Summary
Mitochondrial respiration in the African trypanosome undergoes dramatic developmental stage regulation. This requires co-ordinated control of components encoded by both the nuclear genome and the kinetoplast, the unusual mitochondrial genome of these parasites. As a model for understanding the co-ordination of these genomes, we have examined the regulation and mitochondrial import of a nuclear-encoded component of the cytochrome oxidase complex, cytochrome oxidase subunit VI (COXVI). By generating transgenic trypanosomes expressing intact or mutant forms of this protein, we demonstrate that COXVI is not imported using a conventional cleaved presequence and show that sequences at the N-terminus of the protein are necessary for correct mitochondrial sorting. Analyses of endogenous and transgenic COXVI mRNA and protein expression in parasites undergoing developmental stage differentiation demonstrates a temporal order of control involving regulation in the abundance of, first, mRNA and then protein. This represents the first dissection of the regulation and import of a nuclear-encoded protein into the cytochrome oxidase complex in these organisms, which were among the earliest eukaryotes to possess a mitochondrion.
Introduction
Proteins required for mitochondrial respiration are encoded by both nuclear and mitochondrial genomes. If respiratory activity is regulated in response to a changing cellular environment, then input from each of these genomes must also be regulated (Poyton and McEwen, 1996; Kwast et al., 1998). For nuclear-encoded components, which comprise ≈ 90% of mitochondrial proteins, this regulation may operate at the level of import, as proteins synthesized in the cytoplasm need to be transported and sorted appropriately to their correct location within the mitochondrion. The mechanisms that regulate the nuclear and mitochondrial genomes are therefore central to the understanding of the control of metabolic development.
For a number of reasons, the African trypanosome (Trypanosoma brucei spp.) is an interesting organism in which to investigate the control of mitochondrial development. First of all, trypanosomes are evolutionary ancient, being among the first eukaryotes to possess a mitochondrion (Sogin et al., 1986). Gene expression is also highly unusual in these organisms. Mitochondrial transcripts are post-transcriptionally modified by RNA editing, whereby functional mRNAs are generated by nucleotide addition or deletion, this being templated by short guide RNAs (Stuart et al., 1997). Nuclear genes, organized into polycistronic transcription units (Sutton and Boothroyd, 1986; Tschudi and Ullu, 1988), are devoid of introns (with one identified exception; Mair et al., 2000), and their mRNAs undergo ubiquitous and coupled trans-splicing/polyadenylation reactions (LeBowitz et al., 1993; Ullu et al., 1993; Matthews et al., 1994). Mitochondrial respiration is also highly regulated, being dependent upon life cycle stage. When living in the glucose-rich bloodstream of a mammalian host, the trypanosome mitochondrion is not fully developed and does not contain active Krebs cycle components or undergo oxidative phosphorylation (Priest and Hajduk, 1994a). However, upon differentiation to those forms that inhabit the midgut of their arthropod vector, the tsetse fly, these components become activated (Brown et al., 1973; Overath et al., 1986). Differentiation between these stages can also be performed in culture (Czichos et al., 1986; Matthews and Gull, 1994; Roditi et al., 1989; Ziegelbauer et al., 1990), and each can be subject to genetic manipulation. Thus, life cycle stage-regulated mitochondrial development can be studied in detail.
In yeast and mammalian systems, proteins destined for the mitochondrion typically bear extensive (≈20 amino acids or more) N-terminal signal sequences that are characterized by their relatively basic charge and their potential to form an amphipathic alpha-helix (Neupert, 1997). Recognition of the presequence by components of the mitochondrial inner membrane transport machinery directs the precursor to the mitochondrial matrix, where the signal peptide is removed. Thereafter, the protein may be retained in the matrix or processed further and inserted into the mitochondrial inner membrane (Haucke and Schatz, 1997). Proteins that lack cleaved presequences, such as ADP/ATP carrier protein (Smagula and Douglas, 1988), can also enter the mitochondrion, although the import signals are more complex (reviewed by Tokatlidis and Schatz, 1999). For example, proteins of the inner membrane may possess N-terminal signalling motifs similar to those of cleaved proteins, and these may act in concert with less well-defined signals in other regions of the polypeptide to direct import (Brix et al., 1999). Alternatively, poorly defined signals in the central and C-terminal portion of the protein govern correct mitochondrial sorting (e.g. Hahne et al., 1994; Folsch et al., 1996).
A survey of the N-terminal sequences of nuclear-encoded mitochondrial proteins in kinetoplastids has revealed that they commonly possess signal peptides of only seven to nine amino acids in length (Haüsler et al., 1997). Although unusually short, these signals are able to direct appropriate (although inefficient) mitochondrial localization when assayed in yeast and, conversely, yeast proteins can be correctly imported into trypanosomatid mitochondria (Hauser et al., 1996). A second class of mitochondrial proteins, represented by cytochrome C1, does not possess an obvious cleaved presequence, and it has been proposed that this protein is imported by a non-conservative pathway (Priest et al., 1993). Several levels of control appear to regulate the level of nuclear-encoded mitochondrial components in trypanosomatids, of which the most important seems to be mRNA abundance and protein stability/import (Torri and Hajduk, 1988; Torri et al., 1993; Priest and Hajduk, 1994b). Control at the level of transcription is relatively unimportant because of polycistronic organization of the trypanosome nuclear genome.
Being a key control point in respiratory activity (Capaldi, 1990), the cytochrome oxidase (COX) complex represents an excellent model to understand the regulation of mitochondrial development. The complex comprises > 10 subunits, of which three are encoded by the mitochondrial genome. In trypanosomes, this consists of a concatenated mass of DNA termed the kinetoplast. Different kinetoplast-encoded COX subunits display different levels of RNA editing, such that COX I is never edited during the life cycle, and COX II is edited in the procyclic form but not the bloodstream form, whereas COX III is massively edited in all stages (Benne et al., 1986; Feagin and Stuart, 1988; Feagin et al., 1988). A predicted nuclear-encoded component of this complex has also been identified recently from an analysis of transcripts differentially expressed between life cycle stages of T. brucei (Matthews and Gull, 1998). Specifically, one procyclic-enriched transcript was predicted to encode a protein with homology to the N-terminal peptide sequence of COXVI from Crithidia fasciculata (Speijer et al., 1996; 1997). This is a related kinetoplastid from which the cytochrome oxidase complex has been biochemically purified. A biophysical characterization of the location of COXVI in the cytochrome oxidase complex in mammals predicts that this component lies towards the outer face of the mitochondrial inner membrane (Tomizaki et al., 1999).
Here, we have investigated the development of the cytochrome oxidase complex during trypanosome differentiation by analysis of the regulated synthesis and import for COXVI. We demonstrate that COXVI lacks a cleaved presequence and show that the sequences at the N-terminus of the protein are required for correct mitochondrial localization. Finally, analyses of the expression of endogenous and transgenic COXVI mRNA and protein during differentiation to the procyclic form demonstrates a temporal order of control involving initial regulation in the abundance of mRNA and, later, protein. This represents a first dissection of the regulation and import of nuclear-encoded proteins into the cytochrome oxidase complex in these organisms.
Results
N-terminal processing of COXVI
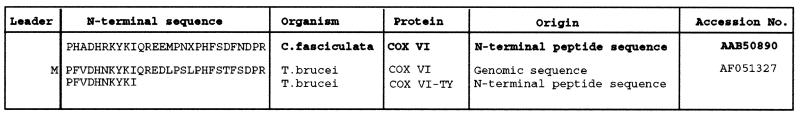
We have previously cloned a transcript encoding the putative COXVI gene of T. brucei (Matthews and Gull, 1998). The N-terminus of the protein predicted to be encoded by this gene demonstrated homology to the N-terminal peptide sequence of purified COXVI from C. fasciculata (Speijer et al., 1996). Analysis of the cDNA sequence for T. brucei COXVI revealed that its predicted methionine initiation codon was positioned one residue upstream of the mature N-terminus of the sequenced C. fasciculata protein. This suggested that T. brucei COXVI sequence might lack an extensive N-terminal cleaved signal sequence (Fig. 1).
Fig. 1. N-terminal sequence comparisons of COXVI.
A comparison between the N-terminal sequence of Crithidia fasciculata COXVI (determined by biochemical purification and amino acid sequencing; Speijer et al., 1996) and the predicted N-terminus of COXVI from T. brucei derived from gene sequencing of its cDNA (Matthews and Gull, 1998). The N-terminal peptide sequence derived by immunoprecipitation of transgenic COXVI in T. brucei COXVI-TY-C-transfected cells is also shown, confirming removal of the N-terminal methionine from the T. brucei COXVI protein.
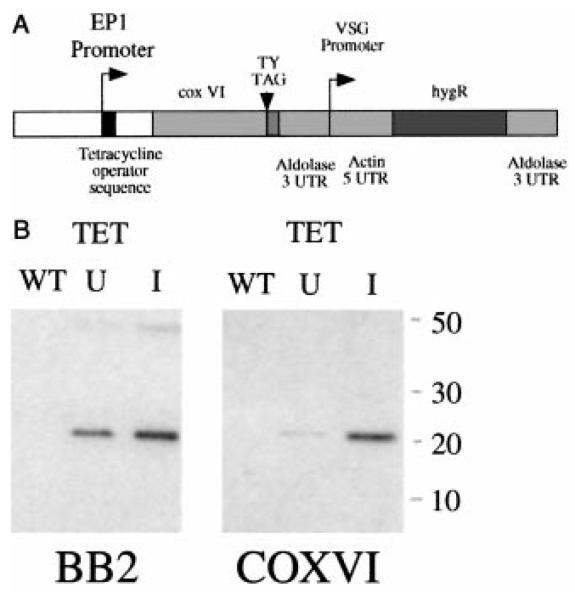
We generated constructs containing a copy of the COXVI gene modified by the incorporation of the sequence encoding a TY epitope tag (Bastin et al., 1996) into the predicted C-terminus of the protein (COXVI-TY-C; Fig. 2A). This was inserted into the expression vector pHD451 (Biebinger et al., 1997), such that the COXVI gene was downstream of an EP1 promoter modified by the incorporation of a tetracycline operator sequence (Wirtz and Clayton, 1995). In cell lines engineered to express the tetracycline repressor, this promoter is responsive to the presence (induced) or absence (uninduced) of tetracycline in the cell culture medium. The COXVI-TY-C construct was integrated into the ribosomal gene intergenic spacer region by transfection of T. brucei procyclic forms, and stably transfected cell lines were selected and cloned. These were then examined for expression of the tagged COXVI protein in the presence or absence of tetracycline by Western blotting with the anti-TY tag antibody BB2. Figure 2B demonstrates that the tagged COXVI protein was readily detected under conditions of tetracycline induction and, to a lesser extent, in the absence of induction. No tagged protein was detected in wild-type cells. The identity of the tagged protein was also verified by probing the same samples with an antibody generated against a COXVI fusion protein. Notice that this antibody does not detect the endogenous COXVI in these whole-cell extracts, although it does so in purified cell fractions (see Figs 4A and 5C). Nevertheless, the transgenic COXVI is clearly detected by this antibody in whole-cell extracts derived from cells grown in either the presence or the absence of tetracycline. This indicates that transgenic COXVI protein expression is somewhat leaky in terms of tetracycline repression and that it is significantly over-expressed (at least fivefold) with respect to wild-type levels of this protein.
Fig. 2. COXVI expression in transgenic procyclic trypanosomes.
A. The trypanosome expression vector pHD451 engineered to express the T. brucei COXVI gene. The gene has been modified by the incorporation of a TY epitope tag immediately before the COXVI stop codon, generating the construct COXVI-TY-C.
B. Expression of COXVI-TY-C in transgenic trypanosomes under the control of tetracycline. Left: a Western blot of either wild-type procyclic trypanosomes (WT) or cells transfected with COXVI-TY-C and cultured in either the absence (U) or the presence (I) of tetracycline. The blot was probed using the anti-tag antibody BB2. Notice that an additional band is detected at approximately twice the size of the COXVI protein when using the BB2 antibody. This band, which is of variable intensity in different experiments, may represent a dimer of transgenic COXVI. Right: the same samples probed with an antibody raised to a T. brucei COXVI fusion protein.
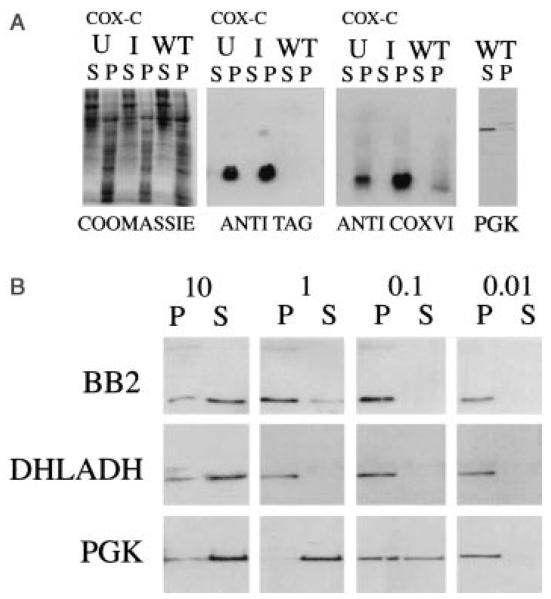
Fig. 4.
A. Cell fractionation of wild type (WT) or COXVI-TY-C (CoxC) expressing trypanosomes in either the presence (I, induced) or the absence (U, uninduced) of tetracycline. The soluble (S) and pellet (P) fractions from cells subjected to freeze–thaw lysis and high-speed centrifugation were probed with either the BB2 antibody or the anti-COXVI antibody. In each case, COXVI localizes to the pellet. A control cyotosolic protein, PGK (Osinga et al., 1985) is detected in the soluble fraction.
B. Crude pellet fractions derived from COXVI-TY-C cells induced with tetracycline were subjected to solubilization in digitonin at concentrations ranging from 0.01 to 10 mg of non-ionic detergent mg−1 trypanosome protein. Soluble (S) protein was separated from pelleted (P) material by centrifugation and blotted to nitrocellulose before being probed with antibodies to the TY epitope, DHLADH or the cytosolic protein PGK.
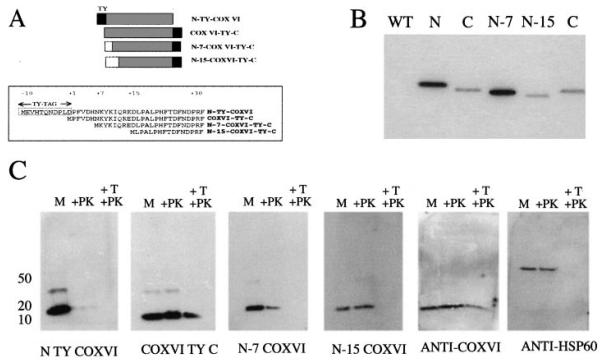
Fig. 5. Expression of COXVI mutant constructs and in vivo import of COXVI into the mitochondrion.
A. Mutations of COXVI. Three COXVI mutants were generated, which either have the TY tag inserted at the N-terminus of the protein (N-TY-COXVI) or have N-terminal deletions of either seven amino acids (N-7 COXVI TY-C) or 15 amino acids (N-15-COXVI TY C). The N-terminal sequence of each predicted protein is shown for each construct.
B. Expression of COXVI-tagged mutants in procyclic-form trypanosomes detected with the anti-TY tag antibody BB2. Notice that the N-tagged protein runs slightly larger than anticipated. The N-7 and N-15 truncations run at the expected size.
C. Mitochondrial localization of tagged and truncated forms of COXVI. Purified mitochondrial vesicles were analysed directly (M lane) or subjected to either proteinase K treatment (+PK) or Triton X-100 and proteinase K treatment (+T, +PK). The respective fractions were probed with the anti-tag antibody BB2, the anti-COXVI antibody or anti-HSP60.
To determine whether the mature N-terminus of the transgenic T. brucei COXVI protein matched that of the peptide sequence of C. fasciculata COXVI, the anti-tag antibody BB2 was used to immunoprecipitate the COXVI-TY-C protein from the induced transgenic cell line. Purified protein was then subjected to sequencing by Edman degradation, and the sequence of the 10 N-terminal amino acids was determined unambiguously. Alignment of this sequence with the predicted cDNA sequence of T. brucei COXVI and with the peptide sequence of COXVI from C. fasciculata demonstrated that the tagged protein lacked the initial methionine encoded in its gene sequence, but there was no removal of a more extensive signal presequence (Fig. 1). This confirmed directly that the COXVI protein does not carry a conventional N-terminal cleaved signal sequence and demonstrated that the abundant TY-tagged COXVI protein within the transgenic cell line could be processed appropriately by the removal of its initiator methionine. This processing would suggest that COXVI does not aggregate upon overexpression but is processed and compartmentalized within the cell.
Import of COXVI into the trypanosome mitochondrion
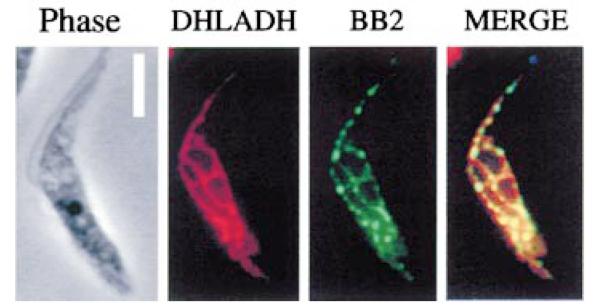
We carried out a number of studies to investigate the localization and import of COXVI into the T. brucei mitochondrion. Initially, an immunofluorescence assay was used to identify the transgenic protein in the cells induced to express COXVI-TY-C. These cells were treated by detergent permeabilization to provide access to mitochondrial proteins (Haüsler et al., 1997). When probed with the anti-tag antibody, BB2, the protein was localized to a discrete network-like structure in the cell, with a branched thread-like staining detectable. Using an antibody to the mitochondrial protein dihydrolipoamide dehydrogenase, we confirmed by co-localization that this pattern precisely matched the position of the trypanosome mitochondrion (Fig. 3). Moreover, the staining observed is consistent with the staining pattern of other mitochondrial proteins under similar fixation conditions (e.g. malate dehydrogenase; Anderson et al., 1998).
Fig. 3. The localization of COXVI in transgenic procyclic trypanosomes.
Immunofluorescence of cells induced to express transgenic COXVI by growth in tetracycline. Left (‘phase’): phase-contrast image of a representative cell; second left (‘DHLADH’): the same cell stained with a rabbit antibody to the mitochondrial protein dihydrolipoamide dehydrogenase. Second right (‘BB2’): the staining pattern using the mouse anti-TY tag antibody, BB2; right (‘merge’): two immunofluorescence images overlaid. The dihydrolipoamide dehydrogenase and transgenic COXVI proteins co-localize. Bar = 5 μm.
We also investigated the localization of COXVI-TY-C by a cell fractionation procedure, involving freeze–thaw cell lysis and high-speed centrifugation (Woods et al., 1989). This generates a soluble (cytosolic) fraction and a pellet derived from the parasite membrane and organelles. Figure 4A shows each cellular fraction derived from either wild-type procyclic-form trypanosomes or COXVI-TY-C cells grown in either the presence or the absence of tetracycline. In each case, abundant tagged protein was detectable only in the pellet fraction, a distribution that matched that of the endogenous COXVI protein detected by the anti-COXVI antibody, but differed from that of a cytosolic marker protein, phosphoglycerate kinase, (PGK; Osinga et al., 1985). Note that the endogenous COXVI protein in these extracts is smaller than the transgenic protein, this being caused by the inclusion of the epitope tag sequence into COXVI-TY-C. To determine that the overexpressed COXVI was not merely forming an insoluble aggregate during cell fractionation, cells expressing the transgenic COXVI were subjected to digitonin fractionation according to the protocol of Haüsler et al. (1997). Figure 4B shows that the majority of the COXVI-TY protein segregated with the soluble fraction at high concentrations of digitonin and with the pellet at lower concentrations. This pattern exactly matched the distribution of the mitochondrial protein DHLADH in the same protein samples and was clearly distinct from the segregation of the soluble cytosolic control protein PGK under the same conditions. Thus, immunofluorescence, crude cellular fractionation and N-terminal processing data demonstrate that the cellular location of COXVI-TY-C protein is consistent with mitochondrial localization. This demonstrates that the transgenic protein is appropriately compartmentalized within the cell despite significant overexpression in comparison with the endogenous protein.
To investigate more precisely the signals and subcellular localization of the T. brucei COXVI protein, we generated constructs encoding a series of mutant forms of COXVI (Fig. 5A). We first generated a protein in which the TY epitope tag sequence was inserted into the extreme N-terminus of COXVI. This was designed to investigate the importance for localization of sequences immediately adjacent to the methionine initiation codon (Fig. 5A; N-TY-COXVI). Two further constructs generated COXVI proteins truncated at the N-terminus. These were designed to investigate the influence of the cluster of basic residues between amino acids 7 and 15 in the COXVI protein sequence (Fig. 5A; MPFVDHNKYKIQRED). Such clusters of charged amino acids are predicted import signals for trypanosomatid mitochondrial proteins with conventional cleaved N-terminal signals (Haüsler et al., 1997) and are also seen in cytochrome C1, which is imported without a cleaved presequence (Priest et al., 1993). Therefore, in construct N-7 COXVI-TY-C, the sequence encoding amino acids 2–7 was deleted from the N-terminus of the COXVI gene, whereas in construct N-15 COXVI-TY-C, the sequence encoding amino acids 2–15 was deleted. Each of these constructs was then used to transfect procyclic-form trypanosomes, and stable cell lines were selected. Figure 5B confirms that proteins of the predicted size were expressed and clearly detectable in the N-7 COXVI-TY-C and N-15 COXVI-TY-C cell lines. In the case of the N-TY-COX-VI cell line, however, the transgenic protein migrated more slowly than the COX-VI-TY-C protein, despite the fact that both predicted proteins should be of near identical size, potentially differing only in the cleaved methionine in the latter. Although we have not been able to establish the basis of this aberrant migration (the construct sequence is as predicted), we have observed with other proteins that an N-terminal TY tag sometimes generates proteins of unexpected migration (our unpublished observations; see also Discussion)
To assess the precise suborganellar location of the various transgenic COXVI proteins, we exploited methodology for trypanosome cell fractionation and mitochondrial purification (Hauser et al., 1996). Each line was induced to express the transgenic protein by growth in the presence of tetracycline, and then 1 × 1010 cells were disrupted by nitrogen bomb cavitation. Reconstituted mitochondrial vesicles were purified from these disrupted cells by Nycodenz step-gradient density centrifugation, with the purified vesicles being isolated from the 18.8–21.7% Nycodenz interface. Purified mitochondrial vesicles were then subjected to three treatments in order to determine the suborganellar localization of the transgenic COXVI proteins. In the first case, intact purified vesicles were not exposed to further treatment, allowing the detection of proteins that associated with the mitochondrion, either internally or externally (M; Fig. 5C). In the second treatment, purified vesicles were exposed to 25 μg ml−1 proteinase K treatment for 15 min. This degrades proteins bound externally to the mitochondrion, but leaves intact those that have been successfully imported (+PK; Fig. 5C). The final treatment was to incubate the mitochondrial vesicles with both proteinase K and 0.2% Triton X-100 (+T, +PK; Fig. 5C). In this case, the detergent induces mitochondrial vesicle lysis, thereby exposing previously protected proteins to protease digestion. At the low concentration of non-ionic detergent used here, it is not expected that COXVI will be fully solubilized, as is borne out by Fig. 4B, in which COXVI was only partially soluble in digitonin at 1 mg mg−1 protein. Those proteins resistant to this treatment are therefore protected by being embedded within or closely associated with mitochondrial membranes themselves.
Figure 5C shows the mitochondrial compartmentalization of each of the transgenic COXVI proteins and, for comparison, the localization of the endogenous COXVI protein (detected by the anti-COXVI antibody). Also shown is the distribution of mitochondrial HSP60, a marker for the mitochondrial matrix (detected by anti-mtHSP60; a kind gift from Dr D. Engman, University of Illinois, Chicago). Figure 5C (‘anti-COXVI’) shows the behaviour of endogenous COXVI under this purification regime: the protein was detected in all three samples and was only partially diminished even after both proteinase K and Triton X-100 treatment. This was the expected pattern for endogenous COXVI, a protein intimately associated with the mitochondrial inner membrane. Similarly, mtHSP60 was resistant to proteinase K digestion in intact vesicles but was completely degraded in the presence of both detergent and proteinase K, the expected pattern for this matrix protein (Fig. 5C, anti-HSP60). These controls confirmed that mitochondrial proteins could be detected effectively in the purified mitochondrial vesicles and that their suborganellar location was accurately discriminated.
When the mutated COXVI proteins were examined, they were found to localize to distinct mitochondrial compartments. Thus, the COXVI-TY-C protein was faithfully imported and detergent/proteinase K resistant, exactly matching the pattern observed for the endogenous COXVI protein. This demonstrated that the TY epitope tag itself did not result in missorting of the COXVI protein within the mitochondrion. When the epitope tag sequence was incorporated into the N-terminus of COXVI, however, the localization of the protein was perturbed; while the protein remained associated with the mitochondrion, it was apparently not imported, being completely susceptible to proteinase K digestion in the presence and absence of Triton-X 100 (N-TY-COXVI; Fig. 5C). In contrast to this severe defect, a more subtle effect was seen when the localization of the N-7 and N-15 mutants of the COXVI protein was examined. In this case, both proteins were found to associate with the mitochondrion and to be imported, being resistant to proteinase K treatment. However, neither mutant protein was resistant to proteinase K treatment in the presence of detergent, demonstrating that their ultimate localization to the inner membrane was disrupted. These results demonstrate that, despite the absence of an extended cleaved presequence, residues at the extreme N-terminus of the COXVI protein contribute to appropriate import and mitochondrial sorting. Nevertheless, as all mutants of COXVI fractionated with this organelle, sequences out-with this extreme N-terminus must also contribute to the association of the COXVI protein with the mitochondrion.
Stage regulation of COXVI
The respiratory activity of the T. brucei mitochondrion exhibits strict stage regulation, being repressed in the bloodstream form and activated during differentiation to the procyclic form. In order to investigate the mechanisms that operate to regulate the expression of COXVI in the bloodstream, we attempted to express the COXVI protein at this inappropriate life cycle stage. The expression construct used for these experiments, pHD451, is driven by the EP-1 promoter, which is less active in bloodstream forms than in procyclic forms (Biebinger et al., 1996). Therefore, we also transfected bloodstream trypano-somes with a control construct, which, in place of COXVI, expresses a gene encoding a protein whose function is unknown but is expressed in both life cycle stages (E. Hendriks and K. Matthews, unpublished observations). In all other respects these constructs were identical.
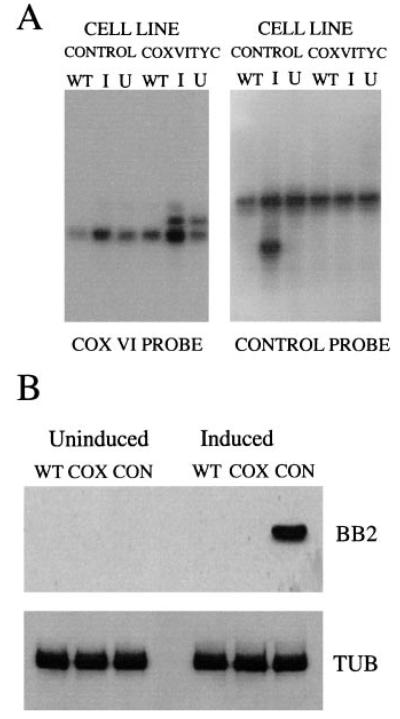
Figure 6A shows that two transcripts can be identified in those bloodstream cell lines transfected with the COXVI-TY-C construct. The lower of these corresponds to the COXVI transcript derived from the genomic copy of the COXVI gene, whereas the upper transcript is derived from the transgenic COXVI gene. This mRNA size difference is a consequence of the use of the 3′ untranslated region (UTR) form of the aldolase gene in the COXVI-TY-C construct rather than the endogenous COXVI-3′ UTR (Biebinger et al., 1997). RNA was also detected from the transgene in the control construct, although there was tight regulation in response to tetracycline in this case (Fig. 6A, right; the transgene is the lower transcript in this case). Although RNA was detectable for both transgenic COXVI and the control transcript, Western blots using the anti-TY tag antibody BB2 detected only the control transgenic protein with no detectable transgenic COXVI protein being generated (Fig. 6B). Thus, the vector used is able to drive significant RNA expression in bloodstream-form trypanosomes and generate protein from a control transcript. However, although COXVI mRNA can be generated, these bloodstream cells do not produce detectable COXVI protein.
Fig. 6. Expression of COXVI-TY-C in bloodstream-form cells.
A. Northern blot of either wild-type bloodstream-form trypanosomes or bloodstream-form trypanosomes transfected with the control construct (‘control’ cell line) or with the COXVI-TY-C construct (‘COXVI-TY-C cell line’) and grown in the presence (I, induced) or absence (U, uninduced) of tetracycline. The left hand blot was probed with a riboprobe detecting COXVI mRNA; the right hand blot was hybridized with a probe recognizing the control mRNA. Left: the transgenic mRNA is larger than the endogenous COXVI mRNA. Right: the transgenic mRNA is smaller than the endogenous control transcript. These differences reflect the differing lengths of the 3′ UTRs for either the COXVI or the control transcript.
B. Protein samples from the same cell lines (‘WT’, wild-type bloodstream forms; ‘cox’, COXVI-TY-C-transfected bloodstream forms or ‘con’, bloodstream forms transfected with the control construct) detected with the anti-TY antibody BB2 (top) or with an antibody to alpha-tubulin (Sherwin et al., 1987). A signal is detected with BB2 only from the trypanosomes transfected with the control construct in the presence of tetracycline.
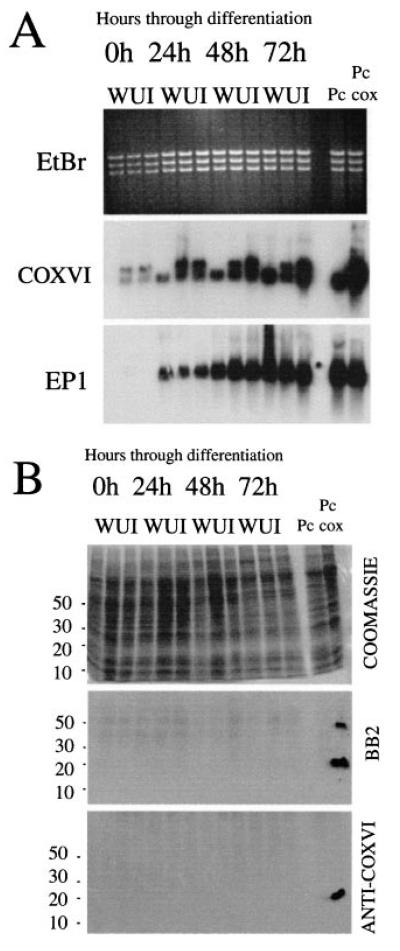
Finally, we followed development of the COXVI mRNA and protein during differentiation to the procyclic form. Figure 7A shows Northern blots of RNA derived from wild-type bloodstream cells and the bloodstream COXVI-TY-C cell line (with or without tetracycline) at 0, 24, 48 and 72 h after the initiation of differentiation by the addition of cis aconitate. Within 24 h, significant upregulation of the COX mRNA was detectable from both the endogenous and the transgenic COXVI gene copy, with transgene mRNA being detected in both the presence and the absence of tetracycline. This expression was further enhanced after 48 h in differentiation conditions and, by 72 h, the COXVI mRNA abundance approached that of the previously established procyclic COXVI-TY-C cell line. Total RNA from each sample and at each time point was also hybridized with a riboprobe detecting the EP1 mRNA, a marker for differentiation to the procyclic form. This demonstrated that the differentiation for each cell line occurred with approximately equal kinetics and efficiency. Thus, mRNAs from both the endogenous and the transgenic gene copies of the COXVI are significantly upregulated during differentiation to the procyclic form.
Fig. 7. Expression pattern of COXVI mRNA and protein during differentiation.
A. Expression of COXVI mRNA during differentiation of monomorphic bloodstream trypanosomes to the procyclic form. Either wild-type (WT) cells or cells transfected with COXVI-TY-C and grown in the presence (I, induced) or absence (U, uninduced) of tetracycline were induced to differentiate to the procyclic form by the addition of cis aconitate and reduction in temperature to 27°C. Note that tetracycline-induced cell lines had been maintained in the presence of tetracycline for > 24 h before the initiation of differentiation. In each case, the endogenous COXVI transcript is detected in addition to the transgenic mRNA. Note that endogenous COXVI mRNA is barely detectable in this sample of the wild-type bloodstream-form trypanosome mRNA. The same RNA samples were also hybridized with a riboprobe detecting the EP1 transcript to confirm equivalent differentiation in each cell line.
B. COXVI protein expression during differentiation between bloodstream and procyclic forms. Protein samples from either wild-type cells (W) or COXVI-TY-C-transfected cells in the presence or absence of tetracycline (‘U’ or ‘I’) were probed with either the anti-TY antibody BB2 or the anti-COXVI antibody. PC, wild-type procyclic-form cells; PC-cox, COXVI-TY-C-transfected procyclic forms grown in the presence of tetracycline.
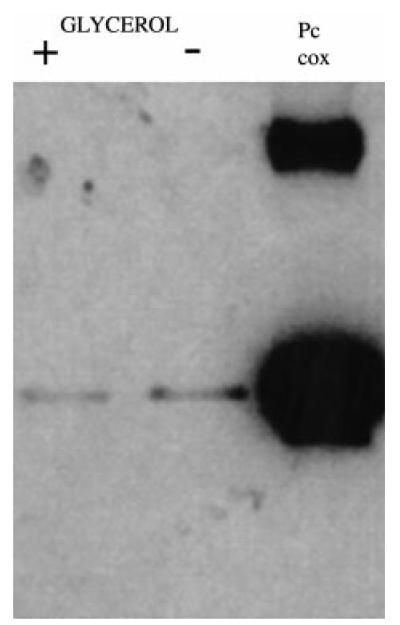
Figure 7B shows Western blots of proteins derived from the same cell cultures used to prepare RNA in Fig. 7A. In this case, COXVI protein was not detected with either BB2 or the anti-COXVI antibody in any of the samples, despite there being abundant COXVI mRNA at the later stages of this time course. This did not result from a mutation in the transgenic COXVI gene or expression construct because two independently generated bloodstream transgenic cell lines also failed to produce COXVI protein under the same conditions (our unpublished observations). To establish whether protein could eventually be detected in the transformed cells, parasites were induced to differentiate in either the presence or the absence of 10 mM glycerol, a regime that has recently been shown to enhance the growth of transformed cells and to regulate a developmental switch from early- to late-form procyclic cells (Vassella et al., 2000). After 5 days under these conditions, the cells had clearly transformed to procyclic forms (on the basis of cellular morphology and their expression of the procyclic-form surface coat procyclin) but were growing only slowly, a common characteristic of recently transformed cell lines under our differentiation conditions (our unpublished observations). Nevertheless, Fig. 8 shows that COXVI protein was detectable in the cell line at this time, albeit at a low level and irrespective of the presence or absence of glycerol. This confirmed that the COXVI-C-TY construct transfected into bloodstream forms was able to generate protein, albeit only after several days in differentiation conditions. Together, these results demonstrate that there is initially regulation of the abundance of COXVI mRNA during transformation to the procyclic form and, thereafter, restriction in the level of protein.
Fig. 8. COXVI protein expression after 5 days in differentiation conditions.
Bloodstream cells, stably transfected with the COXVI-TY-C construct, were subjected to differentiation in SDM 79 either with or without 10 mM glycerol (+ or −). Samples were taken 5 days after initiating differentiation. Transgenic procyclic cells (Pc) containing the COXVI-TY-C construct are shown for comparison.
Discussion
In the African trypanosome, the cytochrome oxidase complex is subject to regulation at many levels. Mitochondrial transcripts are regulated in terms of their abundance and level of stage-specific RNA editing, whereas nuclear transcripts are regulated in terms of mRNA abundance, translation, protein stability and mitochondrial import (Priest and Hajduk, 1994a). Here, we have examined the import and regulation of the COXVI gene, the first identified nuclear-encoded component of this enzyme complex in T. brucei.
The import mechanisms for most mitochondrial proteins in T. brucei are signalled by an unusually short cleaved leader sequence (Haüsler et al., 1997). However, comparison of the genomic sequence of T. brucei cytochrome oxidase VI with the N-terminal peptide sequence of the biochemically purified COXVI from C. fasciculata predicted that this protein would lack a cleaved leader sequence. To test this experimentally, cell lines were generated that overexpressed an epitope-tagged copy of this protein and, by sequencing of the transgenic protein, its mature N-terminus was determined. This confirmed that the N-terminal methionine was specifically missing from the mature transgenic COXVI protein and, moreover, this protein was correctly localized within the mitochondrion as assessed by three criteria: immunofluorescence co-localization with a mitochondrial marker protein; cell fractionation by freeze–thaw and digitonin permeabiliza-tion; and finally, by a mitochondrial vesicle lysis assay. This final assay demonstrated that the transgenic COXVI was not only partitioning with the mitochondrion, but was also imported and protected from protease digestion, implying membrane association. We estimate that the transgenic protein is overexpressed at least fivefold with respect to the endogenous protein, indicating that the import capacity for the trypanosome mitochondrion is considerable. Although similar observations have also been made for the import of transgenic malate dehydrogenase into the T. brucei mitochondrion (Anderson et al., 1998), COXVI is a component of a multisubunit enzyme complex. It seems likely, therefore, that the transgenic protein cannot all be incorporated into the functional enzyme complex. Nevertheless, our studies indicate that transgenic overexpression of mitochondrial components in T. brucei provides an effective route for the dissection of their regulation and mitochondrial import.
When mutant forms of the COXVI protein were expressed in procyclic-form trypanosomes, sequences that are adjacent to the mature N-terminus were found to contribute to its correct localization within the mitochondrion. Thus, incorporation of the epitope tag sequence into the N-terminus of COXVI prevented import, whereas truncation of the N-terminal amino acids perturbed correct sorting. Although the tag had no detectable effect when incorporated into the C-terminus of COXVI and has been used successfully for proteins localized in several cellular compartments in T. brucei (Bastin et al., 1998; Van Hellemond et al., 2000), it may be significant that its sequence is quite rich in amino acids with a negative charge. This contrasts with the positive charge characteristic of amino acids present in the cleaved presequence of T. brucei mitochondrial proteins and the presence of a cluster of positively charged residues in the mature N-terminus of cytochrome C1, which, as with COXVI, is imported without cleavage. A further contributor to the lack of appropriate localization may be processing of the N-terminal methionine. The removal of this residue from COXVI of C. fasciculata and from the C-terminally tagged T. brucei COXVI suggests that this protein is a substrate for a methionine aminopeptidase activity. The specificity of this enzyme is compatible with the penultimate amino acid in COXVI from C. fasciculata (proline), T. brucei (proline) and from T. brucei cytochrome C1 (alanine) but is not compatible with the penultimate residue of the epitope tag sequence (aspartic acid). Although this may contribute to its observed aberrant migration on PAGE gels, we were unable to derive N-terminal sequence from this protein and could not confirm the presence or absence of the N-terminal methionine.
Truncation of sequences at the extreme N-terminus of COXVI resulted in misdirection of the protein, regardless of whether the first seven or 15 amino acids were deleted. This demonstrates that either sequences in the first seven amino acids are required for appropriate sorting or the context of the residues positioned between seven and 15 amino acids is important. Nevertheless, the mutant proteins still partitioned with the mitochondrion, indicating that signals away from the extreme N-terminus also contribute to its targeting. This is a common feature of proteins lacking a cleaved leader sequence, which frequently contain poorly defined internal targeting motifs (Smagula and Douglas, 1988; Brix et al., 1999). Indeed, truncations at the N-terminus of the T. brucei Rieske iron–sulphur protein, which is imported using a cleaved signal sequence, did not prevent its specific binding to isolated mitochondrial vesicles (Priest and Hajduk, 1996).
The absence of a cleaved leader sequence does not seem to be a characteristic of other nuclear-encoded cytochrome oxidase subunits in kinetoplastids. Although no other subunits of this complex have yet been cloned from T. brucei, a cytochrome oxidase subunit IV cDNA sequence has been cloned from C. fasciculata (Speijer et al., 1997), and sequence homologues of this protein and COXX can be identified in the existing genome databases of T. brucei, Trypanosoma cruzi and Leishmania major (Fig. 9). Alignment of these sequences with the N-terminal sequence of their purified counterpart from C. fasciculata indicates that, in each case, these sequences are predicted to encode proteins with a conventional cleaved N-terminal signal sequence, a feature they share with other identified mitochondrial inner membrane proteins in T. brucei (e.g. ATPase subunit 9 and the Rieske iron–sulphur protein; Priest and Hajduk, 1995; Chi et al., 1998). Apparently, therefore, COXVI is imported into the mitochondrion by a mechanism distinct from that of other components of this same complex but similar to that for cytochrome C1. Perhaps significantly, both proteins are relatively hydrophilic and are located on the outer face of the mitochondrial inner membrane, rather than being embedded within it. Consequently, their shared import mechanism may be related to their shared ultimate location.
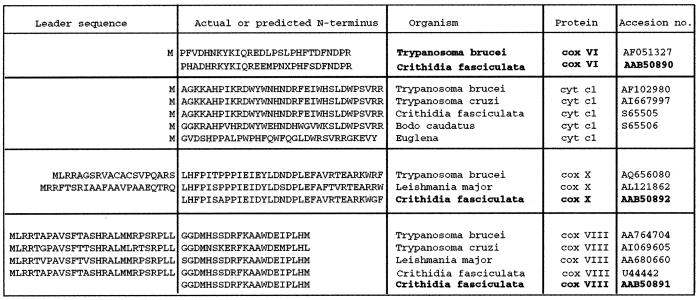
Fig. 9. Mitochondrial respiratory chain components from protozoa.
Comparison of sequences of COXVI, cytochrome C1, COXX and COXVIII from a selection of protozoa (T.b., T. brucei; T.cz, T. cruzi; Leish, Leishmania major, Bc, Bodo caudatus or Eug, Euglena). In each case, sequences are derived from database searches for homologues of the mature N-terminus of cytochrome c oxidase subunits determined by Speijer et al., 1996 (shown in bold type). Cytochrome C1 sequences are derived from Priest et al. (1993).
The mitochondrial activity of T. brucei is subjected to stringent regulation during the course of their life cycle, and development of this activity is an essential component of differentiation to the tsetse midgut procyclic form. In order to begin to address how this activity is controlled during development, we investigated at what levels nuclear COXVI was regulated. We observed that the endogenous and transgene COXVI mRNA levels were upregulated during differentiation, although both were also detectable at low levels in the bloodstream. Furthermore, the transgene could be induced above endogenous levels by growth in the presence of tetracycline in the bloodstream, despite the vector being optimized for expression in the procyclic form. Nevertheless, when the levels of protein expressed from either endogenous or transgenic COXVI genes were examined, it was found that little or none could be detected until several days after the initiation of differentiation. Although the kinetics of the establishment of procyclic cultures can be variable between different trypanosome lines and under different culture conditions, it was clear that, during the first 3 days of differentiation, the abundant COXVI mRNA was not generating abundant COXVI protein and that this was not influenced by the presence of glycerol in the culture medium, a recently identified regulator of some aspects of procyclic form development (Vassella et al., 2000).
A restriction in the ability to generate abundant COXVI protein may operate at the level of translational competence, as observed recently with T. brucei aconitase (Saas et al., 2000). However, signals in the 3′ UTR of the transcript cannot account for the inability to translate COXVI protein, because the aldolase 3′ UTR, which is downstream of the COXVI transgene, was able to support protein expression from a control construct in bloodstream forms. Alternatively, control at the level of protein stability may operate. Priest and Hajduk (1994b) found that the levels of T. brucei cytochrome reductase were restricted at the level of protein stability, and one interpretation was that this was a consequence of the inability of this protein to be imported into the bloodstream-form mitochondrion. However, it has recently been demonstrated that the bloodstream-form mitochondrion is competent for the import of NADH dehydrogenase subunit K, a constitutively expressed mitochondrial protein with a cleaved leader sequence (Bertrand and Hajduk, 2000). We are currently investigating whether COXVI is regulated at the level of translation, import or stability in the bloodstream form.
Experimental procedures
Trypanosome growth in vitro, transfection and differentiation
Bloodstream and procyclic forms of T. brucei rhodesiense EATRO 795 were cultured in HMI-9 medium at 37°C (Hirumi and Hirumi, 1989) or SDM-79 medium at 27°C respectively (Brun and Schönenburger, 1979). Not1 linearized DNA (5 μg) was transfected into 1 × 108 cells in ZFPM+G (Beverley and Clayton, 1993) at 1.5 kV, 25 μF (Bio-Rad gene pulser), and the cells were incubated at 37°C for 18 h to allow recovery of the transfectants. Finally, 5 × 106 cells were plated onto an HMI-9–agarose plate (Carruthers and Cross, 1992) containing 0.3–0.5 μg ml−1 phleomycin and 2 μg ml−1 hygromycin. Selected transfectants were subsequently grown in HMI-9.
For transfection of procyclic forms, 1 × 108 cells in ZPFM were electroporated with 20 μg of linearized plasmid by three pulses at 1.7 kV in a BTX830 electroporator (Kramel Biotech). These cells were then recovered overnight in 10 ml of SDM-79 medium before being diluted to 1 × 105 cells ml−1 and subjected to drug selection in order to isolate stably transfected cell lines. For selection, hygromycin was used at 20 μg ml−1 and phleomycin at 5 μg ml−1. Transformants were cloned by limiting dilution.
Bloodstream trypanosomes were differentiated to procyclic forms by diluting cells to 2 × 106 ml−1 with HMI-9 containing 6 mM cis aconitate (final concentration) and incubating cells at 27°C. The course of differentiation was monitored by immunofluorescence for the procyclic stage-specific antigen procyclin as described previously (Matthews and Gull, 1994). For comparison of the influence of glycerol upon transformation (Vassella et al., 2000), trypanosomes were incubated in SDM-79 with or without 10 mM glycerol.
Plasmid constructions
The trypanosome expression vector pHD 451 (Biebinger et al., 1997) was used to direct COXVI transgene expression in either bloodstream- or procyclic-form parasites that had previously been engineered to express the tetracycline repressor (Wirtz and Clayton, 1995). To integrate the TY epitope tag sequence (Bastin et al., 1996) into the 3′ end of the COXVI gene, the gene was amplified by polymerase chain reaction (PCR) using a 3′ oligonucleotide comprising the COXVI gene 3′ end sequence, the epitope tag followed by a stop codon and a BglII site (COXVI 3′TY; 5′-CCAGATCTTCAGTCAAGTGGATCCTGGTTAGTATGGACCTCCTCATCATATACGGGTTTC-3′). This oligonucleotide was used in combination with an oligonucleotide comprising a HindIII site, a start codon and the extreme N-terminus of the COXVI gene (COXVI 5′; 5′-CCGGATCCATGCCCTTTGTCGATCAC-3′). For the generation of truncated forms of COXVI, the 3′ oligonucleotide, COXVI 3′TY, was used in combination with oligonucleotide COXVI N-7 (5′-CCCAAGCTTATGTACAAGATTCAGAGGGAG-3′) or oligonucleotide COXVI N-15 (5′-CCCAAGCTTATGCCAGCCCTTCCCCATTTC-3′). For construct N-TY-COXVI, the TY tag was inserted into the 5′ end of the COXVI gene by amplification of the gene using the oligonucleotide COXVI 5′TY (5′-CCCAAGCTTATGGAGGTCCATACTAACCAGGATCCACTTGACCCCTTTGTCGATCAC-3′) in combination with a 3′ oligonucleotide COXVI 3′ (5′-GGCTGCAGCTACTCATCATATACGGG-3′). All amplification products were cloned into pHD451 digested with HindIII and BamHI, and the junction sequences were verified in the resulting constructions by DNA sequencing.
RNA isolation and protein isolation; Northern and Western blotting
Trypanosome RNA was prepared using guanidinium hydrochloride–acid phenol as described by Tasker et al. (2000). Northern blots were performed as described previously (Matthews and Gull, 1998). Blots were hybridized at 68°C with riboprobes labelled with digoxygenin (Boehringer Mannheim), and stringency washes were at 68°C in 0.2 × SSC. Blot detection was by chemiluminescence using CDP-star as a reaction substrate.
Proteins were prepared as described by Tasker et al. (2000) and resolved on 12% polyacrylamide gels. Blotted proteins were incubated with either the anti-TY epitope tag monoclonal antibody BB2 (diluted 1:20 from hybridoma culture supernatant) or with a 1:100 dilution of an affinity-purified rabbit anti-COXVI antibody produced against a COXVI full-length fusion protein produced in bacteria. Western blots were processed using a horseradish peroxidase-conjugated secondary antibody and visualized by chemiluminescence (Amersham).
Anti-COXVI antibody generation, affinity purification and immunoprecipitation
For COXVI-TY immunoprecipitation, procyclic cells stably transfected with COXVI-TY-C were induced with tetracycline to a final concentration of 10 μg ml−1 for 18 h. Cells were collected by centrifugation at 1500 g for 10 min and washed twice in PBS before being lysed on ice for 5 min in IP lysis buffer [15 mM Tris-HCI, pH 8.0, 147 mM NaCI, 1 mM MgCI2, 1 mM EGTA, 10 mM β-mercaptoethanol, 0.5% (w/v) sodium deoxycholate, 0.2% (w/v) SDS, 1.0% (v/v) Triton X-100] containing 1 mM phenylmethylsulphonyl fluoride (PMSF). The BB2 antibody was added to the lysate and left to incubate on ice for 2 h. Protein G Sepharose was then added to the lysate, which was incubated at room temperature for 1 h before being collected by centrifugation at 5000 g for 1 min. Protein G Sepharose was washed three times in IP lysis buffer before being resuspended in an equal volume of 2 × Laemmli buffer. Proteins were separated on a 15% SDS polyacrylamide gel and blotted to polyvinylidene difluoride (PVDF) by wet electrophoretic transfer in 10 mM CAPS, 10% (v/v) methanol. Proteins were visualized directly on PVDF by Coomassie staining. The COXVI proteins were excised and sequenced directly from the PVDF filter.
Cell fractionation and purification of mitochondrial vesicles
Crude cell fractionations were prepared as described by Woods et al. (1989). Briefly, 1 × 108 ml−1 cells were centrifuged at 1600 g for 10 min, washed once in PBS and resuspended in 1 ml of MME buffer (10 mM MOPS, pH 6.9, 1 mM EGTA, 1 mM MgS04) before being snap frozen in liquid nitrogen. Rapidly thawed cells were then subjected to ultacentrifugation at 100 000 g for 30 min, and the resulting supernatant and pellet were incubated with 100 μl of boiling Laemmli sample buffer to generate gel-loading samples. Digitonin fractionation was performed exactly as described by Haüsler et al. (1997) using digitonin (Calbiochem) concentrations of 10, 1, 0.1 or 0.01 mg mg−1 trypanosome protein.
Mitochondrial purification was performed according to the protocol of Hauser et al. (1996). For each cell line, 1 I of procyclic cells at mid-log phase growth (5 × 106 ml−1) was concentrated by centrifugation at 5000 g for 5 min at 4°C. The pellet was washed once in 25 ml of PBS before being resuspended in SET buffer (1 × SET is 600 mM sorbitol, 20 mM Tris-CI, pH 7.9, 2 mM EDTA) at a concentration of 1 × 109 cells ml−1. Cells were then incubated for 1 h at 1000 p.s.i., 4°C in a nitrogen cavitation chamber (Parr Instrument). Pressure release from the cavitation chamber resulted in disruption of >95% of cells as assessed by microscopy. Remaining intact cells were then removed by centrifugation at 1500 g for 10 min at 4°C, and the supernatant was centrifuged at 12 000 g to isolate the crude mitochondrial fraction. The resulting pellet was resuspended into one volume of SET containing 50% Nycodenz (Sigma) and 1 mM PMSF, and 0.5 ml of this suspension was overlain with 2.5 ml of 25%, 21.7%, 18.8% and 15% Nycodenz in a Beckman SW40 Ti ultraclear tube. The gradient was centrifuged for 45 min at 284 500 g, 4°C, and mitochondrial vesicles were isolated from the 18.8–21.7% Nycodenz gradient interface. This material was finally washed in SET buffer by centrifugation at 12 000 g in a microfuge at 4°C, and the pellet was stored in SET–50% glycerol, 1 mM PMSF at −80°C. For proteinase K digestion, the pellet from 3 × 109 cell equivalents ml−1 was incubated in SET buffer containing 25 μg ml−1 proteinase K for 15 min at 4°C in the presence or absence of 0.2% Triton X-100. The reaction was terminated by incubation on ice with 1 mM PMSF for 15 min. Finally, the samples were centrifuged at 12 000 g at 4°C for 10 min and resuspended in Laemmli sample buffer with 2 mM PMSF.
Immunofluorescence and protein analysis
Immunofluorescence upon transgenic cells was performed as described by Haüsler et al. (1997) using the antibody BB2 as an undiluted hybridoma culture supernatant or with an antibody to dihydrolipoamide dehydrogenase (a gift from Professor R. Luise Krauth-Siegel, University of Heidelberg) for 1 h. After three washes in PBS, the slides were incubated for 1 h with anti-mouse fluorescein isothiocyanate (FITC)-conjugated secondary antibody diluted 1:50. Finally, the cells were washed in PBS three times and mounted in MOWIOL containing phenylene diamine (1 mg ml−1). Slides were examined on a Zeiss axioscope 2, and images were captured using NIH image 1.55. Figures were processed using Adobe Photoshop 5.0.
Acknowledgements
We thank Professor R. Benne and Dr Andre Schneider for advice at the early stages of this work, Dr Ricky van Deursen, Dr Kostas Tokatlitis for comments on the manuscript, and Professor Stephen Hajduk for excellent and constructive suggestions. We also thank Professor R. Luise Krauth-Siegel for the gift of antibody to DHLADH, and Dr Paul Michels for the gift of antibody to PGK. This work was supported by the BBSRC. K.M. is a Dunkerly Fellow.
References
- Anderson SA, Carter V, Parsons M. Trypanosoma brucei: molecular cloning and stage-regulated expression of a malate dehydrogenase localized to the mitochondrion. Exp Parasitol. 1998;89:63–70. doi: 10.1006/expr.1998.4260. [DOI] [PubMed] [Google Scholar]
- Bastin P, Bagherzadeh A, Matthews KR, Gull K. A novel epitope tag system to study protein targeting and organelle biogenesis in Trypanosoma brucei. Mol Biochem Parasitol. 1996;77:235–239. doi: 10.1016/0166-6851(96)02598-4. [DOI] [PubMed] [Google Scholar]
- Bastin P, Sherwin T, Gull K. Paraflagellar rod is vital for trypanosome motility. Nature. 1998;391:548–548. doi: 10.1038/35300. [DOI] [PubMed] [Google Scholar]
- Benne R, Van Den Burg J, Brakenhoff JPJ, Sloof P, Van Boom JH, Tromp MC. Major transcript of the frameshifted coxll gene from trypanosome mitochondria contains four nucleotides that are not encoded in the DNA. Cell. 1986;46:819–826. doi: 10.1016/0092-8674(86)90063-2. [DOI] [PubMed] [Google Scholar]
- Bertrand KI, Hajduk SL. Import of a constitutively expressed protein into mitochondria from procyclic and bloodstream forms of Trypanosoma brucei. Mol Biochem Parasitol. 2000;106:249–260. doi: 10.1016/s0166-6851(99)00218-2. [DOI] [PubMed] [Google Scholar]
- Beverley SM, Clayton CE. Transfection of Leishmania and Trypanosoma brucei by electroporation. Methods Mol Biol. 1993;21:333–348. doi: 10.1385/0-89603-239-6:333. [DOI] [PubMed] [Google Scholar]
- Biebinger S, Rettenmaier S, Flaspohler J, Hartmann C, PenaDiaz J, Wirtz LE, et al. The PARP promoter of Trypanosoma brucei is developmentally regulated in a chromosomal context. Nucleic Acids Res. 1996;24:1202–1211. doi: 10.1093/nar/24.7.1202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biebinger S, Wirtz LE, Lorenz P, Clayton C. Vectors for inducible expression of toxic gene products in bloodstream and procyclic Trypanosoma brucei. Mol Biochem Parasitol. 1997;85:99–112. doi: 10.1016/s0166-6851(96)02815-0. [DOI] [PubMed] [Google Scholar]
- Brix J, Rudiger S, Bukau B, SchneiderMergener J, Pfanner N. Distribution of binding sequences for the mitochondrial import receptors Tom20, Tom22, and Tom70 in a presequence-carrying preprotein and a non-cleavable preprotein. J Biol Chem. 1999;274:16522–16530. doi: 10.1074/jbc.274.23.16522. [DOI] [PubMed] [Google Scholar]
- Brown RC, Evans DA, Vickerman K. Changes in oxidative metabolism and ultrastructure accompanying differentiation of the mitochondrion in Trypanosoma brucei. Int J Parasitol. 1973;3:691–704. doi: 10.1016/0020-7519(73)90095-7. [DOI] [PubMed] [Google Scholar]
- Brun R, Schönenburger M. Cultivation and in vitro cloning of procyclic forms of Trypanosoma brucei in a semi-defined medium. Acta Trop. 1979;36:289–292. [PubMed] [Google Scholar]
- Capaldi RA. Structure and function of cytochrome-c-oxidase. Annu Rev Biochem. 1990;59:569–596. doi: 10.1146/annurev.bi.59.070190.003033. [DOI] [PubMed] [Google Scholar]
- Carruthers VB, Cross GAM. High-efficiency clonal growth of bloodstream- and insect-form Trypanosoma brucei on agarose plates. Proc Natl Acad Sci USA. 1992;89:8818–8821. doi: 10.1073/pnas.89.18.8818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi TB, Brown SV, Williams N. Subunit 9 of the mitochondrial ATP synthase of Trypanosoma brucei is nuclearly encoded and developmentally regulated. Mol Biochem Parasitol. 1998;92:29–38. doi: 10.1016/s0166-6851(97)00222-3. [DOI] [PubMed] [Google Scholar]
- Czichos J, Nonnengaesser C, Overath P. Trypanosoma brucei: cis-aconitate and temperature reduction as triggers of synchronous transformation of bloodstream to procyclic trypomastigotes in vitro. Exp Parasitol. 1986;62:283–291. doi: 10.1016/0014-4894(86)90033-0. [DOI] [PubMed] [Google Scholar]
- Feagin JE, Stuart K. Developmental aspects of uridine addition within mitochondrial transcripts of Trypanosoma brucei. Mol Cell Biol. 1988;8:1259–1265. doi: 10.1128/mcb.8.3.1259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feagin JE, Abraham JM, Stuart K. Extensive editing of the cytochrome c oxidase III transcript in Trypanosoma brucei. Cell. 1988;53:413–422. doi: 10.1016/0092-8674(88)90161-4. [DOI] [PubMed] [Google Scholar]
- Folsch H, Guiard B, Neupert W, Stuart RA. Internal targeting signal of the BCS1 protein: a novel mechanism of import into mitochondria. EMBO J. 1996;15:479–487. [PMC free article] [PubMed] [Google Scholar]
- Hahne K, Haucke V, Ramage L, Schatz G. Incomplete arrest in the outer-membrane sorts NADH-cytochrome-B(5) reductase to two different submitochondrial compartments. Cell. 1994;79:829–839. doi: 10.1016/0092-8674(94)90072-8. [DOI] [PubMed] [Google Scholar]
- Haucke V, Schatz G. Import of proteins into mitochondria and chloroplasts. Trends Cell Biol. 1997;7:103–106. doi: 10.1016/S0962-8924(96)10052-0. [DOI] [PubMed] [Google Scholar]
- Hauser R, Pypaert M, Haüsler T, Horn EK, Schneider A. In vitro import of proteins into mitochondria of Trypanosoma brucei and Leishmania tarentolae. J Cell Science. 1996;109:517–523. doi: 10.1242/jcs.109.2.517. [DOI] [PubMed] [Google Scholar]
- Haüsler T, Stierhof YD, Blattner J, Clayton C. Conservation of mitochondrial targeting sequence function in mitochondrial and hydrogenosomal proteins from the early-branching eukaryotes Crithidia, Trypanosoma and Trichomonas. Eur J Cell Biol. 1997;73:240–251. [PubMed] [Google Scholar]
- Hirumi H, Hirumi K. Continuous cultivation of Trypanosoma brucei blood stream forms in a medium containing a low concentration of serum protein without feeder cell layers. J Parasitol. 1989;75:985–989. [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Poyton RO. Oxygen sensing and the transcriptional regulation of oxygen-responsive genes in yeast. J Exp Biol. 1998;201:1177–1195. doi: 10.1242/jeb.201.8.1177. [DOI] [PubMed] [Google Scholar]
- LeBowitz JH, Smith HQ, Rusche L, Beverley SM. Coupling of poly(A) site selection and trans-splicing in Leishmania. Genes Dev. 1993;7:996–1007. doi: 10.1101/gad.7.6.996. [DOI] [PubMed] [Google Scholar]
- Mair G, Shi HF, Li HJ, Djikeng A, Aviles HO, Bishop JR, et al. A new twist in trypanosome RNA metabolism: cis-splicing of pre-mRNA. RNA. 2000:6163–6169. doi: 10.1017/s135583820099229x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KR, Gull K. Evidence for an interplay between cell cycle progression and the initiation of differentiation between life cycle forms of African trypanosomes. J Cell Biol. 1994;125:1147–1156. doi: 10.1083/jcb.125.5.1147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews KR, Gull K. Identification of stage-regulated and differentiation-enriched transcripts during transformation of the African trypanosome from its bloodstream to procyclic form. Mol Biochem Parasitol. 1998;95:81–95. doi: 10.1016/s0166-6851(98)00100-5. [DOI] [PubMed] [Google Scholar]
- Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenyl-ation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- Neupert W. Protein import into mitochondria. Annu Rev Biochem. 1997;66:863–917. doi: 10.1146/annurev.biochem.66.1.863. [DOI] [PubMed] [Google Scholar]
- Osinga KA, Swinkels BW, Gibson WC, Borst P, Veeneman GH, Van Boom JH, et al. Topogenesis of microbody enzymes: a sequence comparison of the genes for the glycosomal (microbody) and cytosolic phosphoglycerate kinases of Trypanosoma brucei. EMBO J. 1985;30:3811–3817. doi: 10.1002/j.1460-2075.1985.tb04152.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overath P, Czichos J, Haas C. The effect of citrate/cis-aconitate on oxidative metabolism during transformation of Trypanosoma brucei. Eur J Biochem. 1986;160:175–182. doi: 10.1111/j.1432-1033.1986.tb09955.x. [DOI] [PubMed] [Google Scholar]
- Poyton RO, McEwen JE. Crosstalk between nuclear and mitochondrial genomes. Annu Rev Biochem. 1996;65:563–607. doi: 10.1146/annurev.bi.65.070196.003023. [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. Developmental regulation of mitochondrial biogenesis in Trypanosoma brucei. J Bioenerg Biomembr. 1994a;26:179–191. doi: 10.1007/BF00763067. [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. Developmental regulation of Trypanosoma brucei cytochrome c reductase during bloodstream to procyclic differentiation. Mol Biochem Parasitol. 1994b;65:291–304. doi: 10.1016/0166-6851(94)90080-9. [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. The trypanosomatide Rieske iron-sulfur proteins have a cleaved presequence that may direct mitochondrial import. Biochim Biophys Acta – Mol Cell Res. 1995;1269:201–204. doi: 10.1016/0167-4889(95)00154-6. [DOI] [PubMed] [Google Scholar]
- Priest JW, Hajduk SL. In vitro import of the Rieske iron–sulfur protein by trypanosome mitochondria. J Biol Chem. 1996;271:20060–20069. doi: 10.1074/jbc.271.33.20060. [DOI] [PubMed] [Google Scholar]
- Priest JW, Wood ZA, Hajduk SL. Cytochromes-C(l) of kinetoplastid protozoa lack mitochondrial targeting presequences. Biochim Biophys Acta. 1993;1144:229–231. doi: 10.1016/0005-2728(93)90178-i. [DOI] [PubMed] [Google Scholar]
- Roditi I, Schwarz H, Pearson TW, Beecroft RP, Liu MK, Richardson JP, et al. Procyclin gene expression and loss of the variant surface glycoprotein during differentiation of Trypanosoma brucei. J Cell Biol. 1989;108:737–746. doi: 10.1083/jcb.108.2.737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saas J, Ziegelbauer K, vonHaeseler A, Fast B, Boshart M. A developmentally regulated aconitase related to iron-regulatory protein-1 is localized in the cytoplasm and in the mitochondrion of Trypanosoma brucei. J Biol Chem. 2000;275:2745–2755. doi: 10.1074/jbc.275.4.2745. [DOI] [PubMed] [Google Scholar]
- Sherwin T, Schneider A, Sasse R, Seebeck T, Gull K. Distinct localization and cell cycle dependence of COOH terminally tyrosinolated alpha-tubulin in the microtubules of Trypanosoma brucei bruce. J Cell Biol. 1987;104:439–446. doi: 10.1083/jcb.104.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smagula C, Douglas MG. Mitochondrial import of the ADP/ATP carrier protein in Saccharomyces cerevisiae – sequences required for receptor-binding and membrane translocation. J Biol Chem. 1988;263:6783–6790. [PubMed] [Google Scholar]
- Sogin ML, Elwood HJ, Gunderson JH. Evolutionary diversity of eukaryotic small-subunit rRNA genes. Proc Natl Acad Sci USA. 1986;83:1383–1387. doi: 10.1073/pnas.83.5.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speijer D, Muijsers AO, Dekker H, Dehaan A, Breek CKD, Albracht SPJ, Benne R. Purification and characterization of cytochrome-C-oxidase from the insect trypanosomatid Crithidia fasciculata. Mol Biochem Parasitol. 1996;79:47–59. doi: 10.1016/0166-6851(96)02648-5. [DOI] [PubMed] [Google Scholar]
- Speijer D, Breek CKD, Muijsers AO, Hartog AF, Berden JA, Albracht SPJ, et al. Characterization of the respiratory chain from cultured Crithidia fasciculata. Mol Biochem Parasitol. 1997;85:171–186. doi: 10.1016/s0166-6851(96)02823-x. [DOI] [PubMed] [Google Scholar]
- Stuart K, Allen TE, Heidmann S, Seiwert SD. RNA editing in kinetoplastid protozoa. Microbiol Mol Biol Rev. 1997;61:105–120. doi: 10.1128/mmbr.61.1.105-120.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutton RE, Boothroyd JC. Evidence for trans splicing in trypanosomes. Cell. 1986;47:527–535. doi: 10.1016/0092-8674(86)90617-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasker M, Wilson J, Sarkar M, Hendriks E, Matthews K. A novel selection regime for differentiation defects demonstrates an essential role for the stumpy form in the life cycle of the African trypanosome. Mol Biol Cell. 2000;11:1905–1917. doi: 10.1091/mbc.11.5.1905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokatlidis K, Schatz G. Biogenesis of mitochondrial inner membrane proteins. J Biol Chem. 1999;274:35285–35288. doi: 10.1074/jbc.274.50.35285. [DOI] [PubMed] [Google Scholar]
- Tomizaki T, Yamashita E, Yamaguchi H, Aoyama H, Tsukihara T, Shinzawa-ltoh K, et al. Structure analysis of bovine heart cytochrome c oxidase at 2.8 A resolution. Acta Crystallogr. 1999;55:31–45. doi: 10.1107/S0907444998006362. [DOI] [PubMed] [Google Scholar]
- Torri AF, Hajduk SL. Posttranscriptional regulation of cytochrome c expression during the developmental cycle of Trypanosoma brucei. Mol Cell Biol. 1988;8:4625–4633. doi: 10.1128/mcb.8.11.4625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torri AF, Bertrand KI, Hajduk SL. Protein stability regulates the expression of cytochrome c during the developmental cycle of Trypanosoma brucei. Mol Biochem Parasitol. 1993;57:305–315. doi: 10.1016/0166-6851(93)90206-d. [DOI] [PubMed] [Google Scholar]
- Tschudi C, Ullu E. Polygene transcripts are precursors to calmodulin mRNAs in trypanosomes. EMBO J. 1988;7:455–463. doi: 10.1002/j.1460-2075.1988.tb02833.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullu E, Matthews KR, Tschudi C. Temporal order of RNA-processing reactions in trypanosomes: Rapid trans splicing precedes polyadenylation of newly synthesized tubulin transcripts. Mol Cell Biol. 1993;13:720–725. doi: 10.1128/mcb.13.1.720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- VanHellemond JJ, Neuville P, Schwarz RT, Matthews KR, Mottram JC. Isolation of Trypanosoma brucei CYC2 and CYC3 cyclin genes by rescue of a yeast G(1) cyclin mutant – functional characterization of CYC2. J Biol Chem. 2000;275:8315–8323. doi: 10.1074/jbc.275.12.8315. [DOI] [PubMed] [Google Scholar]
- Vassella E, VandenAbbeele J, Butikofer P, Renggli CK, Furger A, Brun R, Roditi I. A major surface glycoprotein of Trypanosoma brucei is expressed transiently during development and can be regulated post-transcriptionally by glycerol or hypoxia. Genes Dev. 2000;14:615–626. [PMC free article] [PubMed] [Google Scholar]
- Wirtz E, Clayton C. Inducible gene expression in trypanosomes mediated by a prokaryotic repressor. Science. 1995;268:1179–1182. doi: 10.1126/science.7761835. [DOI] [PubMed] [Google Scholar]
- Woods A, Baines AJ, Gull K. Evidence for a M(r) 88 000 glycoprotein with a transmembrane association to a unique flagellum attachment region in Trypanosoma brucei. J Cell Sci. 1989;93:501–508. doi: 10.1242/jcs.93.3.501. [DOI] [PubMed] [Google Scholar]
- Ziegelbauer K, Quinten M, Schwarz H, Pearson TW, Overath P. Synchronous differentiation of Trypanosoma brucei from bloodstream to procyclic forms in vitro. Eur J Biochem. 1990;192:373–378. doi: 10.1111/j.1432-1033.1990.tb19237.x. [DOI] [PubMed] [Google Scholar]