-0 and npr1-1 treated with 1 mM SA, 0.1 mM MeJA, or a combination of both chemicals. Chromatin samples were subjected to immunoprecipitation using an AcH3 antibody. The immunoprecipitated DNA was analyzed for the enrichment of PR-1, PDF1.2, UBQ10 and GAPDH promoter sequences by Q-RT-PCR. The fold increase of immunoprecipitated DNA was calculated relative to the input DNA.
Figure 1B shows that SA treatment resulted in an increase of AcH3 at the PR-1 promoter, confirming previous findings with the SA analog benzothiadiazole S-methyl ester (BTH) as the inducing agent.22 This increase was absent in npr1-1, suggesting that SA-mediated acetylation of histone H3 at the PR-1 promoter is NPR1-dependent. However, the combined treatment with SA and MeJA did not result in AcH3 enrichment at the PR-1 promoter, while the PR-1 gene was normally expressed (Fig. 1A and B). Therefore, the chromatin structure around the PR-1 promoter appears to be sufficiently relaxed to allow access to the transcriptional machinery. This notion is supported by data on the epigenetic regulation of the WRKY70 transcription factor.27 It was suggested that the chromatin structure of WRKY70 target genes, such as PR-1, is sufficiently open to allow a fast response to stimuli. Consequently, by epigenetic regulation of a single transcription factor, the expression of a large number of genes can be affected, thus significantly increasing the regulatory potential of chromatin remodeling.27
In contrast to the PR-1 promoter, AcH3 association at the PDF1.2 promoter did not surmount the fluctuation observed at the promoters of the two constitutively expressed genes, GAPDH and UBQ10 (Fig. 1B). Hence, the negative effect of SA on MeJA-induced expression of PDF1.2 did not correlate with changes in AcH3 histone modification. These data suggest that chromatin remodeling does not play a major role in the regulation of SA/JA cross-talk. Conclusive evidence should be provided by additional experiments and include analysis with antibodies directed against multiple histone modifications.
Acknowledgements
The authors would like to thank Jurriaan Ton and Saskia Van Wees for useful discussions and help with the statistical analysis. This research was supported by grants 813.06.002 and 865.04.002 of the Earth and Life Sciences Foundation (ALW), which is subsidized by The Netherlands Organization of Scientific Research (NWO).
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/6997
References
- 1.Pozo MJ, Van Loon LC, Pieterse CMJ. Jasmonates—signals in plant-microbe interactions. J Plant Growth Regul. 2004;23:211–222. [Google Scholar]
- 2.Van Loon LC, Geraats BPJ, Linthorst HJM. Ethylene as a modulator of disease resistance in plants. Trends Plant Sci. 2006;11:184–191. doi: 10.1016/j.tplants.2006.02.005. [DOI] [PubMed] [Google Scholar]
- 3.Von Dahl CC, Baldwin IT. Deciphering the role of ethylene in plant-herbivore interactions. J Plant Growth Regul. 2007;26:201–209. [Google Scholar]
- 4.Grant M, Lamb C. Systemic immunity. Curr Opin Plant Biol. 2006;9:414–420. doi: 10.1016/j.pbi.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 5.Howe GA. Jasmonates as signals in the wound response. J Plant Growth Regul. 2004;23:223–237. [Google Scholar]
- 6.Vlot AC, Klessig DF, Park S-W. Systemic acquired resistance: the elusive signal(s) Curr Opin Plant Biol. 2008;11:436–442. doi: 10.1016/j.pbi.2008.05.003. [DOI] [PubMed] [Google Scholar]
- 7.Van Wees SCM, Van der Ent S, Pieterse CMJ. Plant immune responses triggered by beneficial microbes. Curr Opin Plant Biol. 2008;11:443–448. doi: 10.1016/j.pbi.2008.05.005. [DOI] [PubMed] [Google Scholar]
- 8.Bostock RM. Signal crosstalk and induced resistance: straddling the line between cost and benefit. Annu Rev Phytopathol. 2005;43:545–580. doi: 10.1146/annurev.phyto.41.052002.095505. [DOI] [PubMed] [Google Scholar]
- 9.Spoel SH, Dong X. Making sense of hormone crosstalk during plant Immune responses. Cell Host Microbe. 2008;3:348–351. doi: 10.1016/j.chom.2008.05.009. [DOI] [PubMed] [Google Scholar]
- 10.Pieterse CMJ, Dicke M. Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci. 2007;12:564–569. doi: 10.1016/j.tplants.2007.09.004. [DOI] [PubMed] [Google Scholar]
- 11.Koornneef A, Verhage A, Leon-Reyes A, Snetselaar R, Van Loon LC, Pieterse CMJ. Towards a reporter system to identify regulators of cross-talk between salicylate and jasmonate signaling pathways in Arabidopsis. Plant Signal Behav. 2008;3:543–546. doi: 10.4161/psb.3.8.6151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koornneef A, Leon-Reyes A, Ritsema T, Verhage A, Den Otter FC, Van Loon LC, Pieterse CMJ. Kinetics of salicylate-mediated suppression of jasmonate signaling reveal a role for redox modulation. Plant Physiol. 2008;147:1358–1368. doi: 10.1104/pp.108.121392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Koornneef A, Pieterse CMJ. Cross-talk in defense signaling. Plant Physiol. 2008;146:839–844. doi: 10.1104/pp.107.112029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Spoel SH, Koornneef A, Claessens SMC, Korzelius JP, Van Pelt JA, Mueller MJ, Buchala AJ, Métraux J-P, Brown R, Kazan K, Van Loon LC, Dong X, Pieterse CMJ. NPR1 modulates cross-talk between salicylate- and jasmonate-dependent defense pathways through a novel function in the cytosol. Plant Cell. 2003;15:760–770. doi: 10.1105/tpc.009159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li J, Brader G, Palva ET. The WRKY70 transcription factor: a node of convergence for jasmonate-mediated and salicylate-mediated signals in plant defense. Plant Cell. 2004;16:319–331. doi: 10.1105/tpc.016980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Li J, Brader G, Kariola T, Palva ET. WRKY70 modulates the selection of signaling pathways in plant defense. Plant J. 2006;46:477–491. doi: 10.1111/j.1365-313X.2006.02712.x. [DOI] [PubMed] [Google Scholar]
- 17.Ndamukong I, Abdallat AA, Thurow C, Fode B, Zander M, Weigel R, Gatz C. SA-inducible Arabidopsis glutaredoxin interacts with TGA factors and suppresses JA-responsive PDF1.2 transcription. Plant J. 2007;50:128–139. doi: 10.1111/j.1365-313X.2007.03039.x. [DOI] [PubMed] [Google Scholar]
- 18.Grant P. A tale of histone modifications. Genom Biol. 2001;2:1–6. doi: 10.1186/gb-2001-2-4-reviews0003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pfluger J, Wagner D. Histone modifications and dynamic regulation of genome accessibility in plants. Curr Opin Plant Biol. 2007;10:645–652. doi: 10.1016/j.pbi.2007.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Devoto A, Nieto-Rostro M, Xie DX, Ellis C, Harmston R, Patrick E, Davis J, Sherratt L, Coleman M, Turner JG. COI1 links jasmonate signalling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 2002;32:457–466. doi: 10.1046/j.1365-313x.2002.01432.x. [DOI] [PubMed] [Google Scholar]
- 21.Zhou C, Zhang L, Duan J, Miki B, Wu K. HISTONE DEACETYLASE19 is involved in jasmonic acid and ethylene signaling of pathogen response in Arabidopsis. Plant Cell. 2005;17:1196–1204. doi: 10.1105/tpc.104.028514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosher RA, Durrant WE, Wang D, Song J, Dong X. A comprehensive structure-function analysis of Arabidopsis SNI1 defines essential regions and transcriptional repressor activity. Plant Cell. 2006;18:1750–1765. doi: 10.1105/tpc.105.039677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Butterbrodt T, Thurow C, Gatz C. Chromatin immunoprecipitation analysis of the tobacco PR-1a- and the truncated CaMV 35S promoter reveals differences in salicylic acid-dependent TGA factor binding and histone acetylation. Plant Mol Biol. 2006;61:665–674. doi: 10.1007/s11103-006-0039-2. [DOI] [PubMed] [Google Scholar]
- 24.Strahl BD, Allis CD. The language of covalent histone modifications. Nature. 2000;403:41–45. doi: 10.1038/47412. [DOI] [PubMed] [Google Scholar]
- 25.Czechowski T, Stitt M, Altmann T, Udvardi MK, Scheible W-R. Genome-wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 2005;139:5–17. doi: 10.1104/pp.105.063743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2-ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 27.Alvarez-Venegas R, Al Abdallat A, Ming G, Alfano JR, Avramova Z. Epigenetic control of a transcription factor at the cross section of two antagonistic pathways. Epigenetics. 2007;2:106–117. doi: 10.4161/epi.2.2.4404. [DOI] [PubMed] [Google Scholar]