Abstract
Subcellular compartmentalisation and the intracellular movement of nuclear receptors are major regulatory steps in executing their transcriptional function. Though significant progress has been made in understanding these regulatory processes in cultured mammalian cells, such results have rarely been confirmed within cells of a living mammal. This article describes a simple, time-efficient approach to study the nuclear versus cytoplasmic accumulation of nuclear receptors and the regions of nuclear receptor proteins that govern subcellular trafficking within hepatocytes of live mice. Pregnane X receptor, a xenobiotic-activated member of the nuclear receptor family, was used to exemplify the approach. Using dual-labeled wild-type and mutant PXR expression constructs, we outline their in vivo delivery, simultaneous cellular expression, visualization and categorical classification within hepatocytes of live mice. Using this approach, we identified three mutants that had an altered subcellular distribution in the presence and absence of a PXR ligand. This novel in vivo method complements the current cell culture-based experimental systems in protein subcellular localisation studies.
Introduction
Intracellular compartmentalisation is a major regulatory step for the function of many proteins. This is especially true for ligand-activated transcription factors such as nuclear receptors (NR) that depend on nuclear localisation to exert transcriptional regulation of their target genes.
Analysis of the factors controlling the movement of nuclear receptors within cells, especially nuclear/cytoplasmic shuffling, is vital for understanding nuclear receptor action and is the subject of intense investigation [Griekspoor et al., 2007]. While cultured mammalian cells are commonly utilised to analyse the subcellular distribution of nuclear receptors and to dissect the structural features that govern their localisation, such results have rarely been confirmed in vivo within cells of a living mammal.
This article describes a simple, time-efficient approach to study the nuclear versus cytoplasmic accumulation of nuclear receptors and the regions of nuclear receptor proteins that govern subcellular trafficking within livers of intact mice. Using the pregnane X receptor (NR1I3; PXR), a xenobiotic-activated member of the nuclear receptor family [Matic et al., 2007] to exemplify the approach, we outline the in vivo delivery, simultaneous expression, visualization and categorical classification of bioengineered, dual-labeled wild-type and mutant PXR proteins within the same hepatocyte in livers isolated from mice following in vivo administration of expression constructs.
Reagents and instruments
pEYFP-C1 and pECFP-C1 vectors (ClonTech, BD Biosciences, Mountain View, CA); pGEM-T vector, EcoRI and XhoI restriction enzymes (Promega, Sydney, Australia); Tissue-Tek® O.C.T. compound (Sakyra Finetek, Torrence, CA); QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, CA); SuperScript III cDNA First-Strand Synthesis System, Prolong Gold mounting agent and Trizol reagent (Invitrogen, Mulgrave, Victoria, Australia); Hoechst 33258 (Sigma, New South Wales, Australia); DNase I (Ambion, Austin, TX, USA); pregnenolone 16α-carbonitrile [PCN; ICN Biomedicals, Aurora, Ohio]; GFP (FL): sc-8334 antibody (Santa-Cruz Biotechnology, Santa Cruz, CA, USA); Immunoprecipitation Kit [Protein G] (Roche, Indianapolis, IN, USA); Supersignal West Femto Maximum Sensitivity Substrate (Pierce Chemical Co., Rockford, ILL, USA).
Biorad mini-PROTEAN 3 cell system and Biorad TRANS-BLOT system (Biorad, Hercules, CA); Shandon cryotome E Cryostat (Thermo Fisher Scientific, Waltham, MA); Olympus BX51 Fluorescent microscope (Olympus, Victoria, Australia); Spot Advanced RT Software Version 3.4 (Diagnostic Instruments, Sterling Heights, MI); Leica DMIRE inverted microscope, Leica HCX Plan Apochromat CS 100X Oil Objective (NA 1.4), HC Plan Fluotar 20 X Ph2 objective (NA 0.5) and Leica TCS2-MP confocal imaging system (Leica Laserteknik, Mannheim, Germany); Argon ion laser and a Coherent Mira tuneable pulsed titanium sapphire laser (Coherent Laser Group, Santa Clara, CA).
Methods
Expression plasmids
The N-terminal Yellow Fluorescent Protein (YFP) tagged mPXR431 and mPXRΔ171-211 (YFP-mPXR431 and YFP-mPXRΔ171-211, respectively) mammalian expression constructs were generated by PCR amplification of mPXR431 and mPXRΔ171-211 cDNA fragments derived from total mouse liver RNA using the following primers: (F); 5-TATTCTCGAG CT(ATG)AGACCTGAGGAGAGC-3' XhoI restriction site, (Start codon) and (R); 5'-TGCGAATTCAGCC(ACT)CAGCCATCTGTGCT-3', (Stop codon), EcoRI restriction site. The primers incorporated a XhoI restriction site and a CT base insertion (to maintain an open reading frame) immediately upstream of the mPXR start codon. An EcoRI restriction site was incorporated downstream of the stop codon. The amplicon was ligated into the pGEM-T vector then digested using EcoRI and XhoI followed by sub-cloning into the EcoRI and XhoI sites of the pEYFP-C1 expression vector. The 46 amino acid C-terminal mPXR431 truncation mutant, YFP-mPXRL385X, was generated by PCR amplification using YFP-mPXR431 as template and the following primers: (F); 5-TATTCTCGAGCT(ATG)AGACCTGAGGAGAGC-3', XhoI restriction site, (Start codon) and (R); 5'-TCCGAATTC(TCA)GAACCTGTGAGCAGGATATGG-3', EcoRI restriction site, (Stop codon). An EcoRI and XhoI digest facilitated insertion of the amplicon into the respective pEYFP-C1 sites. The N-terminal Cyan FP (CFP) tagged mPXR431 (CFP-mPXR431) was generated by EcoRI and XhoI excision of mPXR431 fragment from YFP-mPXR431 and inserting it into the respective pECFP-C1 sites. The C-terminal CFP tagged mPXR431 (mPXR431-CFP) was generated by PCR amplification using YFP-mPXR431 as a template and the following primers; (F); 5-ATCTCGAG CGCCACC(ATG)AGACCTGAGGAGAGCTGG-3' XhoI restriction site, Kozak Translation Sequence, (Start codon) and (R); 5'-GGAATTC GGCCATCTGTGCTGCTAAATAACTCTTGC-3', EcoRI restriction site. These primers incorporated a Kozak translation sequence immediately upstream of the start codon to ensure efficient start of translation. Furthermore, they eliminated the stop codon by replacing it with a single G base, ensuring an open reading frame from mPXR431 into CFP. All other mutants were generated with the QuikChange site-directed mutagenesis kit, using pEYFPC1-mPXR431 or pEYFPC1-mPXRΔ171-211 as a starting template. All PXR expression constructs were verified by sequencing.
Expression of naked plasmid DNA in hepatocytes of a live mouse
Animal experimentation was performed on a protocol approved by Sydney South-west Area Health Service animal welfare committee. Plasmid DNA (pDNA) constructs were delivered into the livers of live mice (n=1-3) via hydrodynamic tail vein injection [Liu et al., 1999; Sueyoshi et al., 2002; Zhang et al., 1999]. Male wild-type FVB strain mice (10-14 weeks old; 25-30g) were anaesthetised with diethyl ether immediately prior to injection. Tail veins were dilated by immersion in 60°C water for ~1-2 min. Ten μg of each pDNA construct was mixed with sterile saline solution (0.9 %w/v) in a volume equivalent to 10% of the total mouse body weight. The solution was administered with a rapid (5-10s) but steady force into the tail vein of the mouse. Twenty-four hours post injection, the mice were culled, liver pieces excised and either snap frozen in liquid nitrogen or immediately embedded in Tissue-Tek® O.C.T. compound on dry ice. Liver tissue was stored at -70°C for subsequent analysis.
Histological processing and visualisation of PXR/fluorescent fusion proteins
Frozen liver sections were cryosectioned at 16μm using a Shandon cryotome E Cryostat, and every fourth consecutive section retained on a glass slide to ensure representation of different cellular regions. Retained sections were air-dried, fixed in ice-cold acetone for 5min then air-dried for a second time. Sections were counterstained with Hoechst 33258 (1μg/ml in PBS/5min), washed in Phosphate Buffered Solution (PBS: Sodium chloride, 145 mM (0.85%) in phosphate buffer (150 mM), air-dried and mounted with Prolong Gold. Samples were cured for 24h at 4°C away from light then sealed with clear nail polish.
Fluorescence was observed and captured (Figure 1) with the Olympus BX51 microscope. For all other studies, liver sections were observed by fluorescence microscopy using a Leica DMIRE inverted microscope and a Leica HCX Plan Apochromat CS 100X Oil Objective, NA 1.4 or a HC Plan Fluotar 20 X Ph2 objective, NA 0.5. Confocal images were then collected using a Leica TCS2-MP confocal imaging system equipped with an argon ion laser and a Coherent Mira tuneable pulsed titanium sapphire laser.
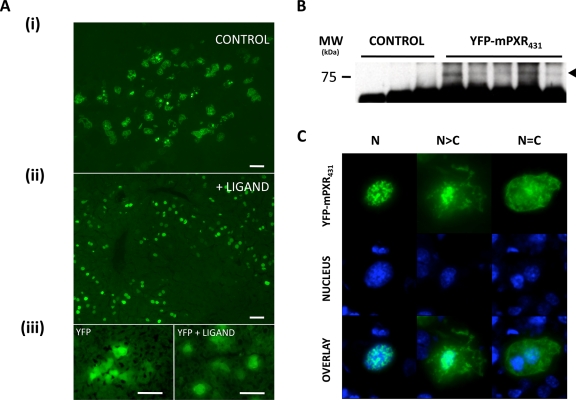
Figure 1. Expression of YFP-mPXR431 in the livers of intact mice.
A. Following the hydrodynamic injection of YFP-mPXR431 expression construct, fluorescence patterns were evident across liver sections of control (i) and ligand (PCN) (ii) treated mice that differed from those of YFP expression alone. B. Immunoprecipitation of YFP-mPXR431 from mouse liver samples indicated a protein band corresponding to the predicted molecular weight of the YFP/PXR fusion protein. C. Subcellular distribution patterns of YFP-mPXR431 expression were categorised into 3 distinct categories. An exclusively nuclear (N), where the fluorescence was confined to the nucleus, a predominantly nuclear (N>C), where a predominant nuclear localisation was evident with some cytoplasmic fluorescence, and an equal distribution (N=C), where nuclear and cytoplasmic fluorescence was evenly distributed across the cell. Nuclei are indicated by blue colour, following Hoechst 33250 staining.
Samples containing Hoechst 33342 were excited using λex = 780 nm (titanium sapphire laser) with pulses in the 100-200 fs range, and detected at λem = 420 to 450 nm. Samples containing CFP were excited using λex = 458 nm (Ar laser), and detected at λem = 470-490 nm. Samples containing YFP were excited using λex = 514 nm (Ar laser), and detected at λem = 530-550 nm. Images were acquired as 8 bit images in at least a 1024 X 1024 pixel array with a scan rate of 400 Hz and line averages were performed at 6-8 scans per image.
Ligand treatment
Five and 20 hours following delivery of PXR/fluorescent protein expression constructs, mice were administered the PXR-specific ligand pregnenolone 16α-carbonitrile (PCN; 80 mg/kg; ip), or dimethylsulfoxide vehicle.
cDNA synthesis
Total RNA was isolated from mouse liver using Trizol reagent and treated with DNase I according to the manufacturer’s protocols. Complementary DNA was synthesized from 5μg of total RNA with SuperScript III cDNA First-Strand Synthesis System using random hexamer primers.
Immunoprecipitation and immunodetection
Immunopurification of tissue-expressed fluorescent proteins was carried out using the GFP (FL): sc-8334 antibody and the Immunoprecipitation Kit [Protein G] according to the manufacturer’s instructions.
Equal volumes of immunoprecipitated YFP-mPXR431-transfected and control tissue samples were resolved on 10% SDS-PAGE using the Biorad mini-PROTEAN 3 cell system. Proteins were transferred to polyvinyldifluoridine (PVDF) membrane using the Biorad TRANS-BLOT system and detected using the GFP (FL): sc-8334 antibody and the Supersignal West Femto Maximum Sensitivity Substrate.
Evaluating the functionality of fluorescent protein recombinants in vivo
The functional validity of in vivo expressed PXR fusion proteins was carried out through confirmation of correct molecular weight using Western blot analysis, and evaluation of ligand-mediated in vivo nuclear localization. Additional validation could be carried out through correlation of target gene transcriptional readout in response to a ligand; however, such analysis needs to be applied in specific knockout mice in order to eliminate the effects of endogenously-expressed receptor.
Results
Establishing and evaluating the in vivo expression system
Following delivery of naked DNA constructs, in vivo hepatic expression of YFP-mPXR431 was visually confirmed by fluorescent microscopy (Figure 1A), and validated by Western analysis to be expressed at ~77kDa corresponding to the expected size of the mPXR/YFP fusion protein (Figure 1B). Hepatocytes of control mice exhibited a variable YFP-mPXR431 subcellular expression pattern spanning both the nuclear and cytoplasmic cellular compartments. To validate the functional integrity of YFP-mPXR431 within the in vivo environment of mouse livers, mice were treated with the mPXR-specific ligand, PCN. As expected, following ligand treatment the majority of hepatocytes exhibited an exclusively nuclear fluorescence pattern (Figure 1Aii), consistent with previous observations of nuclear receptor function following ligand activation [Griekspoor et al., 2007]. Therefore, the YFP-mPXR431 construct used in subsequent experiments could bind ligand and translocate to the nucleus of hepatocytes. Having validated and established the functional and structural integrity of YFP-mPXR431 and the in vivo hepatic expression experimental system, we sought to investigate regional mutations within the PXR protein that could influence its nuclear versus cytoplasmic accumulation.
Approach to the in vivo mutation analysis studies
Due to the extensive physiological factors that could potentially influence the behaviour of transfected mPXR in different hepatocytes of the same mouse, we chose to analyse the behaviour of a bioengineered mutant mPXR protein (MUT) with the wild-type mPXR counterpart (WT; mPXR431) within the same hepatocyte using dual labelling techniques. This approach required mutated and wild-type PXR proteins to be differentially tagged, enabling the direct comparison of their intracellular distribution patterns on a cell-by-cell basis. Thus, we chose to use CFP-mPXR431 and YFP-mPXRMUT for these comparative studies. CFP and YFP are organic fluorescent proteins derived from the Green FP, first identified in the jellyfish Aequorea victoria. YFP and CFP were originally engineered to obtain altered spectral emission properties from GFP, as well as each other. They differ by 4 and 6 amino acids (respectively) from the naturally-occurring GFP. Co-expression of CFP-mPXR431 or mPXR431-CFP together with YFP-mPXR431 revealed identical nuclear-cytoplasmic distributions (data not shown), thus ensuring that differences in fluorescent protein tag orientation at the N- or C-terminus of mPXR or the different amino acids in CFP and YFP did not impact on the subcellular distribution patterns. Once this was established, analysis of mutants proceeded.
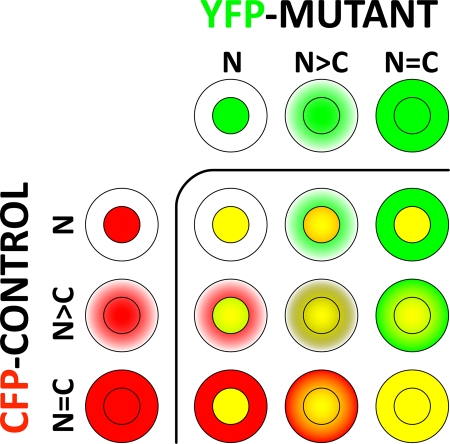
The subcellular distribution of co-expressed CFP-mPXR431/YFP-mPXRMUT proteins was observed and scored in at least 200 cells, encompassing a variety of WT mPXR subcellular distribution states categorised as exclusively nuclear (N); predominantly nuclear (N>C); and equal nuclear/cytoplasmic (N=C) (Figure 1C). These subcellular distribution states of wild-type PXR formed the basis of qualitative distribution reference categories (Figure 2) used to comparatively evaluate the localisation of mPXR mutant protein tagged with CFP within the same cell.
Figure 2. The categorical approach to comparatively investigate the subcellular distribution of co-expressed CFP-mPXR431 (CFP-Control; red colour) and YFP-mPXRMUT (YFP-Mutant; green colour) within the same hepatocyte of the liver.
The subcellular distribution patterns of each expression protein were individually categorised into a N, N>C or N=C category, then cross-referenced within the same hepatocyte to form one of 9 different possibilities representing degrees of differential localisation between the control and mutant protein. Yellow indicates overlapping distribution.
Mutant analysis
A range of C-terminal truncation, internal deletion and single or multiple point mutants of mPXR were bioengineered based on predictive analysis, previous studies with PXR [Kawana et al., 2003; Squires et al., 2004] and comparative homology with domains of other nuclear receptor proteins that determine subcellular distribution [Black et al., 2001; Hsieh et al., 1998; Zelko et al., 2001] (Figure 3).
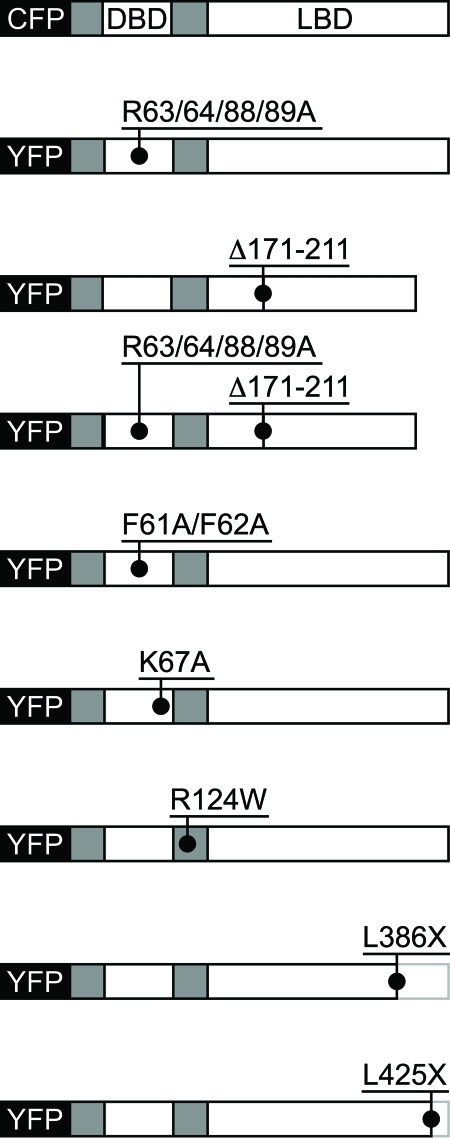
Figure 3. Schematic representation of YFP-mPXR mutants.
C-terminal truncations, internal deletions and single or combinatory point mutants were bioengineered to evaluate their impact on the subcellular distribution patterns of mouse PXR. CFP, Cyan Fluorescent Protein; YFP, Yellow Fluorescent Protein; DBD, DNA Binding Domain; LBD, Ligand Binding Domain. CFP-mPXR431 is the non-mutated wild-type form of murine PXR.
mPXRΔ171-211 is a functional, naturally-occurring mPXR variant that lacks 41 amino acids spanning the hinge/LBD interface [Kliewer et al., 1998]. Although it is a significant mPXR isoform that is conserved in humans [Lamba et al., 2004], the subcellular distribution of mPXRΔ171-211 had not been investigated. Truncations of the C-terminus (mPXRL385X and mPXRL425X) were designed to interfere with the function of xenochemical responsive signal (XRS) and/or the activation function 2 (AF2) domain reported to mediate the nuclear translocation of mPXR [Squires et al., 2004]. The mPXRL385X mutant eliminated 46 amino acids from the C-terminus of mPXR, encompassing the XRS and the AF2 domain, while the 7 amino acid truncation (mPXRL425X) was designed to disrupt AF2 function. The amino acid 61-62 phenylalanine to alanine mutations (i.e., mPXRF61/62A) within the DNA binding domain of mPXR were derived by sequence homology to the glucocorticoid receptor [Black et al., 2001] and the constitutive androstane receptor [Xia and Kemper, 2007], where they have been identified as being critical for nuclear export to the cytoplasm.
Quadruple arginine to alanine mutations incorporated in mPXRR63/64/88/89A and mPXRΔ171-211,R63/64/88/89A were reported to abolish nuclear localisation ability of PXR in addition to the lysine encompassed by the four arginines corresponding to position 67 of mPXR [Kawana et al., 2003; Squires et al., 2004]. Finally, a search for putative cytoplasmic to nuclear translocation sequences of mPXR using PredictNLS [Cokol et al., 2000] revealed a putative NLS encompassing amino acids 123KRKKREK129. The arginine at position 67 was point mutated to tryptophan to disrupt the NLS through steric hindrance. mPXR mutants were analysed in ligand-treated mice to study mPXR when fully activated by a xenobiotic ligand, as well as in control mice. Confocal microscopy with appropriate discriminatory filters for YFP and CFP (Figure 4) indicated three YFP-mPXR mutants had a substantial change in subcellular distribution patterns, exhibited as increased cytoplasmic presence when compared with the wild-type CFP-mPXR431 reference distributions within the same cell.
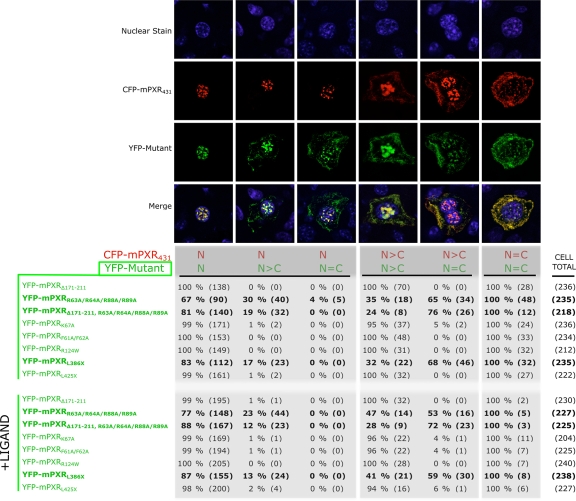
Figure 4. Altered subcellular distribution of YFP-tagged mutant PXR compared with CFP-tagged wild-type PXR.
Ranges of subcellular distribution categories were identified (see Figure 2) in a number of cells. Categories indicating decreased cytoplasmic presence of the mutant (with reference to the control) were not identified. Investigations were carried out in both control and ligand-treated mice. Categorical counts are represented as percentage of the total number of cells exhibiting a single CFP reference state (i.e., N, N>C or N=C) based on cell number. YFP-mPXRR63/64/88/89A, YFP-mPXRΔ171-211, R63/64/88/89A and YFP-mPXRL386X showed a substantial alteration in the subcellular distribution, indicative of impaired intracellular localisation ability.
The quad mutant YFP-mPXRR63A/R64A/R88A/R89A (which has 4 mutated arginine residues adjacent to the zinc fingers within the DNA binding domain of mPXR) accumulated in the cytoplasm of 34% (i.e., 30% N>C plus 4% N=C) of the same hepatocytes that displayed an exclusively nuclear (N) distribution of wild-type CFP-mPXR431. The effect of mutating these 4 residues also altered the distribution of YFP-mPXR protein in cells displaying N>C distribution of CFP-mPXR431, indicating 65% of these hepatocytes displayed an N=C accumulation of YFP-mPXRR63A/R64A/R88A/R89A. The same four mutations generated in the mPXR variant (mPXRΔ171-211 variant) retained an increased cytoplasmic presence of mPXRΔ171-211 in reference to the CFP-mPXR431 wild-type. YFP-mPXRΔ171-211, R63A/R64A/R88A/R89A was seen as N>C in 19% and as N=C in 76% of hepatocytes showing a N and a N>C distribution of CFP-mPXR431, respectively. The truncation mutant YFP-mPXRL386X, which lacks 25 amino acid residues at the C-terminus, was found to be distributed as N>C in 17% and N=C in 68% of hepatocytes that indicated the wild-type CFP-mPXR431 as N and N>C distribution, respectively. Examination of livers from mice dual transfected with other mutant PXR constructs did not detect an appreciable number of hepatocytes (<5%) exhibiting an altered distribution of mutant YFP-mPXR, as compared with wild-type CFP-mPXR431 proteins.
As expected, in mice treated with the potent mPXR-specific ligand PCN, the overall distribution of wild-type CFP-mPXR431 shifted to a more nuclear accumulation in most hepatocytes (data not shown). However, mutant YFP-mPXR proteins that displayed increased cytoplasmic accumulation in untreated mice also had altered distribution in the presence of PCN (i.e., N>C: YFP-mPXRR63A/R64A/R88A/R89A 23%, YFP-mPXRΔ171-211, R63A/R64A/R88A/R89A 12% and mPXRL386X 13% in hepatocytes indicating a N distribution of CFP-mPXR431; N=C: YFP-mPXRR63A/R64A/R88A/R89A 53%, YFP-mPXRΔ171-211, R63A/R64A/R88A/R89A 72% and mPXRL386X 59% in hepatocytes indicating a N> distribution of CFP-mPXR431). Therefore, within the same hepatocytes of mice livers with ligand-activated wild-type mPXR, these mutant forms of PXR distribute differently.
Discussion
Investigating the in vivo subcellular distribution of nuclear receptors (NR) is often seen as a daunting task. Frequently, immunohistochemical analysis is employed to analyse the distribution of endogenously-expressed NR, however this approach is highly dependent on the availability and/or specificity of the applied antibody. Even so, it is now becoming more evident that nuclear receptors can have multiple alternate forms (isoforms), which can be structurally identical for all but a few amino acids. These protein variants may have alternate subcellular distribution patterns and/or functional properties. As such, the existence of identical antibody epitopes shared between multiple nuclear receptor isoforms, some of which are yet to be discovered, may mask the subcellular distribution pattern of the targeted protein. Therefore, immunohistochemically-derived data needs to be critically assessed when applied to subcellular distribution studies.
Additional complications arise when in vivo analysis of mutant variants is desired. Such analysis generally requires targeted expression of mutant proteins via generation of transgenic mice or production of viral expression vectors, both of which are time consuming and laborious. These compounding factors usually preclude in vivo studies in favour of cell culture-based systems, which allow transfection and expression of isoform-specific nuclear receptor expression constructs and careful manipulation of the experimental environment. While these immortalised cell lines can be informative, they lack the complex physiological interactions between different cell types and tissues in the context of a whole organism. This is especially true for proteins such as nuclear receptors, whose action is modulated by endogenously-synthesised ligands potentially produced elsewhere within the body.
Our studies outline a simple, time-efficient approach to study the subcellular distribution of nuclear receptors and their protein variants in vivo. Using the hydrodynamic tail vein injection, we achieved efficient hepatic expression of fluorescently-labeled mPXR constructs (Figure 1A), that could be rapidly visualized, processed and analysed within 48hrs of initial injection time.
Initial observations (Figure 1) confirmed the structural and functional integrity of the expressed proteins and the in vivo experimental system. Consistent with the model of nuclear receptor action, ligand-treated mice showed YFP-mPXR431 proteins focused to the nucleus. Interestingly, the YFP-mPXR431 subcellular distribution within the same liver of control mice showed considerable heterogeneity between nuclear and cytoplasmic cellular compartments of hepatocytes. These observations, although novel, were not surprising and could potentially be attributed to i) known functional differences in liver zonation patterns and heterogeneous complexity of the liver as an organ [Malarkey et al., 2005], ii) the dynamic nature of subcellular nuclear-cytoplasmic shuttling [Kumar et al., 2006] and iii) the ligand promiscuity of PXR, which potentiates activation by a range of endogenous and exogenous compounds [Matic et al., 2007]. Facing such cell-to-cell variability in the subcellular distribution of YFP-mPXR431 within livers of mice, we adopted a cell-by-cell comparative approach, evaluating the mPXR protein (MUT) with the wild-type mPXR counterpart (WT; mPXR431) within the same hepatocytes. This ensured mutant variants were analysed under the same physiological conditions as the reference proteins. Pivotal to this approach was the use of differential but very closely related CFP and YFP organic tags fused to the reference or mutated PXR protein counterparts (Figure 3). As such, a 1:1 ratio of tag to protein was guaranteed and could be visualised at 100% specificity (for the expressed protein) by using appropriate fluorescent microscopy filters. The choice of the CFP and YFP protein tags has an additional benefit of making the expressed fusion proteins immediately amenable to protein interaction analysis via Fluorescent Energy Resonance Transfer [Griekspoor et al., 2007], thereby providing an additional level of study.
Utilising a categorical approach based on the variable distribution states of mPXR431 (Figure 2), we successfully identified three mPXR431 mutants with altered subcellular distribution behaviour in reference to the control mPXR431 (Figure 4). Interestingly, only a percentage of the total cells analysed for the altered distribution mutants indicated differential distribution compared with wild-type PXR, suggesting the in vivo distribution of PXR proteins is likely regulated by additional endogenous factors. Such observations emphasize the importance of physiological in vivo factors present in the whole organism, which are likely absent in cell culture-based systems.
In summary, we report an efficient and generally applicable technique to analyse protein function in an in vivo model. Utilising the CFP and YFP as differential protein tags, we simultaneously visualised and subsequently analysed multiple protein variants inside the same cell in an intact organ of a living organism. In addition, the same samples have the potential to be analysed for protein-protein interactions between the co-expressed CFP and YFP fusion proteins through the use of Fluorescent Energy Resonance Transfer. We also describe an innovative categorical approach applicable in the described in vivo system for studying the subcellular distribution of nuclear receptors. The described technique is not limited to studies of nuclear receptor subcellular distribution, but is applicable to many other proteins to conveniently analyse differential proteins, protein variants or bioengineered protein alterations within livers of intact animals. This quick and efficient in vivo approach complements current cell culture-based experimental systems to provide valuable in vivo observations of nuclear receptor protein function.
Acknowledgments
We would like to acknowledge the generous support of Prof. Chris Liddle and members of the Storr Liver Unit, Westmead Millennium Institute and the assistance of the Electron Microscopy Unit, University of Sydney. Arran Painter and Maria Tsoli for helping to restrain the mice. MM was the recipient of an Australian Postgraduate Award. This work was supported by NHMRC grant 352419 to GRR and the Oncology Trust Fund, Concord RG Hospital.
Abbreviations
- CFP
cyan fluorescent protein
- M
mouse
- PCN
pregnenolone 16α-carbonitrile
- PXR
pregnane X receptor
- YFP
yellow fluorescent protein
References
- Black B. E., Holaska J. M., Rastinejad F., Paschal B. M. DNA binding domains in diverse nuclear receptors function as nuclear export signals. Curr Biol. 2001;11:1749–58. doi: 10.1016/s0960-9822(01)00537-1. [DOI] [PubMed] [Google Scholar]
- Cokol M., Nair R., Rost B. Finding nuclear localization signals. EMBO Rep. 2000;1:411–5. doi: 10.1093/embo-reports/kvd092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griekspoor A., Zwart W., Neefjes J., Michalides R. Visualizing the action of steroid hormone receptors in living cells. Nucl Recept Signal. 2007;5:e003. doi: 10.1621/nrs.05003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh J. C., Shimizu Y., Minoshima S., Shimizu N., Haussler C. A., Jurutka P. W., Haussler M. R. Novel nuclear localization signal between the two DNA-binding zinc fingers in the human vitamin D receptor. J Cell Biochem. 1998;70:94–109. [PubMed] [Google Scholar]
- Kawana K., Ikuta T., Kobayashi Y., Gotoh O., Takeda K., Kawajiri K. Molecular mechanism of nuclear translocation of an orphan nuclear receptor, SXR. Mol Pharmacol. 2003;63:524–31. doi: 10.1124/mol.63.3.524. [DOI] [PubMed] [Google Scholar]
- Kliewer S. A., Moore J. T., Wade L., Staudinger J. L., Watson M. A., Jones S. A., McKee D. D., Oliver B. B., Willson T. M., Zetterstrom R. H., Perlmann T., Lehmann J. M. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Kumar S., Saradhi M., Chaturvedi N. K., Tyagi R. K. Intracellular localization and nucleocytoplasmic trafficking of steroid receptors: an overview. Mol Cell Endocrinol. 2006;246:147–56. doi: 10.1016/j.mce.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Lamba V., Yasuda K., Lamba J. K., Assem M., Davila J., Strom S., Schuetz E. G. PXR (NR1I2): splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol Appl Pharmacol. 2004;199:251–65. doi: 10.1016/j.taap.2003.12.027. [DOI] [PubMed] [Google Scholar]
- Liu F., Song Y., Liu D. Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 1999;6:1258–66. doi: 10.1038/sj.gt.3300947. [DOI] [PubMed] [Google Scholar]
- Malarkey D. E., Johnson K., Ryan L., Boorman G., Maronpot R. R. New insights into functional aspects of liver morphology. Toxicol Pathol. 2005;33:27–34. doi: 10.1080/01926230590881826. [DOI] [PubMed] [Google Scholar]
- Matic M., Mahns A., Tsoli M., Corradin A., Polly P., Robertson G. R. Pregnane X receptor: promiscuous regulator of detoxification pathways. Int J Biochem Cell Biol. 2007;39:478–83. doi: 10.1016/j.biocel.2006.08.017. [DOI] [PubMed] [Google Scholar]
- Squires E. J., Sueyoshi T., Negishi M. Cytoplasmic localization of pregnane X receptor and ligand-dependent nuclear translocation in mouse liver. J Biol Chem. 2004;279:49307–14. doi: 10.1074/jbc.M407281200. [DOI] [PubMed] [Google Scholar]
- Sueyoshi T., Moore R., Pascussi J. M., Negishi M. Direct expression of fluorescent protein-tagged nuclear receptor CAR in mouse liver. Methods Enzymol. 2002;357:205–13. doi: 10.1016/s0076-6879(02)57680-1. [DOI] [PubMed] [Google Scholar]
- Xia J., Kemper B. Subcellular trafficking signals of constitutive androstane receptor: evidence for a nuclear export signal in the DNA-binding domain. Drug Metab Dispos. 2007;35:1489–94. doi: 10.1124/dmd.107.016493. [DOI] [PubMed] [Google Scholar]
- Zelko I., Sueyoshi T., Kawamoto T., Moore R., Negishi M. The peptide near the C terminus regulates receptor CAR nuclear translocation induced by xenochemicals in mouse liver. Mol Cell Biol. 2001;21:2838–46. doi: 10.1128/MCB.21.8.2838-2846.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang G., Budker V., Wolff J. A. High levels of foreign gene expression in hepatocytes after tail vein injections of naked plasmid DNA. Hum Gene Ther. 1999;10:1735–7. doi: 10.1089/10430349950017734. [DOI] [PubMed] [Google Scholar]