Abstract
Cas ligand with multiple Src homology (SH) 3 domains (CMS) is an ubiquitously expressed signal transduction molecule that interacts with the focal adhesion protein p130Cas. CMS contains three SH3 in its NH2 terminus and proline-rich sequences in its center region. The latter sequences mediate the binding to the SH3 domains of p130Cas, Src-family kinases, p85 subunit of phosphatidylinositol 3-kinase, and Grb2. The COOH-terminal region contains putative actin binding sites and a coiled-coil domain that mediates homodimerization of CMS. CMS is a cytoplasmic protein that colocalizes with F-actin and p130Cas to membrane ruffles and leading edges of cells. Ectopic expression of CMS in COS-7 cells resulted in alteration in arrangement of the actin cytoskeleton. We observed a diffuse distribution of actin in small dots and less actin fiber formation. Altogether, these features suggest that CMS functions as a scaffolding molecule with a specialized role in regulation of the actin cytoskeleton.
Tumor cells are characterized by their acquired capacity of uncontrolled proliferation and an increased ability to migrate and invade healthy tissue. Transformation of normal mammalian cells into an invasive tumor results in alterations of cell matrix responses by integrins and loss of polarity (reviewed in refs. 1 and 2). Most integrins are transmembrane linkers, which mediate the interaction between the cytoskeleton and the extracellular matrix. Their activation leads to the assembly of signaling complexes, known as focal adhesions and focal contacts, and subsequently to the activation of intracellular signaling cascades (3, 4).
p130Cas, a docking protein that is tyrosine-phosphorylated in response to a variety of extracellular stimuli, such as growth factors, cell–cell interaction, and cell–matrix interaction, appears to play a critical role in the integrin-linked formation of focal complexes (5–8). In p130Cas-deficient mice, a prominent feature is heart failure caused by the disruption of the Z discs in cardiomyocytes (9). Z discs are structures similar to focal adhesions, and their role is to organize the actin cytoskeleton (10). Moreover, recent work has indicated a role for phosphorylated p130Cas in cell migration (11).
p130Cas is composed of an NH2-terminal Src homology (SH) 3 domain and regions with SH3 and SH2-binding motifs. It initially was identified as highly phosphorylated protein in v-src and v-crk transformed fibroblasts (12–14), and a role in transformation has been suggested (15). In these cells, the constitutive phosphorylation of p130Cas results in the permanent recruitment of SH2 containing signaling proteins into focal complexes (16, 17). The consequence is the constitutive propagation of downstream signals, resulting in changes in the cytoskeleton, induction of migration, and enhanced invasive potential of carcinoma cells (11, 18). Indeed, by using p130Cas (−/−) mouse embryonic fibroblasts, Honda et al. (9) demonstrated that expression of activated Src did not induce a fully transformed phenotype.
To understand the growth regulatory pathway of p130Cas we used the yeast-two hybrid system in search for interacting molecules (19). We identified CMS (p130Cas ligand with multiple SH3 domains) as a direct binding protein of p130Cas. CMS is a multifunctional adapter type molecule, which is localized to the cytoplasm, membrane ruffles, and leading edges of cells. Its structure and the colocalization with F-actin and p130Cas suggests a function as scaffolding protein involved in the dynamic regulation of the actin cytoskeleton.
MATERIALS AND METHODS
Cells and Reagents.
Human 293T kidney epithelial (293T) cells and COS-7 cells were grown in DMEM containing 10% FCS. mAbs and antisera used were: anti-glutathione S-transferase (GST), Santa Cruz Biotechnology: anti-flag (M2), Sigma; anti-p130Cas, anti-focal adhesion kinase (FAK), and anti-Csk, Transduction Laboratories, Lexington, KY; antiphosphotyrosine (4G10) and anti-Fyn, Upstate Biotechnology; anti-Abl, PharMingen; anti-Src (327), a gift from J. Brugge (Harvard Medical School, Boston); and anti-Yes and anti-CMS polyclonal antisera (raised against a peptide containing the third SH3 domain) (Covance, Denver, PA). Rhodamine phalloidin was purchased from Molecular Probes.
Yeast-Based Assays.
The yeast two-hybrid placenta library screen and the yeast two-hybrid interaction assay have been described (19). Briefly, the SFY526 yeast cells were cotransformed with the GAL4 DNA binding CMS/P-CC fusion construct and a CMS deletion construct fused to the GAL4 DNA activation domain, DL1, DL2, or DL3 and analyzed for interaction of the two transformed constructs in a β-galactosidase assay.
Isolation of Full-Length CMS cDNA.
A 5′ rapid amplification of complementary DNA ends was performed by using the Marathon cDNA Amplification Kit (CLONTECH). PCR products were cloned into the pCR-Blunt vector and subjected to DNA sequencing. Independent clones (1.5 × 106) derived from a human kidney cDNA library cloned in to the λgt10 vector (kindly provided by M. Stoffel, The Rockefeller University, New York) were screened. The CMS cDNA probe was labeled with [α-32P]dCTP (NEN) by using the readyprime labeling system (Amersham Pharmacia). Membranes were hybridized overnight at 65°C in hybridization buffer (1% BSA/7% SDS/1 mM EDTA/0.25 M Na2HPO4, pH 7.2/20 μg/ml salmon sperm DNA). The filters were washed three times at 65°C in washing buffer (0.1% SDS/1 mM EDTA/20 mM Na2HPO4, pH 7.2) for 10–15 min each. The cDNAs were subcloned in pBSK+(Stratagene) and subjected to sequencing.
Plasmid Construction.
Flag-tagged constructs were: full-length CMS, CMS/TH (TH, two-hybrid clone; encoding the cDNA fragment isolated in the yeast two-hybrid screen), and CMS/3SH3 (containing only the three SH3 domains) were generated by PCR and cloned in-frame into the pFLAG-CMV-2 vector. A truncated CMS construct, GST-CMS/P-CC, (lacking all three SH3 domains), was generated by standard PCR and cloned into the pEBG vector containing a GST tag. GST fusion protein constructs for the three SH3 domains of CMS were constructed as follows: The EcoRI DNA fragment encoding the first SH3 domain [CMS-SH3(1)] was cloned in-frame into the pGEX-4T-3 vector. The cDNA fragments encoding the second SH3 domain [CMS-SH3(2)] and the third SH3 domain [CMS-SH3(3)] of CMS were generated by standard PCR. The amplification products were cloned into the pGEX-2TK vector. The GST fusion protein construct for the SH3 domains of p130Cas has been described (19). The CMS deletion constructs DL1, DL2, and DL3 were cloned into pACT2 (CLONTECH), and CMS/P-CC was cloned into pAS2–1 (CLONTECH). All PCR products and all junctions were verified by DNA sequencing.
Analysis of RNA Expression.
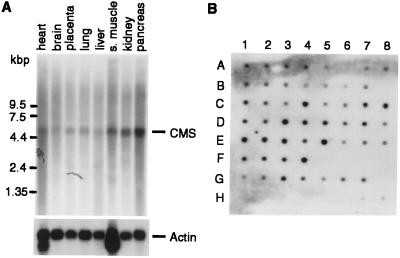
A Multi-Tissue Northern Blot and a Human RNA Master Blot (CLONTECH) were hybridized with a 32P-labeled cDNA probe of 1.1 kbp corresponding to the CMS clone isolated in the yeast two-hybrid screen (shown in Fig. 1) under high stringency conditions. The labeling method of the probe and the hybridization conditions were the same as described above. The blots were reprobed with β-actin.
Figure 1.
Schematic representation of the human CMS and protein sequence. (A) Alignment of human cDNA clones isolated in the yeast two-hybrid screen, 5′ rapid amplification of complementary DNA ends (RACE) and λgt11 cDNA library screen with respect to the full-length CMS cDNA. PR, proline-rich region. (B) Predicted protein sequence of CMS. Sequence numbers are shown on the left. The SH3 domains are underlined. Proline-rich sequences are marked in bold, the CC domain is marked in italics, and the putative actin binding sites are bold underlined. IP, immunoprecipitation. (C) In vitro interaction of p130Cas and CMS. 293T cells were transiently transfected with the flag-tagged CMS yeast TH clone (CMS-TH) together with the CasSH3 domain or full-length Cas (both GST tagged) or the vector alone (GST control). Cell lysates were immunoprecipitated with anti-flag antibody or glutathione Sepharose beads, and precipitates were subjected to SDS/PAGE and probed with anti-GST antibody and anti-flag antibody, respectively. The upper blot was striped and reprobed with anti-flag antibody. (D) Direct interaction of CMS with p130Cas. Five hundred micrograms of protein from 293T cells expressing the various CMS peptides or C3G used as a positive control was immunoprecipitated with the antibodies indicated above. Blots were probed for binding with 32P-labeled GST-p130CasSH3 domain.
Transfection, Western Blotting (WB), in Vitro Binding Assay, and Far WB.
293T cells were transiently transfected with 3 μg of cDNA per 60-mm dish or 6 μg cDNA per 100-mm dish by using the modified calcium phosphate transfection system (Stratagene). Thirty-six hours after transfection, the cells were washed once with cold PBS and lysed with lysis buffer. One milligram of total protein was used for protein association experiments. Cell lysis, immunoprecipitation, and WB was performed as described (19). Expression and affinity purification of GST-fusion peptides, in vitro binding assays, and Far WB were carried out as described (19).
Kinase Assay.
CMS was immunoprecipitated with anti-flag mAb, and precipitates were washed three times. The kinase reaction was performed for 30 min at 30°C in 30 μl of kinase buffer (30 mM Hepes, pH 7.4/10 mM MgCl2/1 mM DTT/25 mM ATP) containing 10 μCi of [γ-32P]ATP. The reaction was terminated by adding 5 μl of sample buffer and boiled for 5 min, and the samples were separated by SDS/PAGE. Proteins were transferred onto Immobilon-P transfer membrane and exposed to autoradiography films (Kodak).
Immunocytochemistry and Confocal Microscopy Analysis on Transiently Transfected Cells.
COS-7 cells (3 × 103) were plated on poly-d-lysine-coated coverslips (Biocoat Cellware, Becton Dickinson) overnight and transfected with 1 μg plasmid. If indicated, 24 h after transfection, the cells were serum-starved for 14–18 h. Cells plated on poly-d-lysine-coated coverslips were either left untreated or treated with 10 nM phorbol 12-myristate 13-acetate for 10 min. Cells were fixed with 3% formaldehyde in PBS for 30 min and permeabelized with 0.2% Triton X-100 in PBS for 5 min, and unspecific binding sites were blocked with 50 mM glycine in PBS for 10 min. Cells were stained with the mAb M2 (3 μg/ml) for CMS or with anti-p130Cas antibody (1:200 dilution). After four washes, cells were incubated with a mix of FITC-conjugated donkey anti-mouse IgG (1:300 dilution) and rhodamine-labeled phalloidin (Molecular Probes) (1:300 dilution) for visualizing actin filaments. Cells were inspected by confocal laser scanning microscopy using a Zeiss Axioplan2 instrument.
RESULTS
CMS Is a Protein Containing Domains Common to Adapter-Type Molecules.
A partial cDNA clone of CMS was isolated from a human placenta library in a yeast two-hybrid screen by using the SH3 domain of p130Cas as bait. Additional overlapping clones were isolated by 5′ rapid amplification of complementary DNA ends by using human placenta RNA as template or by Southern hybridization of a human kidney cDNA library cloned in λgt11. Conceptional translation revealed a protein of 639 aa with a deduced molecular mass of approximately 70 kDa (Fig. 1). The analysis of the amino acid sequence revealed that CMS contains in its NH2 terminus three SH3 domains followed by a proline-rich region containing binding sites for SH3 domains. Putative actin binding sites and a coiled-coil (CC) domain are located at the COOH terminus of the protein, which also contains a putative leucine zipper motif. The coils program (20) was used to predict the probability of a CC structure in CMS. Besides the described domains, the amino acid sequence shows no significant homology to other proteins listed in the available databases (data not shown), therefore all structural features indicate that CMS is a unique adapter-type molecule.
CMS and p130Cas Interact in Vitro and in Vivo.
To confirm the interaction of p130Cas with the TH clone of CMS, we analyzed its in vivo binding in mammalian cells by transiently coexpressing CMS and p130Cas in 293T cells. When we cotransfected the flag-tagged CMS/TH clone together with the GST-tagged SH3 domain of p130Cas or the full-length p130Cas, we were able to detect CMS in anti-GST immunoprecipitates of the p130Cas proteins (Fig. 1C). Conversely, we also could detect the SH3 domain of p130Cas and p130Cas full-length (after longer exposure) in anti-flag immunoprecipitates (Fig. 1C), indicating that the interaction of p130Cas and CMS takes place in mammalian cells in vivo. To determine whether the interaction of CMS and p130Cas was direct or mediated by other proteins, a Far WB analysis was carried out by using the 32P-labeled GST-p130CasSH3 domain as probe. As shown in Fig. 1D, the p130CasSH3 domain bound specifically to overexpressed flag-tagged CMS and CMS/TH, respectively as confirmed by probing with anti-flag antibody (not shown). The direct interaction of the p130CasSH3 domain with C3G was found previously (19) and used as positive control in this experiment.
CMS mRNA Is Ubiquitously Expressed in Adult and Fetal Human Tissues.
To assess the expression profile of CMS mRNA, we analyzed various adult and fetal human tissues by Northern blotting (Fig. 2). We detected a prominent transcript of ≈5.4 kb in all tissues examined. The CMS transcript is widely expressed in fetal and adult human tissues. In most of the cases there was no clear tissue-specific difference in expression levels. However, we noticed slightly higher expression levels (2-fold) in the placenta (F4), colon (C4), kidney (E1), pancreas (D1), and thymus (E5). In contrast, lower expression levels (50% reduction) were observed in the aorta (C2), skeletal muscle (C3), bladder (C5), and uterus (C6) with respect to their β-actin levels (not shown) (Fig. 2B).
Figure 2.
Tissue distribution of CMS mRNA. (A) Northern blot analysis of poly(A)+ RNA from adult human tissues by using the 32P-labeled CMS cDNA clone isolated in the two-hybrid screen. Hybridization analysis of the same Northern blot by using a 32P-labeled β-actin cDNA probe (Lower). (B) Relative expression levels of CMS in different human tissues and developmental stages. Adult tissue: A1, whole brain; A2, amygdala; A3, caudate nucleus; A4, cerebellum; A5, cerebral cortex; A6, frontal lobe; A7, hippocampus; A8, medulla oblongata; B1, occipital lobe; B2, putamen; B3, substantia nigra; B4, temporal lobe; B5, thalamus; B6, nucleus accumbeus; B7, spinal cord; C1, heart; C2, aorta; C3, skeletal muscle; C4, colon; C5, bladder; C6, uterus; C7, prostate; C8, stomach; D1, testis; D2, ovary, D3, pancreas, D4, pituitary gland; D5, adrenal gland; D6, thyroid gland; D7, salivary gland; D8, mammary gland; E1, kidney; E2, liver; E3, small intestine; E4, spleen; E5, thymus; E6, peripheral leukocyte; E7, lymph node; E8, bone marrow; F1, appendix; F2, lung; F3, trachea; F4, placenta. Fetal tissue: G1, brain; G2, heart; G3, kidney; G4, liver; G5, spleen; G6, thymus; G7, lung. Controls (100 ng each): H1, yeast total RNA; H2, yeast tRNA; H3, Escherichia coli rRNA; H4, E. coli DNA; H5, poly(rA); H6, human C0t 1 DNA; H7, human DNA; H8, human DNA (500 ng).
CMS Induces Vesicle Formation and Colocalizes with p130Cas and F-Actin to Membrane Ruffles.
Immunofluorescence staining and confocal microscopy of CMS-expressing cells were performed to analyze CMS’s intracellular localization and distribution. Because of the lack of a specific anti-CMS antibody, we examined the localization of flag-tagged CMS constructs transiently expressed in COS-7 cells. In randomly growing cells, CMS is localized in the cytoplasm and to the leading edges of these cells (Fig. 3). We observed this expression pattern also in resting, serum-starved cells and CMS-expressing cells plated on fibronectin-coated slides (data not shown). Interestingly, more than 80% of the CMS cells contained vesicles in various numbers and sizes, and the distribution of CMS in the cytoplasm was concentrated in small dots (Fig. 3B). CMS was found to be localized to the surface of these vesicle. In contrast, cells that expressed truncated forms of CMS consisting exclusively of the three SH3 domains (CMS/3SH3) or consisting of the CMS-TH fragment (lacking the first 279 aa) did not induce vesicle formation. Moreover, both truncated peptides were more diffusely distributed in the cytoplasm. These results suggested that the staining pattern in cells expressing full-length CMS was specific and that both the NH2 and COOH terminus are necessary for the vesicle formation and sublocalization of CMS. In addition, we noticed less actin fiber formation in randomly growing CMS-expressing cells. The staining of F-actin in CMS expressing cells appeared to be brighter compared with untransfected cells, and in transfected cells the actin was concentrated in small dots similarly to the staining pattern observed for CMS. Frequently, actin clustering adjacent to the nucleus also could be observed in CMS cells.
Figure 3.
CMS is localized in the cytoplasm and colocalizes with actin and p130Cas to membrane ruffles. (A and B) COS-7 cells grown in medium containing 10% FBS, plated on poly-d-lysine coverslips, were transfected with the flag-tagged CMS constructs CMS, CMS/TH, and CMS/3SH3 as indicated. Cells were fixed and stained with rhodamine-labeled phalloidin (lane 1) or with anti-flag antibody (lanes 3 and 4). Merged images are shown in lane 2. Actin clustering adjacent to the nucleus (B, lane 1 marked with an arrow). Vesicles in CMS cells are indicated by arrows (B, lane 3). (C and D) Membrane ruffle induction in COS-7 cells transfected with flag-tagged CMS or GST-tagged p130Cas. Serum-starved cells were treated with 10 nM phorbol 12-myristate 13-acetate (PMA) for 10 min, fixed, and subjected to immunofluorescence analysis by using the anti-flag or p130Cas antibody (lane 3). Actin filaments were visualized by staining with rhodamine-labeled phalloidin (lane 1). The cells were analyzed by confocal microscopy (×250 and ×630). Yellow-orange color in merged image shows colocalization of CMS and p130Cas with actin (lane 2, marked with arrows). (Bar, 20 μm.)
CMS and p130Cas colocalize in Cos-7 cells expressing both molecules (not shown). To investigate whether CMS can translocate to membrane ruffles, serum-starved cells expressing full-length CMS or p130Cas were treated overnight with phorbol 12-myristate 13-acetate (PMA). After PMA treatment, cells showed prominent membrane ruffles that contained both CMS and p130Cas (Fig. 3 C and D). Taken together, these findings demonstrated that both CMS and p130Cas colocalize with actin in membrane ruffles and leading edges of cells.
CMS Associates and Is Phosphorylated by Tyrosine Kinases.
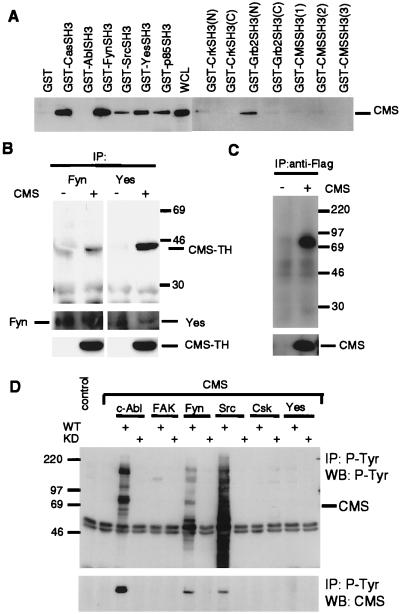
To further investigate the signaling potential of CMS, we analyzed the ability of selected SH3-containing proteins to bind to the COOH-terminal region of CMS, which includes the proline-rich region indicated in Fig. 1. This in vitro binding study clearly demonstrated that besides the SH3 domain of p130Cas, the SH3 domains of the tyrosine kinases Fyn, Src, and Yes, the SH3 domain of the p85 subunit of phosphatidylinositol 3-kinase, and the NH2-terminal SH3 domain of Grb2 bound to the adapter protein CMS. On the other hand, CMS proline-rich sequences did not bind to the SH3 domains of c-Abl, Crk, and the three SH3 domains of CMS (Fig. 4A). We also confirmed the binding of endogenous Fyn and Yes to the CMS-TH clone when it was overexpressed in 293T cells (Fig. 4B).
Figure 4.
Analysis of SH3-containing proteins for their interaction with CMS. (A) GST-fusion peptides coupled to glutathione Sepharose beads were incubated with lysates of 293T cells expressing flag-tagged CMS. Precipitates were analyzed by SDS/PAGE and WB with anti-flag antibody. (B) In vivo interaction of CMS with the tyrosine kinases Fyn and Yes. Lysates of 293T cells expressing flag-tagged CMS-TH or control vector were incubated with anti-Fyn and anti-Yes antibody coupled to protein A/G agarose beads. Immunoprecipitates (IP) were analyzed in SDS/PAGE and WB with anti-flag antibody. (C) Kinase activity is associated with CMS. Lysates of 293T cells expressing flag-tagged CMS or vector control were incubated with anti-flag antibody coupled to protein A/G agarose, and immunoprecipitates (IP) were subjected to a kinase assay. (D) Tyrosine phosphorylation of CMS. 293T cell lysates expressing flag-tagged CMS together with the indicated tyrosine kinases [wild type (WT) or kinase dead (KD)] were immunoprecipitated (IP) with antiphoshotyrosine antibody (4G10) and analyzed by WB with antiphosphotyrosine antibody and anti-CMS polyclonal serum. Total cell lysates were analyzed for expression of the transfected plasmids (not shown).
Based on this observation, we were interested in analyzing whether CMS is associated with endogenous kinase activity. A kinase assay performed on CMS immunoprecipitates resulted in a phosphorylated band of ≈80 kDa corresponding to the size of the flag-tagged CMS (Fig. 4C). To investigate whether CMS can serve as a substrate for tyrosine kinases, the tyrosine phosphorylation of CMS transiently coexpressed with c-Abl, FAK, Fyn, c-Src, Yes, or Csk was analyzed. As shown in Fig. 4D, CMS could be detected in anti-phosphotyrosine precipitates of CMS lysates of cells coexpressing c-Abl, Fyn, or c-Src. Conversely, tyrosine phosphorylation of CMS also could be seen in anti-flag immunoprecipitates corresponding to overexpressed CMS (data not shown). However, no tyrosine phosphorylation of CMS was observed in cells coexpressing FAK, Csk, or Yes.
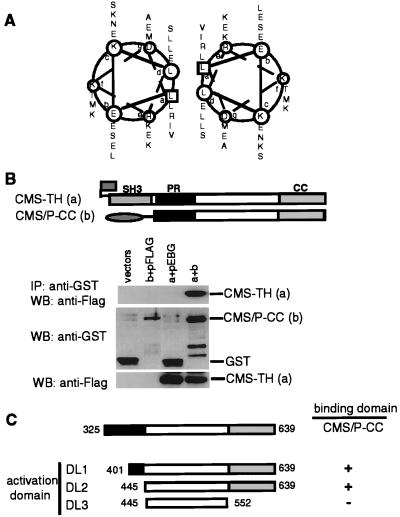
Homodimerization of CMS via Its Putative Leucine Zipper Motif.
As described above, the structural analysis of CMS revealed a CC domain in its COOH terminus containing a putative leucine zipper motif as shown by the helical wheel analysis in Fig. 5A. The analysis of the secondary structure using the ssp program (21) suggested that the region from Glu-607 to Ala-635 should be helical. Leucine zipper motifs have been implicated in homotypic and heterotypic protein–protein interactions. To evaluate the potential of the CMS COOH terminus to homodimerize, we tested whether truncated forms of CMS differently tagged interact in vivo. A GST-tagged construct, consisting only of the COOH terminal half of CMS (lacking all three SH3 domains, GST-CMS/P-CC), was coexpressed with the flag-tagged CMS-TH construct in 293T cells. The peptides were tested for complex formation in a GST pull-down assay. As shown in Fig. 5B (lane 4), the flag-tagged peptide coprecipitated with GST-CMS/P-CC and not with GST alone (lane 3).
Figure 5.
Dimerization of CMS is mediated through the COOH-terminal CC region. (A) Helical wheel representation of the putative leucine zipper in the CC region of CMS. (B) 293T cells were cotransfected with either empty vectors, GST vector, and flag-tagged CMS-TH (a), FLAG vector and GST-tagged CMS/P-CC (b), or CMS-TH and CMS/P-CC. Immunoprecipitates (IP) were analyzed by WB using anti-flag antibody (Upper). The expression levels were analyzed in total cell lysates by WB. (C) Mapping of the dimerization site in CMS. CMS target constructs for expression in yeast were introduced in vector pACT2 and transfected together with the bait CMS/P-CC cloned in pAS2–1. Transformants were grown on plates lacking leucine and tryptophane. Three single colonies were analyzed for interaction in a β-galactosidose filter assay (19). −, no interaction. +, interaction.
To further analyze the sequence requirements for CMS homodimerization, we assayed various CMS deletion constructs for their ability to interact in a yeast two-hybrid assay. The CMS deletion constructs (DL1, DL2, and DL3) were cloned into the yeast expression vector pACT2, expressed as GAL4 activation domain fusion peptide, and tested for binding the CMS/P-CC peptide expressed as GAL4 DNA-binding domain fusion peptides (Fig. 5C). We found homotypic interaction between the CMS/P-CC constructs and DL1 and DL2. In contrast, the construct DL3 that lacked the COOH terminus that contains the CC domain failed to interact with the CMS/P-CC construct. These results indicated that indeed CMS has the ability to homodimerize and for that it uses the CC domain in its COOH terminus. In contrast, we did not detect intramolecular or intermolecular binding via the SH3 domains and PXXP binding.
DISCUSSION
p130Cas, a family member of structurally related docking molecules (22–24), is expressed in a broad variety of tissues. By using the yeast two-hybrid system, we have identified a molecule, CMS, as potential p130Cas interacting protein. By using in vitro and in vivo studies, we clearly showed that p130Cas and CMS interact via binding between the SH3 domain of p130Cas and PXXP motifs of CMS, and that this interaction is direct. It has been shown that amino acids immediately adjacent to the proline residues are important for specificity (25, 26). Recently, we identified the PpK/RPp motif as consensus binding sequence for the SH3 domain of p130Cas (19). Located in the center part of CMS is a proline-rich region containing five core PXKP motifs that could serve as binding sites for the SH3 domain of p130Cas.
Human CMS is composed of 639 aa, and similarly to p130Cas, is ubiquitously expressed. In transiently transfected epithelial cells, we have demonstrated that CMS resides in the cytoplasm and accumulates in membrane ruffles and leading edges of migratory cells, where it colocalizes with F-actin. Furthermore, the most prominent phenotype observed in CMS-expressing cells was the accumulation of multiple large vesicles in the cytoplasm of CMS-expressing COS-7 cells. Interestingly, we found that both the NH2-teminal and the COOH-terminal region of CMS are necessary for the formation of vesicles and its dot-like distribution in the cytoplasm. This finding suggests that CMS acts as an adapter protein that forms larger complexes with other proteins. Indeed, CMS contains many potential protein binding sites, the most notable being the three adjacent SH3 domains in its NH2 terminus. Remarkably, all SH3 domains are most related to each other, to various myosins (especially the unconventional myosin 1B) (27), to the newly identified Rac target POSH (28), and to PIX, a p65PAK- interacting exchange factor (29). Interestingly, these are proteins that are structural or signaling components of the cytoskeleton. Among the SH3 domains of CMS, the highest identity (48.2%) was observed between the second and the third SH3 domain, suggesting that they may bind to the same or related ligands. While this report was in preparation, the mouse homologue of CMS, 86% identical and named CD2AP, has been reported as a protein that binds to the cytoplasmic tail of the adhesion molecule CD2 (30). The authors have shown that the interaction of the NH2-terminal SH3 domain with CD2 is necessary for CD2 clustering and cytoskeletal polarization of T cells. Because the expression of CMS is not restricted to immune cells, it is likely that CMS has different binding partners in other tissues. In addition, we showed that CMS contains a functional leucine zipper that can mediate its homodimerization. It is also possible that the leucine zipper of CMS binds to other proteins with higher affinity and thus contributes to the assembly of larger protein complexes as described for HEF, a p130Cas family member (23). Another indication that CMS can interact with a number of different proteins simultaneously is given by the consensus binding sites for SH3 domains located in its center region. We could show that, besides p130Cas, proteins with different functions, Src-family protein tyrosine kinases, the p85 subunit of the phosphatidylinositol 3-kinase, and the Grb2 adapter protein, bind to these sites.
The finding that p130Cas and CMS interact in vivo and their overlapping intracellular localization suggests that both molecules act in concert. Notably, in our experiments, p130Cas overexpression led to the accumulation of vesicles similarly to CMS (not shown). It has been shown that p130Cas contributes to the dynamic regulation of the actin cytoskeleton, inducing membrane ruffling, and positively regulating cell migration, which depends on the small G protein Rac (11). Cytoskeletal rearrangements and membrane ruffling have been linked to the activation state of Rac (31). In confluent cells activated Rac induced actin reorganization at the plasma membrane to form membrane ruffles, whereas in subconfluent cells it mostly induced the accumulation of large vesicles (31). The accumulation of large vesicles that we observed in CMS-expressing cells suggests that CMS could be a component of Rac signaling pathways.
The structure of CMS formed by proline-rich sequences that bind a large array of proteins and the presence of three SH3 domains suggest a function for CMS as scaffolding protein. Also, CMS contains at least four putative actin binding sites similar to the LKKTET motifs found in a number of actin binding proteins, including α-actinin, tropomyosin, villin, dematin, and actobindin (32), and in the small actin-sequestering protein thymosin β4 (33). Comparably, the yeast protein Bem1p, which contains two SH3 domains, appears to coordinate rearrangements of the cytoskeleton during cell polarization in response to mating factor and vegetative growth. It binds to Cdc24, a guanine nucleotide exchange factor for Cdc42, a close family member of Rac (34). Bem1p also has been shown to interact directly with Ste20, the yeast homologue of the serine/threonine kinase p65PAK as well as with actin, and it is thought to link membrane proteins to the cytoskeleton (35).
Several lines of evidence suggest that tyrosine phosphorylation is important in Rac signaling. Growth factors, such as platelet-derived growth factor, epidermal growth factor, and insulin, not only activate their receptor tyrosine kinases, they also rapidly induce Rac-dependent membrane ruffling and the tyrosine phosphorylation of p130Cas (5, 31, 36). It has been shown that Src-family kinases are responsible for the phoshorylation of proteins involved in the dynamic regulation of the cytoskeleton, including paxillin, vinculin, talin, and cortactin (37–40). Furthermore, the activation of c-Abl in response to integrin stimulation also has been demonstrated (41). Interestingly, we have demonstrated that CMS is a putative substrate for the tyrosine kinases Src, Fyn, and c-Abl. However, the physiological conditions under which CMS becomes tyrosine-phosphorylated have yet to be defined. We failed to induce tyrosine phosphorylation of CMS by treatment of 293T cells with epidermal growth factor or cell spreading on fibronectin, but we observed a moderate increase in tyrosine phosphorylation of CMS after 15-min treatment with cytochalasin D at concentrations that disrupt the cortical actin cytoskeleton (unpublished results).
In summary, we have identified CMS as a p130Cas binding molecule that may regulate the temporal and spatial assembly of larger signaling complexes and link membrane proteins to the actin cytoskeleton.
Acknowledgments
We are grateful to M. Sudol for GST-SrcSH3, GST-FynSH3, and GST-YesSH3, T. Shishido for GST-AblSH3, T. Ouchi for GST-p85SH3, A. Wong for GST-Grb2SH3, S. Hanks for FAK, B. Mayer for c-Abl, A Monteiro for c-Yes, T. Kurosaki for Fyn, and M. Matsuda for C3G. We also thank T. Qian for assistance with the confocal microscope. This work was supported by National Institutes of Health Grant CA44356, a fellowship from the Medical Research Council of Canada to M.-M.G., and a Norman and Rosita Winston Fellowship to S.I.
ABBREVIATIONS
- CMS
Cas ligand with multiple Src homology 3 domains
- SH
Src homology
- GST
glutathione S-transferase
- TH
two-hybrid
- CC
coiled-coil
- WB
Western blot
- FAK
focal adhesion kinase
Footnotes
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF146277).
References
- 1.Keely P, Parise L, Juliano R. Trends Cell Biol. 1998;8:101–106. doi: 10.1016/s0962-8924(97)01219-1. [DOI] [PubMed] [Google Scholar]
- 2.Clezardin P. Cell Mol Life Sci. 1998;54:541–548. doi: 10.1007/s000180050182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clark E A, Brugge J S. Science. 1995;268:233–239. doi: 10.1126/science.7716514. [DOI] [PubMed] [Google Scholar]
- 4.Howe A, Aplin A E, Alahari S K, Juliano R L. Curr Opin Cell Biol. 1998;10:220–231. doi: 10.1016/s0955-0674(98)80144-0. [DOI] [PubMed] [Google Scholar]
- 5.Ojaniemi M, Vuori K. J Biol Chem. 1997;272:25993–25998. doi: 10.1074/jbc.272.41.25993. [DOI] [PubMed] [Google Scholar]
- 6.Ingham R J, Krebs D L, Barbazuk S M, Turck C W, Hirai H, Matsuda M, Gold M R. J Biol Chem. 1996;271:32306–32314. doi: 10.1074/jbc.271.50.32306. [DOI] [PubMed] [Google Scholar]
- 7.Vuori K, Ruoslahti E. J Biol Chem. 1995;270:22259–22262. doi: 10.1074/jbc.270.38.22259. [DOI] [PubMed] [Google Scholar]
- 8.Nojima Y, Morino N, Mimura T, Hamasaki K, Furuya H, Sakai R, Sato T, Tachibana K, Morimoto C, Yazaki Y. J Biol Chem. 1995;270:15398–15402. doi: 10.1074/jbc.270.25.15398. [DOI] [PubMed] [Google Scholar]
- 9.Honda H, Oda H, Nakamoto T, Honda Z, Sakai R, Suzuki T, Saito T, Nakamura K, Nakao K, Ishikawa T, et al. Nat Genet. 1998;19:361–365. doi: 10.1038/1246. [DOI] [PubMed] [Google Scholar]
- 10.Burridge K, Fath K. BioEssays. 1989;10:104–108. doi: 10.1002/bies.950100403. [DOI] [PubMed] [Google Scholar]
- 11.Klemke R L, Leng J, Molander R, Brooks P C, Vuori K, Cheresh D A. J Cell Biol. 1998;140:961–972. doi: 10.1083/jcb.140.4.961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sakai R, Iwamatsu A, Hirano N, Ogawa S, Tanaka T, Mano H, Yazaki Y, Hirai H. EMBO J. 1994;13:3748–3756. doi: 10.1002/j.1460-2075.1994.tb06684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Matsuda M, Mayer B J, Fukui Y, Hanafusa H. Science. 1990;248:1537–1539. doi: 10.1126/science.1694307. [DOI] [PubMed] [Google Scholar]
- 14.Kanner S B, Reynolds A B, Wang H C, Vines R R, Parsons J T. EMBO J. 1991;10:1689–1698. doi: 10.1002/j.1460-2075.1991.tb07693.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Auvinen M, Paasinen-Sohns A, Hirai H, Andersson L C, Holtta E. Mol Cell Biol. 1995;15:6513–6525. doi: 10.1128/mcb.15.12.6513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burnham M R, Harte M T, Richardson A, Parsons J T, Bouton A H. Oncogene. 1996;12:2467–2472. [PubMed] [Google Scholar]
- 17.Nievers M G, Birge R B, Greulich H, Verkleij A J, Hanafusa H, van Bergen en Henegouwen P M. J Cell Sci. 1997;110:389–399. doi: 10.1242/jcs.110.3.389. [DOI] [PubMed] [Google Scholar]
- 18.Nakamoto T, Sakai R, Honda H, Ogawa S, Ueno H, Suzuki T, Aizawa S, Yazaki Y, Hirai H. Mol Cell Biol. 1997;17:3884–3897. doi: 10.1128/mcb.17.7.3884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kirsch K H, Georgescu M M, Hanafusa H. J Biol Chem. 1998;273:25673–25679. doi: 10.1074/jbc.273.40.25673. [DOI] [PubMed] [Google Scholar]
- 20.Lupas A, Van Dyke M, Stock J. Science. 1991;252:1162–1164. doi: 10.1126/science.252.5009.1162. [DOI] [PubMed] [Google Scholar]
- 21.Solovyev V V, Salamov A A. Comput Appl Biosci. 1994;10:661–669. doi: 10.1093/bioinformatics/10.6.661. [DOI] [PubMed] [Google Scholar]
- 22.Ishino M, Ohba T, Sasaki H, Sasaki T. Oncogene. 1995;11:2331–2338. [PubMed] [Google Scholar]
- 23.Law S F, Estojak J, Wang B, Mysliwiec T, Kruh G, Golemis E A. Mol Cell Biol. 1996;16:3327–3337. doi: 10.1128/mcb.16.7.3327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Alexandropoulos K, Baltimore D. Genes Dev. 1996;10:1341–1355. doi: 10.1101/gad.10.11.1341. [DOI] [PubMed] [Google Scholar]
- 25.Yu H, Chen J K, Feng S, Dalgarno D C, Brauer A W, Schreiber S L. Cell. 1994;76:933–945. doi: 10.1016/0092-8674(94)90367-0. [DOI] [PubMed] [Google Scholar]
- 26.Sparks A B, Rider J E, Hoffman N G, Fowlkes D M, Quillam L A, Kay B K. Proc Natl Acad Sci USA. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Vargas M, Voigt H, Sansonetti P, Guillen N. Mol Biochem Parasitol. 1997;86:61–73. [PubMed] [Google Scholar]
- 28.Tapon N, Nagata K, Lamarche N, Hall A. EMBO J. 1998;17:1395–1404. doi: 10.1093/emboj/17.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Manser E, Loo T H, Koh C G, Zhao Z S, Chen X Q, Tan L, Tan I, Leung T, Lim L. Mol Cell. 1998;1:183–192. doi: 10.1016/s1097-2765(00)80019-2. [DOI] [PubMed] [Google Scholar]
- 30.Dustin M L, Olszowy M W, Holdorf A D, Li J, Bromley S, Desai N, Widder P, Rosenberger F, van der Merwe P A, Allen P M, Shaw A S. Cell. 1998;94:667–677. doi: 10.1016/s0092-8674(00)81608-6. [DOI] [PubMed] [Google Scholar]
- 31.Ridley A J, Paterson H F, Johnston C L, Diekmann D, Hall A. Cell. 1992;70:401–410. doi: 10.1016/0092-8674(92)90164-8. [DOI] [PubMed] [Google Scholar]
- 32.Vandekerckhove J, Vancompernolle K. Curr Opin Cell Biol. 1992;4:36–42. doi: 10.1016/0955-0674(92)90056-i. [DOI] [PubMed] [Google Scholar]
- 33.Van Troys M, Dewitte D, Goethals M, Carlier M F, Vandekerckhove J, Ampe C. EMBO J. 1996;15:201–210. [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson J, Zheng Y, Bender L, Myers A, Cerione R, Bender A. J Cell Biol. 1994;127:1395–1406. doi: 10.1083/jcb.127.5.1395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chenevert J, Corrado K, Bender A, Pringle J, Herskowitz I. Nature (London) 1992;356:77–79. doi: 10.1038/356077a0. [DOI] [PubMed] [Google Scholar]
- 36.Ridley A J, Hall A. Cell. 1992;70:389–399. doi: 10.1016/0092-8674(92)90163-7. [DOI] [PubMed] [Google Scholar]
- 37.Thomas S M, Soriano P, Imamoto A. Nature (London) 1995;376:267–271. doi: 10.1038/376267a0. [DOI] [PubMed] [Google Scholar]
- 38.Sefton B M, Hunter T, Ball E H, Singer S J. Cell. 1981;24:165–174. doi: 10.1016/0092-8674(81)90512-2. [DOI] [PubMed] [Google Scholar]
- 39.Pasquale E B, Maher P A, Singer S J. Proc Natl Acad Sci USA. 1986;83:5507–5511. doi: 10.1073/pnas.83.15.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wu H, Reynolds A B, Kanner S B, Vines R R, Parsons J T. Mol Cell Biol. 1991;11:5113–5124. doi: 10.1128/mcb.11.10.5113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lewis J M, Baskaran R, Taagepera S, Schwartz M A, Wang J Y. Proc Natl Acad Sci USA. 1996;93:15174–15179. doi: 10.1073/pnas.93.26.15174. [DOI] [PMC free article] [PubMed] [Google Scholar]