Abstract
A multiple criteria approach is presented, that is used to perform a comparative analysis of four recently developed combinatorial libraries to drugs, Molecular Libraries Small Molecule Repository (MLSMR) and natural products. The compound databases were assessed in terms of physicochemical properties, scaffolds and fingerprints. The approach enables the analysis of property space coverage, degree of overlap between collections, scaffold and structural diversity and overall structural novelty. The degree of overlap between combinatorial libraries and drugs was assessed using the R-NN curve methodology, which measures the density of chemical space around a query molecule embedded in the chemical space of a target collection. The combinatorial libraries studied in this work exhibit scaffolds that were not observed in the drug, MLSMR and natural products collections. The fingerprint-based comparisons indicate that these combinatorial libraries are structurally different to current drugs. The R-NN curve methodology revealed that a proportion of molecules in the combinatorial libraries are located within the property space of the drugs. However, the R-NN analysis also showed that there are a significant number of molecules in several combinatorial libraries that are located in sparse regions of the drug space.
INTRODUCTION
The advent of combinatorial chemistry and high-throughput screening (HTS) technologies coupled with the development of computational tools has given rise to large compound databases. Such databases can be used for a wide variety of tasks such as the development and exploration of structure-activity relationships1 and identification of polypharmacology.2 For example, the National Institutes of Health (NIH) Molecular Library Initiative3 through the Molecular Libraries Screening Center Network (MLSCN) generates large and diverse screening datasets publicly available in PubChem.4 Other publicly available databases like ZINC5 also enable the retrieval of millions of compounds for diverse purposes. These databases along with the combinatorial libraries6, 7 and natural products continue to be a key component in the drug discovery processes.8
It is widely accepted that the comparison of compound collections is important in library selection and design.9 When designing new libraries or screening existing libraries in approaches such as mixture-based screening,6, 10, 11 conventional high-throughput,12 structure-based or high-content screening,13 it is relevant to consider the chemical space coverage of the new compounds, the structural novelty (i.e., are the compounds accessing unexplored regions of chemical space?) and the pharmaceutical relevance (i.e., are the new regions of chemical space pharmaceutically or biological relevant?).14, 15
Comparing the chemical space of compound collections is not a trivial task since it is very dependent on the method used and the structural representation of the compounds.16, 17 Traditionally, compound databases have been compared using physicochemical properties (including drug-like descriptors), scaffolds or fingerprints. However, these comparisons usually focus on only one or two criteria that do not always necessarily provide a comprehensive assessment of the structural and property diversity. It is also important to note that each of these methods has its own advantages and disadvantages. For example, the use of whole molecule properties (holistic properties) has the advantage of being intuitive and straightforward to interpret. In fact, the drug-like18 and lead-like criteria,19 and Congreve’s ‘rule of 3′ for fragment-based lead-discovery20 have been formulated using only the physicochemical properties. However, physicochemical properties do not provide information regarding the structural patterns and molecules with different chemical structures can have the same or similar physicochemical properties. Similar to physicochemical descriptors, chemotypes or scaffolds are straightforward to interpret and enable easy communication with medicinal chemists and biologists. For example, scaffold analysis has lead to concepts that are widely used in medicinal chemistry and drug discovery such as “scaffold hopping”21 and “privileged structures”.22, 23 One of the disadvantages of the scaffold or framework analysis is the lack of information regarding structural similarity due to the side chains and the inherent similarity or dissimilarity of the scaffolds themselves. An obvious solution is the analysis not only of the molecular frameworks but also the side chains, functional groups and other sub-structural analysis strategies.24
Molecular fingerprints are widely used and have been successfully applied to a number of chemoinformatic and computer-aided drug design applications.25–27 A disadvantage of some fingerprints is that they are more difficult to interpret. Also, it is well known that chemical space will depend on the types of fingerprints used.16 Using multiple fingerprints and representations to derive consensus conclusions (e.g., consensus activity cliffs) have been proposed as a solution.28
In this work, four in-house combinatorial libraries are compared to known drugs, natural products and molecules from the Molecular Libraries Small Molecule Repository (MLSMR). The combinatorial libraries have been developed and published recently by our group and are part of an on-going effort to develop positional scanning libraries for the rapid identification and ranking of active library scaffolds.6 A number of compounds obtained from these libraries have shown activity in a variety of assays such as ones related to the treatment of pain and central nervous system (CNS) indications, X-linked inhibitor apoptosis protein (XIAP), methicillin-resistant S. aureus, and vaccinia virus-infected cells to name a few recent examples.6 The analysis and comparison is based on a three-fold criteria namely physicochemical properties, cyclic systems (also referred to in the literature as frameworks or scaffolds), and multiple fingerprints including MACCS keys,29 graph-based three point pharmacophores (GpiDAPH3) and typed graph distance (TGD).30 The use of these multiple criteria allowed us to obtain a more comprehensive analysis of the density, coverage of chemical space, scaffold content, diversity and structural similarity of combinatorial libraries compared to other collections, particularly the known drugs. In addition to characterizing the libraries in terms of their structural features, we also investigated the overlap of the combinatorial libraries with drugs. This analysis was performed using the R-NN curve technique published elswhere.31 This technique is a general approach for the characterization of the density of space of a compound collection, in a specified chemical space and in this work addressed the following question: Are the compounds in the combinatorial libraries located in very dense regions or very sparse regions of the drugs?
METHODS
Datasets
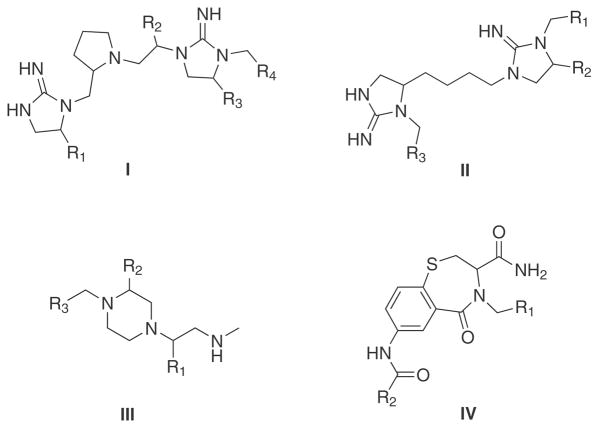
The core template of the four combinatorial libraries is presented in Figure 1 and the number of compounds per library is shown in Table 1. Note the wide range of compounds from ~3,500 to more than 700,000. The number of diversity position spans from two in library IV to four in library I. Table 1 also presents the number of molecules in the drugs, natural products and MLSMR collections considered in this work. The collection of drugs was obtained from DrugBank32 as collected in the ZINC database.5 Natural products were obtained from ZINC. At the time of download, the natural products available at ZINC contained compounds from seven vendors that advertise their compounds as being natural products or natural product derivatives. Molecules from ZINC were used “as is” without further processing. The MLSMR collection was obtained from PubChem and processed with Molecular Operating Environment (MOE)30 program by disconnecting group I metals in simple salts and keeping the largest fragment.
Figure 1.
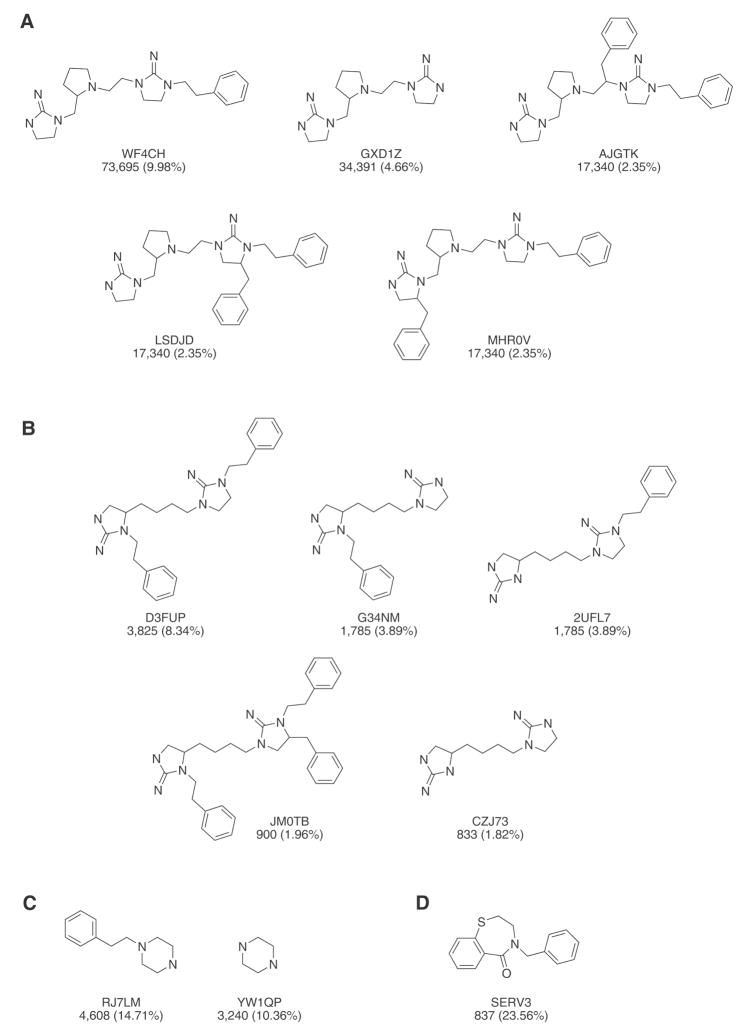
Core templates of combinatorial libraries covered in this study. Library I is made up of 738,192 compounds (R1, R2, and R3 = 26 substituents, and R4 = 42 substituents). Library II is made up of 45,864 compounds (R1 = 42 substituents, R2 = 26 substituents, and R3 = 42 susbtituents). Library III is made up of 31,320 compounds (R1 = 29 susbtituents, R2 = 27 susbtituents, R3 = 40 susbtituents). Library IV is made up of 3,552 compounds (R1 = 48 substituents, R2 = 74 substituents).
Table 1.
Compound Databases Considered in this Study
Representations and comparison metrics
Compound collections were analyzed and compared based on physicochemical properties, scaffolds, fingerprints and the R-NN curve analysis as described herein:
Physicochemical properties
The following properties were computed with MOE program: molecular weight (MW), number of rotatable bonds (RB) (the bonds were considered rotatable if they satisfied the criteria of: bond order of 1; not a ring; and at least two heavy neighbors), hydrogen bond acceptors (HBA), hydrogen bond donors (HBD), topological polar surface area (TPSA), and the octanol/water partition coefficient (SlogP). To obtain a visual representation of the property space,17 a principal component analysis (PCA) was carried out in Spotfire 9.1.133 considering all six of the above mentioned physicochemical properties. For the PCA analysis and property distribution analysis the molecule with Substance ID: 11532938 was excluded from the MLSRM because of its high weight (MW = 3,358). Prior to analysis, the descriptor values for the target dataset (i.e. DrugBank collection) were scaled using the median and interquartile ranges. Each descriptor for the combinatorial libraries were then centered and scaled using the median and the interquartile range of the corresponding descriptors for the target collection.
R-NN curve analysis
The R-NN curve method is described in detail elsewhere31 and herein we provide only a brief overview of the algorithm and its implementation for the current study. Given a target collection of molecules, characterized by a set of descriptors representing the chemical space, we can take a query molecule and identify the number of neighbors lying within a sphere of radius R, centered on the query molecule. Clearly, the number of neighbors within the sphere will increase with a larger radius. We evaluate the maximum pairwise distance in the target collection and then perform the above procedure for multiple values of R, ranging from 1% to 100% of the maximum pairwise radius. The result of this is a set of nearest neighbor counts versus radii, and is termed the R-NN curve. When plotted, these curves exhibit a sigmoidal shape. More importantly, if the query molecule is located in a sparse region of the target collection, the lower tail will be relatively flat and long. On the other hand, if the query is in a very dense region of the target collection, the lower tail will be very short. Thus, the form of the R-NN curve characterizes the spatial location of the query point with respect to the target collection. In order to generate a summation of the R-NN curves for many query compounds, we evaluate the Rmax(S) value, which is the value of R, at which the linear, lower tail of the curve transitions to the initial exponentially increasing region of the curve. Small values of Rmax(S) indicate that the molecule is located in a dense region of the target space, whereas large values indicate that it is located in a sparse region. By plotting the Rmax(S) values for an entire query dataset, we can obtain an intuitive summary of the spatial distribution of the query molecules with respect to the target collection.
Given that the method requires multiple nearest neighbor calculations for any given query molecule, it can become computationally intensive, especially for large target collections. To enhance the performance of this procedure, we employed a relational database coupled with a spatial index. Specifically, we generated the descriptors for the target collection and loaded them into a Postgres database. We then generated an R-tree index.34 This is a spatial data structure similar in concept to the B-tree.35 Briefly, an R-tree divides an n-dimensional space into a series of hierarchical hypercubes (termed minimum bounding rectangle or MBR). Thus, the entire space is represented as a top-level MBR, which can be then be divided into two ore more smaller MBR’s enclosing different portions of the space. Each of these MBR’s can be further divided and so on. The key feature of this index is that points in the original space that are close to each other will lie in the same bounding rectangle. The use of such an index, allows one to perform near neighbor searches extremely rapid, since given a query point, one identifies the MBR that contains the query point. One can then perform a linear search over the points in the MBR (which is usually much smaller than the whole collection) to identify the nearest neighbors. With the target collection loaded, we then queried the database using the descriptor values of the query molecules along with the query radii. The advantage of this approach is that it can scale to target collections of millions of compounds. It should be noted that the use of the R-tree index imposes a constraint on the nature of the nearest neighbor queries. In particular, the R-tree index works with (hyper) rectangular regions, rather than spheres. As a result, the original R-NN query is now performed by asking how many neighbors lie within a hypercube of side 2R, centered on the query point. Clearly, this query will result in a number of extra neighbors being included, compared to a query based on a hyperspherical region. However, our experiments indicated that the increased number of neighbors did not significantly change the results and so we did not perform the extra step of culling extra neighbors.
It is important to note that the results of the R-NN curve analysis are dependent on the chemical space that the molecules are embedded in. As a result one will obtain different spatial distributions of the compounds and hence different values of Rmax(S). Furthermore, the descriptors for the target collection are autoscaled followed by appropriately scaling the query descriptors (using the means and standard deviations of the target descriptors). Given these observations comparison of the same dataset in two different chemical spaces should only be considered on a qualitative level and rigorous, quantitative conclusions will, in general, not be valid. It is also important to note that the descriptors used to characterize the datasets should be as orthogonal as possible. A closely related point is that high-dimensional chemical spaces may not be suitable for such an analysis. In such cases, the bulk of the dataset will be relegated to the “corners” of the chemical space – in effect forcing a clustering of the compounds, which can obscure any clustering due to actual molecular structure or property features.
Scaffolds – molecular frameworks
In this work the scaffolds were defined as the cyclic systems implemented in the chemotype methodology developed by Johnson and Xu and were computed with the program Molecular Equivalence Indices.36, 37 In this approach the cyclic systems are obtained from the entire molecules by iteratively removing all vertices of degree one (e.g., by removing the side chains) and defining the molecular framework of the molecule.37 In fact the cyclic systems are comparable to the frameworks of Bemis and Murcko (“defined as the union of ring systems and linkers in a molecule”).38 A code of five characters, named a chemotype identifier or chemotype code, is assigned to each cyclic system using a unique naming algorithm.36 This methodology has been widely applied to construct a hierarchical classification of compound collections,39 assess scaffold-hopping efficiency of topological and pharmacophore pair descriptors,40 identify structural features that distinguish chemical libraries,37 database shaving,41 and to the construction of an annotated compound library directed to nuclear receptors.42 It is important to mention that the chemotypes employed in this study are not the only ones that can be used. However, classifying and comparing molecular databases based on cyclic systems (i.e., molecular frameworks) is a well known approach to compare the structures of compound databases. An advantageous feature of using cyclic systems to compare databases is that they represent equivalence classes and molecules classified in a given cyclic system do not lie in any other chemotype class.39
For each database the most frequent cyclic systems were identified. The number of cyclic systems was recorded along with the number of cyclic systems containing only one compound (e.g., singletons). The number of singletons accounts for the number of compounds containing its own scaffold and thus provides information regarding the scaffold diversity in the collection. It is important to point out that in the scaffold analysis the structural similarity of the cyclic systems is ignored. Therefore, in the present study we also included a comparison of databases using other criteria such as fingerprints (vide infra). We also like to stress here that the total number of cyclic systems in the database depends not only on the inherent scaffold diversity but also on the size of the database.
Fingerprints
Compound collections were analyzed in terms of three different types of fingerprints namely MACCS keys,29 GpiDAPH3 and TGD as implemented in MOE.30 In order to measure the internal molecular diversity of each collection, pair-wise similarities were computed for 5 random samples of 1,000 compounds each. Previous studies have suggested that random samples of 1,000 molecules are representative of the molecular diversity.43 The combinatorial libraries, natural products and MLSMR were also compared to drugs by means of the multi-fusion similarity (MFS) maps.44 A MFS map is a method developed recently for the visual characterization and comparison of compound databases and is based on data-fusion similarity measures. The fusion data is plotted in two dimensions, where the ordinate represents the max-fusion values and the abscissa the mean-fusion values. Each point in the map is associated with a specific molecule in the test set, and its position is determined by the corresponding fusion values computed with respect to molecules in the reference set.44 The MFS maps can be further characterized by the corresponding distributions of the max- and mean-fusion values.17 This approach has recently been used to explore structure-activity relationships of compounds obtained from mixture-based combinatorial libraries45 and to compare combinatorial libraries.17
RESULTS AND DISCUSSION
Physicochemical properties
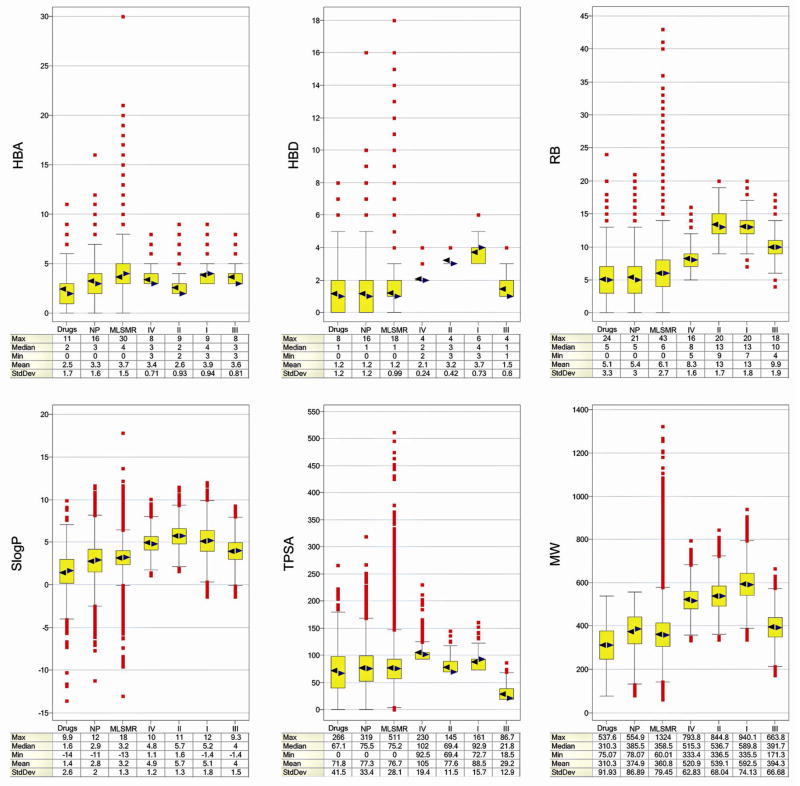
Figure 2 presents the distribution of the six physicochemical properties described in Methods as box plots implemented in Spotfire 9.1.1.33 The three important molecular properties of size, flexibility, and molecular polarity are described by MW; RB; and SlogP, TPSA, HBA and HBD respectively. The six descriptors used here have been used recently to compare the property space covered by a virtual collection and reference databases.46 In these plots, the yellow boxes enclose data points with values within the first and third quartile; the black and blue triangles denote the mean and median of distributions, respectively; and the lines above and below indicate the upper and lower adjacent values.47 The red squares indicate the data points with values beyond the upper and lower adjacent values (i.e., outliers). At the bottom of the box plots, a summary of the maximum, minimum, median, mean and standard deviations of the distributions are presented. For the analysis in Figure 2, the molecule with SubstanceID: 11532938 was excluded from the MLSRM because of the very high weight of this molecule (MW = 3,358).
Figure 2.
Box plots for the physicochemical properties. The yellow boxes encloses data points with values within the first and third quartile; the black and blue triangles denote the mean and median of distributions, respectively; the lines above and below indicate the upper and lower adjacent values. The red squares indicate outliers.
According to Figure 2, library II has a distribution of HBA similar to drugs as reflected by the median and mean values although with a narrower distribution as can be deduced from the standard deviation. Library IV also has a distribution of HBA similar to natural products. Libraries I and III have larger values of HBA than drugs but similar values to MLSMR compounds (also with narrower distributions). The four combinatorial libraries in Figure 1 have, in general, more HBD, RB and larger SlogP values than drugs, natural products and MLSMR compounds with the exception of III, which has a distribution of HBD similar to that of drugs. Library II has median and mean TPSA values similar to drugs and natural products but with less standard deviation. Libraries IV and I tend to have larger TPSA and MW values than drugs, natural products and MLSRM. Library III shows lower values of TPSA than the other collections (Figure 2). The higher hydrophobicity of combinatorial libraries in Figure 1 resembles the higher hydrophobicity reported for diversity oriented (DOS) libraries.15
Natural products contained in the ZINC database have similar distribution of HBA, HBD and RB as that of drugs. The distribution of SlogP values shows that natural products are slightly more hydrophobic than drugs and overall have a slightly larger MW as previously observed for other collections of natural products.48, 49 Compounds in MLSMR have the same median and mean distribution of HBD as drugs and natural products (1 and 1.2, respectively) although with outliers (HBD > 10). The MLSMR compounds have more HBA, RB and larger SlogP values than natural products and drugs. Regarding MW, the MLSMR compounds are slightly bulkier than drugs, with natural products MW in between these collections. Also, MLSMR have a large number of outliers with very high MW.
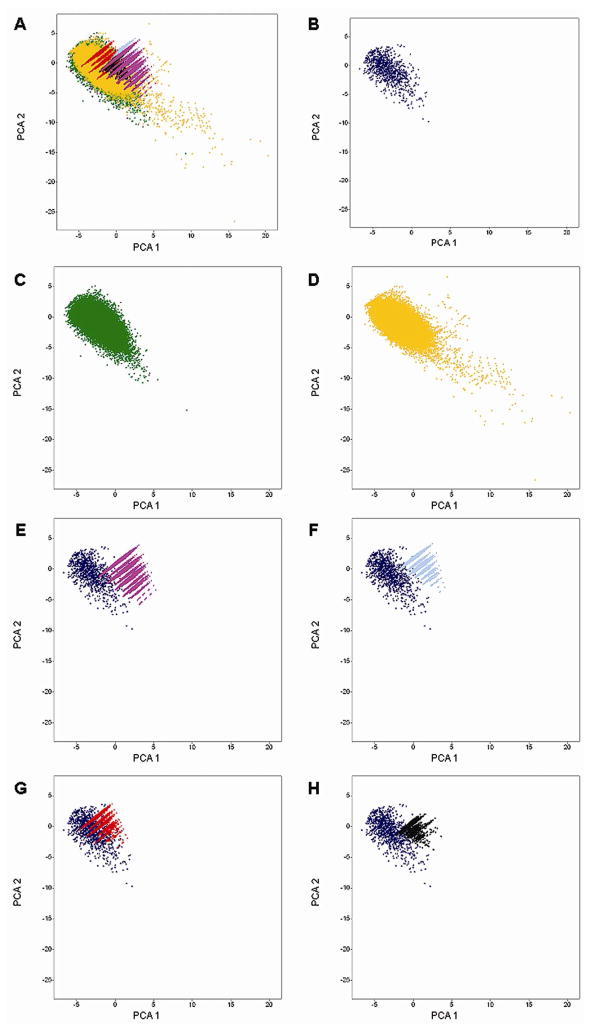
The six physicochemical properties were subject to PCA after Z-scaling. Figure 3 depicts an approximation of the property space as defined by these properties. The first two principal components (PC) with eigenvalues 3.205 and 1.858, respectively account for 84.47% of the variance (components with eigenvalues less than 1.0 were not considered). Figure 3A shows all databases in the same space. For the sake of clarity, Figures 3B – 3H shows each compound collection separately but within the same coordinates as Figure 3A. Table 2 summarizes the corresponding loadings and eigenvalues for the first four PC. Note that for the first PC, the larger loadings correspond to MW followed by RB, whereas, for the second PC the largest loading corresponds to SlogP. Visualization of the property space in Figure 3 reveals that the natural products cover an area of the property space similar to drugs but also includes additional areas. Similar observations have been made previously.15, 49, 50 Similar to natural products, the MLSMR covers a broad sample of the property space occupied by drugs but also including other regions.
Figure 3.
Property space of seven libraries obtained by PCA of six (autoscaled) molecular descriptors. The first two PC account for 84.47% of the variance. The loadings are summarized in Table 2. (A) All libraries; (B) drugs; (C) natural products; (D) MLSMR; (E) drugs and library I; (F) drugs and library II; (G) drugs and library III; and (H) drugs and library IV.
Table 2.
Loadings for the First Four Principal Components of the Property Space of Seven Librariesa
| Principal Component | PC 1 | PC 2 | PC 3 | PC 4 |
|---|---|---|---|---|
| Eigenvalue | 3.205 | 1.858 | 0.451 | 0.246 |
| Cumulative eigenvalue (%) | 53.47 | 84.47 | 91.99 | 96.09 |
|
| ||||
| HBA | 0.273 | −0.532 | −0.636 | −0.411 |
| HBD | 0.477 | −0.129 | 0.635 | −0.132 |
| RB | 0.493 | 0.207 | 0.114 | −0.472 |
| SlogP | 0.270 | 0.593 | −0.383 | 0.284 |
| TPSA | 0.349 | −0.501 | 0.024 | 0.693 |
| MW | 0.510 | 0.235 | −0.178 | 0.175 |
See corresponding PCA plot in Figure 3.
Figures 3E–H show different degrees of overlap for the combinatorial libraries with drugs. Library III in Figure 3G shows a significant degree of overlap with the space of drugs. However, a fraction of library III also samples regions sparsely covered by drugs. Figures 3E, 3F and 3H show that combinatorial libraries I, II and IV cover regions of the property space that are sparsely populated by drugs. The areas in the property space with few drugs represent areas that are biologically relevant as revealed by the presence of some drugs but may not have been sufficiently explored. In addition, libraries I and II sample unexplored regions of the drug space. Coverage of regions unexplored by drugs has previously been reported for DOS libraries.15 Molecules in these areas, while potentially unlikely to make drugs by themselves, are valuable in drug discovery as chemical probes in order to better understand the structure-activity relationships associated with unknown targets. Comparing libraries I, II and, to a lesser extent, library IV with MLSMR (Figure 3A) reveals that part of the combinatorial libraries cover regions in the property space not covered by MLSMR.
Although Figure 3 provides a useful idea of the molecules distribution in the property space, PCA represents a visual approximation of such space. A more quantitative assessment of the degree of overlap of compound collections, which avoids dimension reduction (and therefore loss of information), was evaluated using the R-NN curves. Results of the R-NN methodology for the comparison of combinatorial libraries and drugs are described ahead.
Overlap between combinatorial libraries and drugs – R-NN analysis
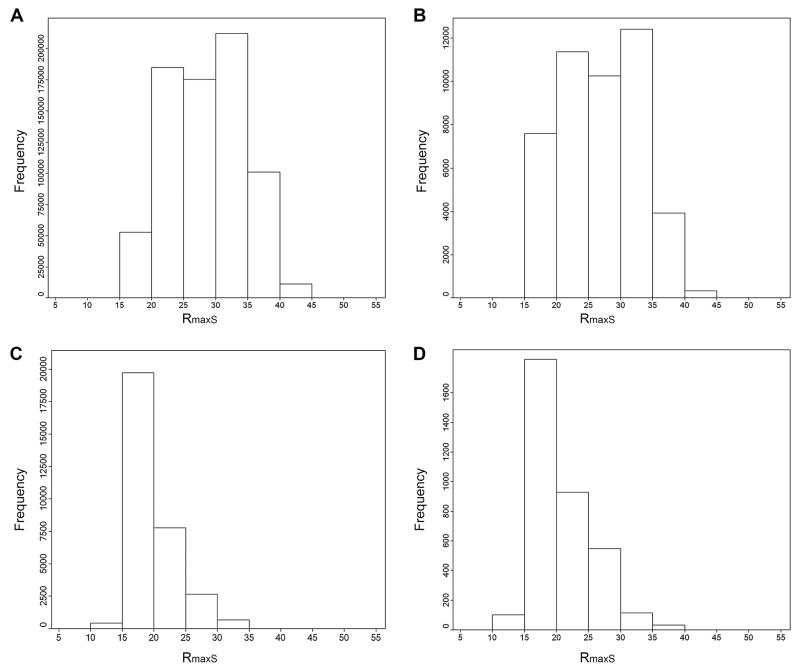
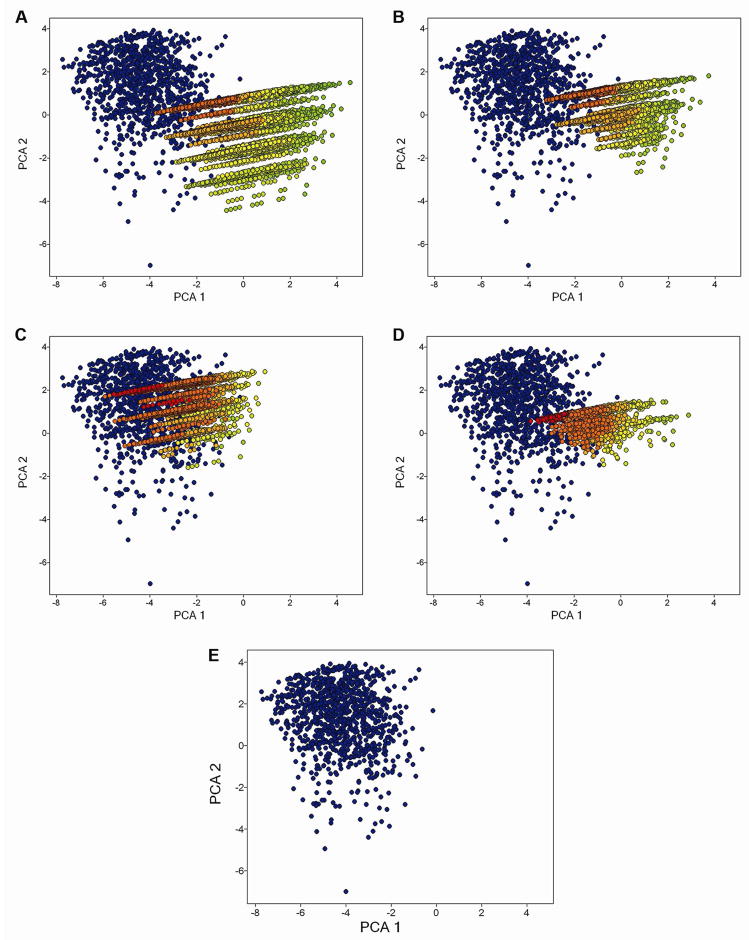
Here we consider the overlap of the combinatorial libraries with the DrugBank dataset in the physicochemical descriptor space. Given the sizes of the libraries, we summarize the Rmax(S) of the individual compounds in terms of histograms. Figure 4C shows the histogram of the Rmax(S) for library III. It is clear that most of library III compounds are located mainly within the space of the DrugBank collection. In fact, the Rmax(S) histogram for III peaks at approximately 18 (Figure 4C). In this space, the library appears to be relatively compactly distributed, with only 29 (0.8%) compounds exhibiting an Rmax(S) > 35. This is illustrated further in Figure 5C by mapping the Rmax(S) values into a visualization of the property space obtained from a PCA of the six physicochemical descriptors. It is worth noting that the property space depicted in Figure 5 was obtained by considering compounds from drugs and the four combinatorial libraries only. The first two PC (eigenvalues 1.317 and 1.018, respectively) account for 86.87% of the variance. PC with eigenvalues lower than 1.0 were not considered. The Rmax(S) values are represented in a continuous scale from red (small Rmax(S) values) to green (large Rmax(S) values). In Figure 5C it is clear that most of the library III compounds are red-to-orange in color with few yellow-to-green ones. Noteworthy, the Rmax(S) values are derived from all six dimensions and do not involve any dimensionality reduction as in the PCA plot.
Figure 4.
Histograms of Rmax(S) values for the combinatorial libraries. The target collection was DrugBank. The plots were generated using the six scaled physicochemical descriptors. (A) Library I; (B) library II; (C) library III and (D) library IV.
Figure 5.
Property space of drugs and four combinatorial libraries (820,418 molecules total) obtained by PCA of six scaled molecular descriptors. The first two PC account for 86.87% of the variance. Drugs are colored in blue. Combinatorial libraries are color-coded by the Rmax(S) value using a continuous scale from red (low Rmax(S) value) to green (high Rmax(S) value). Each panel depicts a different database: (A) drugs and library I; (B) drugs and library II; (C) drugs and library III; (D) drugs and library IV and (E) drugs.
The histogram of the Rmax(S) for library I (Figure 4A) indicates that a large number of molecules are within the space of the DrugBank collection (626,168 (84.9%), those with Rmax(S) < 35). In contrast to library III, this library contains a number of outlying compounds. More specifically, 32 compounds exhibit an Rmax(S) > 50. Figure 5A depicts the Rmax(S) values mapped into a visualization of the property space obtained with PCA. While there are molecules colored red-to-orange (small Rmax(S) values) within the area of drugs, there is also a large number of molecules colored yellow-to-green (large Rmax(S) values).
The histogram of the Rmax(S) for library II (Figure 4B) reveals that a significant proportion is within the space of the DrugBank collection (41,608 (90.7%) those with Rmax(S) < 35). As with the other libraries, there are few outliers (324 compounds have an Rmax(S) > 40, though no compounds exhibit an Rmax(S)>50). Figure 5B shows the Rmax(S) values mapped into a visualization of the property space.
Library IV is nearly wholly located within the DrugBank space (3,523 (99%) those with Rmax(S) < 35) (Figure 4D). The largest value of Rmax(S) is 41 and only 1 molecule exhibits an Rmax(S) > 40. Figure 5D shows the Rmax(S) values mapped into the PCA. It is clear that a number of molecules in library IV colored in red-to-orange are within the space of drugs. It is also clear that there are also a number of molecules in sparse regions of the DrugBank collection.
In summary, it can be concluded from Figure 4 that combinatorial library III is more within the property space of DrugBank molecules as defined by the properties used here. Library I followed by II has a larger fraction of molecules located in sparse regions (“holes”) of the DrugBank collection. Library IV represents a borderline case between libraries I, II and III.
R-NN analysis for actual and randomized data sets
While considering the overlap between the datasets, it is also useful to compare the R-NN analysis for these datasets with one performed on a randomized dataset, in other words a computational control experiment. As an example, we consider the DrugBank dataset, characterized by the physicochemical descriptors. We generated six new descriptors using random uniform numbers in the ranges of each original descriptor. If we then perform an R-NN analysis of the random dataset and plot the histogram of the Rmax(S) values we see that it is approximately centered upon Rmax(S) = 50, as one might expect if the points are uniformly distributed within the space (Figure S1 in Supporting Information). It should be noted that this analysis is not completely rigorous, since the random values were not obtained from a true multivariate distribution. When the original DrugBank dataset is used to generate the histogram, the distribution of Rmax(S) values is significantly different (Figure S2 in Supporting Information). Another interesting property that can be evaluated is the maximum observed pairwise observed distance versus the theoretically maximum distance (i.e., the distance between two hypothetical points whose coordinates would be the minimum values of the descriptors and the maximum values of the descriptors, respectively). For the DrugBank dataset characterized using physicochemical descriptors these values are 9.20 and 13.61 respectively. For the random version of this dataset, the values are 6.87 and 8.48 respectively. These values suggest that the distribution of compounds in the physicochemical space of the DrugBank dataset are unsurprisingly, not distributed uniformly. One could further empirically evaluate a p-value for these measurements. Given the focus of the paper on the analysis of combinatorial libraries we do not further consider further analysis of the R-NN method itself.
Scaffolds – molecular frameworks
Table 3 summarizes the number of cyclic systems in each database and the fraction of cyclic systems relative to the number of molecules in the data set. The number and fraction of singletons (e.g., cyclic systems with only one molecule) are also reported. Drugs collected in DrugBank have the largest proportion of cyclic systems and the largest proportion of singletons (39.7% and 23%, respectively) followed by the MLSMR (29.4% and 14.1%, respectively) and natural products (17.9% and 7.2%, respectively). This suggests a larger degree of cyclic system diversity in drugs as compared to MLSMR and natural products. The surprising lower scaffold diversity of natural products may be explained by the contents of the natural products collection available in the ZINC database (which also includes natural products derivatives). Not surprisingly, the four combinatorial libraries have a much lower proportion of cyclic systems and singletons than the other databases (Table 3) suggesting a lower framework diversity. Interestingly, the 738,192 compounds in library I are distributed in only 2,250 cyclic systems and just a small proportion of the molecules are singletons with their own cyclic system (Table 3). In fact, out of the four combinatorial libraries and all other collections, library I has the lowest proportion of cyclic systems and singletons (0.3% and 0.0018%, respectively), even though it is the largest library (Table 1).
Table 3.
Scaffold Analysis: Cyclic Systems and Singletons Cyclic Systems
| Library | Cyclic systems |
Singletons |
||
|---|---|---|---|---|
| Number | Percentage relative to library size | Percentage relative to library size | Percentage relative to cyclic systems | |
| Drugs | 591 | 39.7 | 23.0 | 58.0 |
| Natural products | 15,968 | 17.9 | 7.2 | 40.1 |
| MLSMR | 73,551 | 29.4 | 14.1 | 48.1 |
| I | 2,250 | 0.3 | 0.0018 | 0.6 |
| II | 1,620 | 3.5 | 0.4 | 10.4 |
| III | 472 | 1.5 | 0.05 | 3.2 |
| IV | 336 | 9.5 | 5.3 | 55.7 |
Table 3 also presents the fraction of singletons relative to the number of cyclic systems. Notably, roughly half of the cyclic systems in the drugs, MSLMR and library IV are singletons (58.0%, 48.1% and 55.7%, respectively). In contrast, the cyclic systems for library I have a low proportion of singletons (0.6%).
To note, neither the intra-cyclic system structural similarity nor the similarity due to the side chains is considered here. However, structural similarity using fingerprints described later in this paper further confirmed the large structural diversity in the drugs collection.
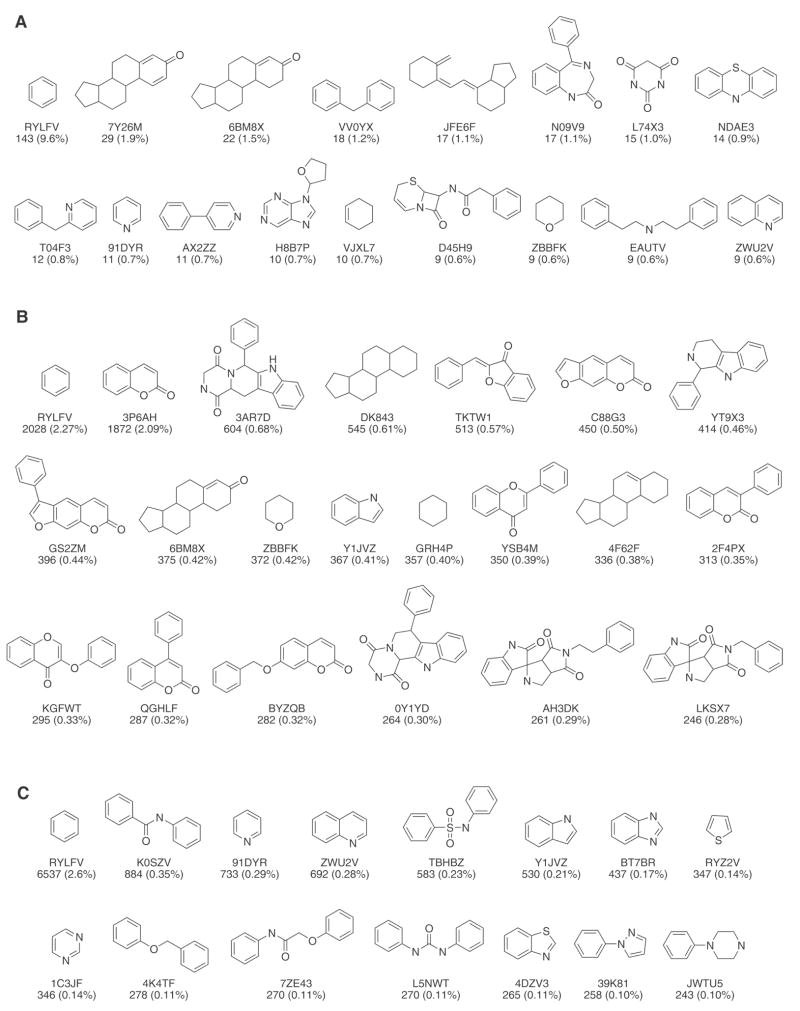
Figure 6 depicts the most frequent cyclic systems in the drugs, natural products and MLSMR collections along with the corresponding chemotype identifier. All cyclic systems with a frequency of at least 9 molecules for drugs and a frequency of more than 240 molecules for natural products and MLSMR are shown. Acyclic structures, not indicated in Figure 6, account for a relatively large proportion of structures in each collection: 92 (6.2%) in drugs; 639 (0.7%) in natural products and 460 (0.2%) in MLSMR.
Figure 6.
Most frequent cyclic systems (molecular frameworks) found in (A) drugs, (B) natural products and (C) MLSMR collections studied in this work. Chemotype identifier, frequency and percentage are displayed.
For drugs in DrugBank the 17 cyclic systems in Figure 6, in addition to the acyclic structures, account for 31% of the database. The most frequent cyclic system is benzene as previously reported for other collection of drugs.38 In fact, benzene is also the most frequent cyclic system in natural products and MLSMR (Figure 6). For natural products considered here, the 21 cyclic systems in Figure 6 in addition to the acyclic systems accounts for 13% of the collection. For the MLSMR the 15 cyclic systems (Figure 6) along with the acyclic structures accounts for 5% of the database.
Other than benzene, several other cyclic systems were found common in at least two of the databases. For example, tetrahydropyran (chemotype identifier ZBBFK) is common to drugs and natural products (population of 0.60% and 0.42%, respectively); pyridine (chemotype identifier 91DYR) is common to drugs and MLSMR (population of 0.70% and 0.29%, respectively); and indole (chemotype identifier Y1JVZ) is common to natural products and MLSMR compounds (population of 0.41% and 0.21%, respectively) (Figure 6).
Figure 7 presents the most frequent cyclic systems of the combinatorial libraries considered in this work. The corresponding chemotype identifier is also shown. Together the depicted cyclic systems retrieve ~20% of each combinatorial library. Overall a very small number of cyclic systems account for ~1/5 of each database: 5 cyclic systems for libraries I and II, 2 cyclic systems for library III and 1 for library IV (Figure 7). Interestingly, the 5 cyclic systems shown in Figure 7 accounts for 22% of the library I, and the same amount of cyclic systems accounts for 20% of the library II, which is a much smaller library compared to I. Note also the different nature of the scaffolds (e.g., molecular frameworks) of the combinatorial libraries, particularly I and II as compared to the scaffolds of drugs, natural products and MLSRM (Figure 6). Noteworthy, none of the cyclic systems in Figure 7 are common to drugs, MLSMR and natural products.
Figure 7.
Most frequent cyclic systems (molecular frameworks) derived from combinatorial libraries (A) I, (B) II, (C) III and (D) IV. Chemotype identifier, frequency and percentage are shown. Cyclic systems shown recover ~20% of each combinatorial library.
Fingerprints
Library diversity
Table 4 summarizes the mean, standard deviation and median molecular similarity of each collection considering three different structural representations namely MACCS keys, GpiDAPH3 and TGD as described in Methods. In a previous work, to define consensus activity cliffs we have shown the significance of using multiple structural representations to get generalized (i.e., consensus) solutions.28 To compute the intra-molecular similarity of each library we considered 5 random samples of 1,000 molecules each. The distribution of similarities within each random sample was very similar per library for the three fingerprints (see Supporting Information), indicating that random samples of 1,000 molecules are good representatives of the molecular diversity as previously suggested.43 Noteworthy, these observations hold true not only for the combinatorial libraries but also for drugs, natural products and MLSMR collections. Similar conclusions were obtained by analyzing the physiochemical properties distributions of random samples with 1,000 compounds (also summarized in Supporting Information).
Table 4.
Molecular Diversity of Compound Collections Using Different Molecular Representations
| Librarya | MACCS | GpiDAPH3 | TGD | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Mean | Stdev | Median | Mean | Stdev | Median | Mean | Stdev | Median | |
| Drugs | 0.311 | 0.126 | 0.300 | 0.134 | 0.154 | 0.000 | 0.521 | 0.133 | 0.528 |
| Natural products | 0.392 | 0.139 | 0.388 | 0.235 | 0.157 | 0.261 | 0.616 | 0.122 | 0.627 |
| MLSMR | 0.400 | 0.112 | 0.395 | 0.230 | 0.147 | 0.259 | 0.690 | 0.098 | 0.697 |
| I | 0.797 | 0.073 | 0.793 | 0.703 | 0.078 | 0.702 | 0.938 | 0.035 | 0.940 |
| II | 0.778 | 0.078 | 0.774 | 0.746 | 0.068 | 0.746 | 0.926 | 0.061 | 0.945 |
| III | 0.781 | 0.081 | 0.774 | 0.564 | 0.114 | 0.565 | 0.894 | 0.060 | 0.900 |
| IV | 0.782 | 0.073 | 0.776 | 0.651 | 0.085 | 0.642 | 0.932 | 0.036 | 0.933 |
Random sets with 1,000 molecules each except drugs (complete library).
Overall, similarity values in Table 4 obtained with TGD are higher than similarity values computed with MACCS keys. While values calculated with GpiDAPH3 had the lowest similarity of the three fingerprints. This is related to the intrinsic resolution of the fingerprints.
Molecular diversity considering the different fingerprints indicates that drugs from DrugBank are the most diverse, with the lowest mean and median similarities for MACCS keys, GpiDAPH3 and TGD. The second most diverse databases are natural products considered in this study and MLSMR with similar mean and median similarities for MACCS keys and GpiDAPH3, respectively. Not surprisingly, combinatorial libraries have the largest mean and median similarities for the three fingerprints (with the lowest standard deviation in the three cases) indicating less diversity than drugs, natural products and MLSMR (Table 4). According to MACCS keys, the four combinatorial libraries have approximately the same diversity. In comparison, library I is slightly less diverse than the others (i.e., median similarity of 0.79 vs. 0.77 for other combinatorial libraries with a standard deviation of 0.07). According to GpiDAPH3, which seem to have higher resolution than MACCS keys, out of the four combinatorial libraries, III is the most diverse (median similarity of 0.56 and standard deviation of 0.11) and I and II the least (median similarity of 0.70 and 0.75, respectively with standard deviation of 0.08 and 0.07, respectively). Similar conclusions are obtained considering the TGD fingerprints, although with higher similarity values probably due to the lesser resolution of this type of fingerprints (vide supra). Interestingly, the collection of drugs also showed the largest cyclic systems diversity (vide supra). Similar conclusions can be drawn for natural products and MLSMR. As opposed to these collections, combinatorial libraries have not only low cyclic system diversity but also low structural diversity considering the entire molecules. In other words, combinatorial libraries are packed more densely within the chemical space (vide supra).
MFS maps
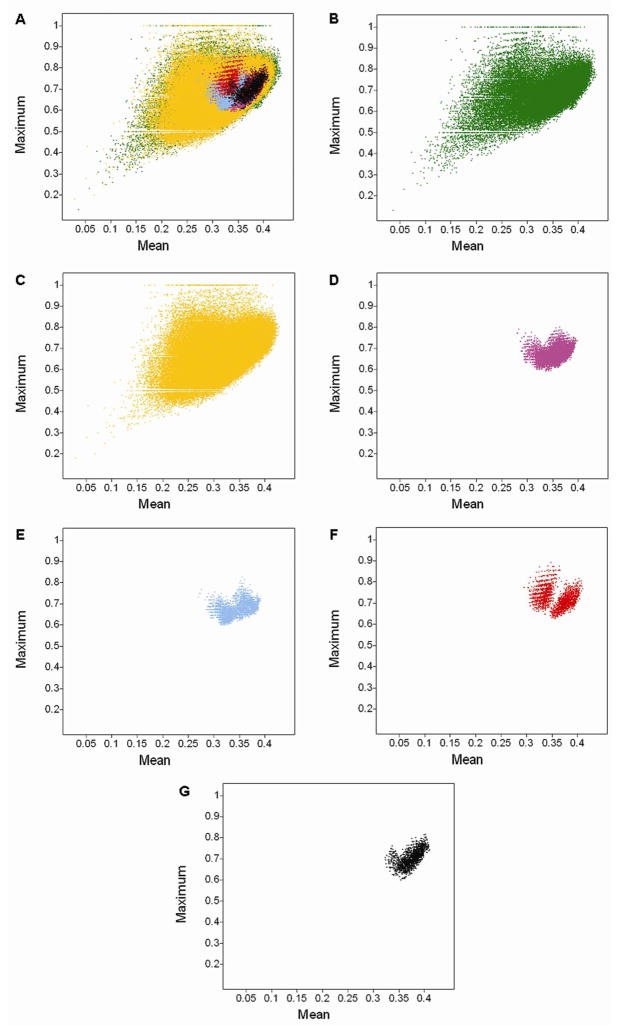
The four combinatorial libraries were also compared to drugs by means of the MFS maps using MACCS keys and GpiDAPH3. Natural products and MLSMR were included in the comparison. TGD was not considered in this analysis because of the lower resolution (vide supra). As summarized in Methods, and detailed elsewhere,44 MFS maps characterize the relationship of test molecules (e.g., combinatorial libraries) to a set of reference molecules (e.g., drugs). Figure 8 depicts the MFS maps comparing the four combinatorial libraries, natural products and MLSMR to drugs using MACCS keys. A quantitative characterization of the MFS maps is presented in Table 5. The table summarizes the maximum and mean similarities distributions of each database17 for MACCS keys and GpiDAPH3. The low mean similarity for all compounds in the libraries for MACCS keys (maximum mean similarity < 0.45) and GpiDAPH3 (maximum mean similarity < 0.29) (Table 5) indicates the large diversity of the reference set (i.e., drugs).44
Figure 8.
Multi-fusion similarity maps comparing six compound collections (test sets) to drugs (reference set) using MACCS keys. (A) All libraries; (B) natural products; (C) MLSMR; (D) library I; (E) library II; (F) library III; (G) library IV. A quantitative characterization of the plots is presented in Table 5.
Table 5.
Distribution of Maximum and Mean Similarities of Six Libraries to Drugs
| MACCS keys | |||||||
|---|---|---|---|---|---|---|---|
| Maximum similarity | |||||||
| Library | Max | Q3a | Median | Q1b | Min | Mean | Stdev |
| Natural products | 1.00 | 0.74 | 0.69 | 0.65 | 0.13 | 0.70 | 0.08 |
| MLSMR | 1.00 | 0.72 | 0.67 | 0.62 | 0.18 | 0.67 | 0.07 |
| I | 0.80 | 0.69 | 0.67 | 0.65 | 0.59 | 0.67 | 0.03 |
| II | 0.83 | 0.69 | 0.66 | 0.65 | 0.60 | 0.67 | 0.03 |
| III | 0.89 | 0.74 | 0.72 | 0.69 | 0.63 | 0.72 | 0.04 |
| IV | 0.81 | 0.73 | 0.70 | 0.67 | 0.60 | 0.70 | 0.04 |
| Mean similarity | |||||||
| Natural products | 0.44 | 0.38 | 0.35 | 0.30 | 0.04 | 0.33 | 0.06 |
| MLSMR | 0.43 | 0.35 | 0.32 | 0.29 | 0.03 | 0.32 | 0.04 |
| I | 0.40 | 0.36 | 0.35 | 0.33 | 0.28 | 0.35 | 0.02 |
| II | 0.39 | 0.36 | 0.33 | 0.32 | 0.27 | 0.34 | 0.02 |
| III | 0.41 | 0.38 | 0.35 | 0.33 | 0.29 | 0.35 | 0.02 |
| IV | 0.41 | 0.38 | 0.37 | 0.36 | 0.32 | 0.37 | 0.02 |
|
| |||||||
| GpiDAPH3 | |||||||
| Maximum similarity | |||||||
| Library | Max | Q3a | Median | Q1 b | Min | Mean | Stdev |
|
| |||||||
| Natural products | 1.00 | 0.58 | 0.53 | 0.49 | 0.00 | 0.54 | 0.09 |
| MLSMR | 1.00 | 0.55 | 0.51 | 0.47 | 0.00 | 0.51 | 0.07 |
| I | 0.57 | 0.51 | 0.49 | 0.47 | 0.33 | 0.48 | 0.03 |
| II | 0.59 | 0.55 | 0.53 | 0.51 | 0.39 | 0.53 | 0.03 |
| III | 0.60 | 0.51 | 0.48 | 0.45 | 0.28 | 0.48 | 0.05 |
| IV | 0.58 | 0.52 | 0.49 | 0.48 | 0.42 | 0.50 | 0.03 |
| Mean similarity | |||||||
| Natural products | 0.28 | 0.21 | 0.17 | 0.13 | 0.00 | 0.17 | 0.06 |
| MLSMR | 0.28 | 0.17 | 0.14 | 0.10 | 0.00 | 0.14 | 0.05 |
| I | 0.19 | 0.16 | 0.16 | 0.15 | 0.10 | 0.16 | 0.01 |
| II | 0.20 | 0.17 | 0.17 | 0.16 | 0.13 | 0.17 | 0.01 |
| III | 0.19 | 0.14 | 0.12 | 0.10 | 0.00 | 0.12 | 0.03 |
| IV | 0.21 | 0.17 | 0.14 | 0.12 | 0.07 | 0.14 | 0.03 |
Q3: third quartile.;
Q1: first quartile.
From the MFS maps in Figure 8, it is observed that there are no molecules in the combinatorial libraries with maximum similarity to drugs equal or greater than 0.90. Only library III has a few molecules (158) with maximum MACCS keys similarity greater than 0.85 to any of the drugs in DrugBank. This reveals the overall structural novelty represented in the combinatorial libraries as compared to drugs. Concerning structural novelty, similar conclusions can be derived from the molecular framework analysis (vide supra). In contrast, the natural products and MLSMR have a significant number of molecules that are very similar to drugs. There are 1,617 compounds in the natural products collection with maximum MACCS keys similarity to any of the drug equal or greater than 0.90 and 399 molecules with similarity of 1.0. (Similarity of 1.0 does not necessarily mean that the molecules are identical and could be an effect of the resolution of the MACCS keys). Concerning the MLSMR, there are 1,115 molecules with MACCS keys similarity to drugs equal or greater than 0.90 and 383 compounds with similarity of 1.0.
From Figure 8 and Table 5, it is clearly observed that the widespread distribution of data points in the MFS maps for natural products and MLSMR are in agreement with the larger structural diversity of these two collections (Table 4). In fact, natural products and MLSMR have the largest standard deviation and range of maximum and mean MACCS keys and GpiDAPH3 similarities (Table 5). In contrast, the more constrained distribution of maximum and mean MACCS keys and GpiDAPH3 similarities of the four combinatorial libraries are in agreement with the more restricted diversity of the combinatorial libraries (e.g., high density) as discussed in terms of property and scaffold diversity (vide supra). Among the combinatorial libraries, III has slightly higher maximum and mean MACCS key similarities to drugs as deduced from Figure 8 and Table 5. According to GpiDAPH3, library II shows slightly higher similarity to drugs than other combinatorial libraries.
CONCLUSIONS
In this study we present a comprehensive assessment of a novel series of combinatorial libraries using diverse criteria including physicochemical properties, scaffolds and three different fingerprints. Combinatorial libraries were compared to a collection of drugs, compounds from MLSMR and natural products (as implemented in the ZINC database). Analysis of the property space by means of PCA shows that combinatorial libraries studied here have different degree of overlap with the property space of drugs. In addition, combinatorial libraries occupy regions of the property space that are poorly or not populated by the MLSMR and natural products. Comparison of the combinatorial libraries with drugs using the R-NN curve methodology provided a quantitative analysis of the degree of overlap between these collections. The R-NN analysis revealed that a large proportion of molecules of several libraries are located within the property space of the collection of drugs. In agreement with the visualization of the property space obtained with PCA, the R-NN analysis also showed that there is a large proportion of molecules in sparse regions (“holes”) of the DrugBank collection mainly molecules from library I (pyrrolidine bis-cyclic guanidine library) followed by compounds in library II (bis-cyclic guanidine library). The scaffold analysis indicated that frequent molecular frameworks in the combinatorial libraries are not common to drugs, natural products and the MLSRM collection. In addition, the fingerprints-based comparisons using MFS maps indicate that combinatorial libraries are structurally different to drugs. The low scaffold and fingerprint-based diversity of combinatorial libraries evaluated in this work represent an advantageous feature of screening these collections, especially using fast and low cost screening approaches (e.g., positional scanning synthetic combinatorial libraries), since the high density coverage of the chemical space would increase the potential of detecting activity cliffs and be beneficial to better understand the structure-activity relationships associated with novel targets.
During the course of this work it was found that random samples of combinatorial libraries containing 1,000 molecules resemble the property and inter-molecular similarity distribution of the complete library. This observation is useful to develop large combinatorial-based libraries for virtual screening and other chemoinformatic applications.
Supplementary Material
Acknowledgments
Authors are grateful to Dr. Karina Martínez-Mayorga, Dr. Mark Johnson and Dr. Andreas Bender for helpful discussions and to the anonymous reviewers for their comments. We also acknowledge Kyle Kryak for technical assistance. This work was supported by the State of Florida, Executive Office of the Governor’s Office of Tourism, Trade, and Economic Development. Authors are also grateful to the National Institute on Drug Abuse (DA019620) and to the Multiple Sclerosis National Research Institute for partial funding. We thank Dr. Mark Johnson, Pannanugget Consulting, LLC, for providing the program MEQI.
Footnotes
Supporting Information Available: Properties distribution and similarity analysis of random sets (Tables S1 and S2, respectively). Histogram of Rmax(S) values for the DrugBank collection with respect to itself using six random and actual physicochemical descriptors (Figures S1 and S2, respectively). This material is available free of charge via the Internet at http://pubs.acs.org.
REFERENCES AND NOTES
- 1.Scior T, Bernard P, Medina-Franco JL, Maggiora GM. Large Compound Databases for Structure-Activity Relationships Studies in Drug Discovery. Mini-Rev Med Chem. 2007;7:851–860. doi: 10.2174/138955707781387858. [DOI] [PubMed] [Google Scholar]
- 2.Hopkins AL. Network Pharmacology: The Next Paradigm in Drug Discovery. Nat Chem Biol. 2008;4:682–690. doi: 10.1038/nchembio.118. [DOI] [PubMed] [Google Scholar]
- 3.Austin CP, Brady LS, Insel TR, Collins FS. Molecular Biology: NIH Molecular Libraries Initiative. Science. 2004;306:1138–1139. doi: 10.1126/science.1105511. [DOI] [PubMed] [Google Scholar]
- 4.PubChem. [accessed Feb 21, 2009]; Available at: http://pubchem.ncbi.nlm.nih.gov.
- 5.Irwin JJ, Shoichet BK. ZINC - a Free Database of Commercially Available Compounds for Virtual Screening. J Chem Inf Model. 2005;45:177–182. doi: 10.1021/ci049714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Houghten RA, Pinilla C, Giulianotti MA, Appel JR, Dooley CT, Nefzi A, Ostresh JM, Yu YP, Maggiora GM, Medina-Franco JL, Brunner D, Schneider J. Strategies for the Use of Mixture-Based Synthetic Combinatorial Libraries: Scaffold Ranking, Direct Testing, in Vivo, and Enhanced Deconvolution by Computational Methods. J Comb Chem. 2008;10:3–19. doi: 10.1021/cc7001205. [DOI] [PubMed] [Google Scholar]
- 7.Kennedy JP, Williams L, Bridges TM, Daniels RN, Weaver D, Lindsley CW. Application of Combinatorial Chemistry Science on Modern Drug Discovery. J Comb Chem. 2008;10:345–354. doi: 10.1021/cc700187t. [DOI] [PubMed] [Google Scholar]
- 8.Harvey AL. Natural Products in Drug Discovery. Drug Discovery Today. 2008;13:894–901. doi: 10.1016/j.drudis.2008.07.004. [DOI] [PubMed] [Google Scholar]
- 9.Fitzgerald SH, Sabat M, Geysen HM. Diversity Space and Its Application to Library Selection and Design. J Chem Inf Model. 2006;46:1588–1597. doi: 10.1021/ci060066z. [DOI] [PubMed] [Google Scholar]
- 10.Pinilla C, Appel JR, Borras E, Houghten RA. Advances in the Use of Synthetic Combinatorial Chemistry: Mixture-Based Libraries. Nat Med. 2003;9:118–122. doi: 10.1038/nm0103-118. [DOI] [PubMed] [Google Scholar]
- 11.Houghten RA, Pinilla C, Appel JR, Blondelle SE, Dooley CT, Eichler J, Nefzi A, Ostresh JM. Mixture-Based Synthetic Combinatorial Libraries. J Med Chem. 1999;42:3743–3778. doi: 10.1021/jm990174v. [DOI] [PubMed] [Google Scholar]
- 12.Macarron R. Critical Review of the Role of HTS in Drug Discovery. Drug Discovery Today. 2006;11:277–279. doi: 10.1016/j.drudis.2006.02.001. [DOI] [PubMed] [Google Scholar]
- 13.Carpenter AE. Image-Based Chemical Screening. Nat Chem Biol. 2007;3:461–465. doi: 10.1038/nchembio.2007.15. [DOI] [PubMed] [Google Scholar]
- 14.Fitzgerald SH, Sabat M, Geysen HM. Survey of the Diversity Space Coverage of Reported Combinatorial Libraries. J Comb Chem. 2007;9:724–734. doi: 10.1021/cc0601579. [DOI] [PubMed] [Google Scholar]
- 15.Shelat AA, Guy RK. The Interdependence between Screening Methods and Screening Libraries. Curr Opin Chem Biol. 2007;11:244–251. doi: 10.1016/j.cbpa.2007.05.003. [DOI] [PubMed] [Google Scholar]
- 16.Shanmugasundaram V, Maggiora GM, Lajiness MS. Hit-Directed Nearest-Neighbor Searching. J Med Chem. 2005;48:240–248. doi: 10.1021/jm0493515. [DOI] [PubMed] [Google Scholar]
- 17.Medina-Franco JL, Martínez-Mayorga K, Giulianotti MA, Houghten RA, Pinilla C. Visualization of the Chemical Space in Drug Discovery. Curr Comput -Aided Drug Des. 2008;4:322–333. [Google Scholar]
- 18.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Experimental and Computational Approaches to Estimate Solubility and Permeability in Drug Discovery and Development Settings. Adv Drug Delivery Rev. 1997;23:3–25. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 19.Teague SJ, Davis AM, Leeson PD, Oprea T. The Design of Leadlike Combinatorial Libraries. Angew Chem, Int Ed. 1999;38:3743–3748. doi: 10.1002/(SICI)1521-3773(19991216)38:24<3743::AID-ANIE3743>3.0.CO;2-U. [DOI] [PubMed] [Google Scholar]
- 20.Congreve M, Carr R, Murray C, Jhoti H. A ‘Rule of Three’ for Fragment-Based Lead Discovery? Drug Discovery Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 21.Schneider G, Neidhart W, Giller T, Schmid G. Scaffold-Hopping by Topological Pharmacophore Search: A Contribution to Virtual Screening. Angew Chem, Int Ed. 1999;38:2894–2896. [PubMed] [Google Scholar]
- 22.Evans BE, Rittle KE, Bock MG, DiPardo RM, Freidinger RM, Whitter WL, Lundell GF, Veber DF, Anderson PS, et al. Methods for Drug Discovery: Development of Potent, Selective, Orally Effective Cholecystokinin Antagonists. J Med Chem. 1988;31:2235–2246. doi: 10.1021/jm00120a002. [DOI] [PubMed] [Google Scholar]
- 23.Mason JS, Morize I, Menard PR, Cheney DL, Hulme C, Labaudiniere RF. New 4-Point Pharmacophore Method for Molecular Similarity and Diversity Applications: Overview of the Method and Applications, Including a Novel Approach to the Design of Combinatorial Libraries Containing Privileged Substructures. J Med Chem. 1999;42:3251–3264. doi: 10.1021/jm9806998. [DOI] [PubMed] [Google Scholar]
- 24.Villar HO, Hansen MR, Kho R. Substructural Analysis in Drug Discovery. Curr Comput -Aided Drug Des. 2007;3:59–67. [Google Scholar]
- 25.Johnson MA, Maggiora GM, editors. Concepts and Applications of Molecular Similarity. John Wiley & Sons; New York: 1990. [Google Scholar]
- 26.Bender A, Glen RC. Molecular Similarity: A Key Technique in Molecular Informatics. Org Biomol Chem. 2004;2:3204–3218. doi: 10.1039/B409813G. [DOI] [PubMed] [Google Scholar]
- 27.Willett P. Similarity-Based Virtual Screening Using 2D Fingerprints. Drug Discovery Today. 2006;11:1046–1053. doi: 10.1016/j.drudis.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 28.Medina-Franco JL, Martínez-Mayorga K, Bender A, Marin R, Giulianotti M, Pinilla C, Houghten R. Characterization of Activity Landscapes Using 2D and 3D Similarity Methods: Consensus Activity Cliffs. J Chem Inf Model. 2009 doi: 10.1021/ci800379q. [DOI] [PubMed] [Google Scholar]
- 29.MACCS Structural Keys. MDL Information Systems Inc.; San Leandro, CA: [Google Scholar]
- 30.Molecular Operating Environment (MOE), version 2007. Chemical Computing Group Inc.; Montreal, Quebec, Canada: [(accessed Feb 21, 2009)]. Available at: http://www.chemcomp.com. [Google Scholar]
- 31.Guha R, Dutta D, Jurs PC, Chen T. R-NN Curves: An Intuitive Approach to Outlier Detection Using a Distance Based Method. J Chem Inf Model. 2006;46:1713–1722. doi: 10.1021/ci060013h. [DOI] [PubMed] [Google Scholar]
- 32.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. Drugbank: A Knowledgebase for Drugs, Drug Actions and Drug Targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Spotfire, version 9.1.1. TIBCO Software, Inc.; Somerville, MA: [(accessed Feb 21, 2009)]. Available at: http://spotfire.tibco.com. [Google Scholar]
- 34.Guttman A. SIGMOD Conference. ACM Press; 1984. R-Trees: A Dynamic Index Structure for Spatial Searching; pp. 47–57. [Google Scholar]
- 35.Cormen TH, Leiserson CE, Rivest RL, Stein C. Introduction to Algorithms. 2. MIT Press; 2001. Binary Search Trees. [Google Scholar]
- 36.Xu Y, Johnson M. Algorithm for Naming Molecular Equivalence Classes Represented by Labeled Pseudographs. J Chem Inf Comput Sci. 2001;41:181–185. doi: 10.1021/ci0003911. [DOI] [PubMed] [Google Scholar]
- 37.Xu YJ, Johnson M. Using Molecular Equivalence Numbers to Visually Explore Structural Features That Distinguish Chemical Libraries. J Chem Inf Comput Sci. 2002;42:912–926. doi: 10.1021/ci025535l. [DOI] [PubMed] [Google Scholar]
- 38.Bemis GW, Murcko MA. The Properties of Known Drugs. 1. Molecular Frameworks. J Med Chem. 1996;39:2887–2893. doi: 10.1021/jm9602928. [DOI] [PubMed] [Google Scholar]
- 39.Medina-Franco JL, Petit J, Maggiora GM. Hierarchical Strategy for Identifying Active Chemotype Classes in Compound Databases. Chem Biol Drug Des. 2006;67:395–408. doi: 10.1111/j.1747-0285.2006.00397.x. [DOI] [PubMed] [Google Scholar]
- 40.Renner S, Schneider G. Scaffold-Hopping Potential of Ligand-Based Similarity Concepts. ChemMedChem. 2006;1:181–185. doi: 10.1002/cmdc.200500005. [DOI] [PubMed] [Google Scholar]
- 41.Schreyer SK, Parker CN, Maggiora GM. Data Shaving: A Focused Screening Approach. J Chem Inf Comput Sci. 2004;44:470–479. doi: 10.1021/ci030025s. [DOI] [PubMed] [Google Scholar]
- 42.Cases M, Garcia-Serna R, Hettne K, Weeber M, van der Lei J, Boyer S, Mestres J. Chemical and Biological Profiling of an Annotated Compound Library Directed to the Nuclear Receptor Family. Curr Top Med Chem. 2005;5:763–772. doi: 10.2174/1568026054637665. [DOI] [PubMed] [Google Scholar]
- 43.Agrafiotis DK. A Constant Time Algorithm for Estimating the Diversity of Large Chemical Libraries. J Chem Inf Comput Sci. 2001;41:159–167. doi: 10.1021/ci000091j. [DOI] [PubMed] [Google Scholar]
- 44.Medina-Franco JL, Maggiora GM, Giulianotti MA, Pinilla C, Houghten RA. A Similarity-Based Data-Fusion Approach to the Visual Characterization and Comparison of Compound Databases. Chem Biol Drug Des. 2007;70:393–412. doi: 10.1111/j.1747-0285.2007.00579.x. [DOI] [PubMed] [Google Scholar]
- 45.Martínez-Mayorga K, Medina-Franco JL, Giulianotti MA, Pinilla C, Dooley CT, Appel JR, Houghten RA. Conformation-Opioid Activity Relationships of Bicyclic Guanidines from 3d Similarity Analysis. Bioorg Med Chem. 2008;16:5932–5938. doi: 10.1016/j.bmc.2008.04.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Fink T, Reymond JL. Virtual Exploration of the Chemical Universe up to 11 Atoms of C, N, O, F: Assembly of 26.4 Million Structures (110.9 Million Stereoisomers) and Analysis for New Ring Systems, Stereochemistry, Physicochemical Properties, Compound Classes, and Drug Discovery. J Chem Inf Model. 2007;47:342–353. doi: 10.1021/ci600423u. [DOI] [PubMed] [Google Scholar]
- 47.The upper adjacent value is defined as the largest observation that is less than or equal to the upper whisker limit. The upper whisker limit is the third quartile plus 1.5 times the interquartile range (third quartile minus first quartile). The lower adjacent value is the smallest observation that is greater than or equal to the lower whisker limit, which is the first quartile minus 1.5 times the interquartile range.
- 48.Lee ML, Schneider G. Scaffold Architecture and Pharmacophoric Properties of Natural Products and Trade Drugs: Application in the Design of Natural Product-Based Combinatorial Libraries. J Comb Chem. 2001;3:284–289. doi: 10.1021/cc000097l. [DOI] [PubMed] [Google Scholar]
- 49.Feher M, Schmidt JM. Property Distributions: Differences between Drugs, Natural Products, and Molecules from Combinatorial Chemistry. J Chem Inf Comput Sci. 2003;43:218–227. doi: 10.1021/ci0200467. [DOI] [PubMed] [Google Scholar]
- 50.Kong DX, Li XJ, Zhang HY. Where Is the Hope for Drug Discovery? Let History Tell the Future. Drug Discovery Today. 2009;14:115–119. doi: 10.1016/j.drudis.2008.07.002. [DOI] [PubMed] [Google Scholar]
- 51.Nefzi A, Ong NA, Giulianotti MA, Ostresh JM, Houghten RA. Solid Phase Synthesis of 1,4-Benzothiazepin-5-One Derivatives. Tetrahedron Lett. 1999;40:4939–4942. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.