Abstract
Purpose of review
This review is to highlight the most current mutation-targeted therapeutic approaches and provide insights into new developments for treating primary immunodeficiencies.
Recent findings
Significant progress in mutation-targeted treatment was achieved in the past year with the identification and characterization of a translational read-through compound, PTC124. PTC124 demonstrates a new class of nontoxic bioavailable small drugs. Antisense oligonucleotide-mediated techniques such as splicing redirection, exon skipping, and mismatch repair have been successfully used to correct splicing, frameshift, and missense mutations, respectively. Delivery of antisense oligonucleotides to mammalian cells, including primary leukocytes and neurons, saw great progress during the past year. Recent advances for other approaches to correct frameshift and missense mutations are also considered.
Summary
Primary immunodeficiencies are monogenic disorders. The characterization and classification of disease-causing mutations facilitate the design and development of new mutation-targeted treatments. To date, using ataxia–telangiectasia (A–T) as a model primary immunodeficiency, the most promising advances have been with chemicals that read through various premature stop codons as well as with antisense oligonucleotides that mask aberrant splice sites. These principles can now be applied to other primary immunodeficiencies.
Keywords: antisense oligonucleotides, mutation-targeted therapy, pharmacogenetic treatment, primary immunodeficiencies, read-through
Introduction
Primary immunodeficiencies (PIDs) are genetically inherited disorders characterized by deficiencies of distinct components of the innate or adaptive immune systems, the clinical hallmark being an increased susceptibility to infections. With great progress having been made in delineating the innate and adaptive immune systems, novel forms of PIDs have been recognized and their molecular bases have been characterized [1,2]. More than 120 distinct genes have been identified to date, accounting for more than 150 different forms of PIDs [3•]. The characterization of distinct genes and their mutations for each PID further defines the word ‘primary’ and prompts both the design of more accurate diagnostic tools and the new opportunities for therapy aimed at correcting the genetic errors at the DNA or RNA level, before their ramifications become systemic.
Current available treatments for PIDs include intravenous immunoglobulins (IVIGs) [4], stem cell transplantation [5–7], and gene therapy [8,9]. However, each treatment approach has drawbacks. Stem cell transplantation may lead to life-threatening graft-versus-host disease (GVHD) if poorly matched donors are used, which is true more than 80% of the time [6]. Gene therapy is currently limited by technical difficulties and the risks of side effects of genomic manipulation, such as leukemia [10,11]. Novel approaches to treatment of PIDs need to be explored. Pharmacogenetic agents modify gene expression after the problem has become systemic. In this review, we highlight mutation-targeted therapies with chemicals that mitigate mutational pathology at the nuclear level. Although the principles of mutation-targeted therapy have developed primarily from non-PID genetic diseases, they can now be applied here as well.
Types of mutations and mutation-targeted therapy
Genetic mutations can be categorized into four major groups: nonsense, splicing, frameshift, and missense. These have been observed in almost all genes studied (Fig. 1). Nonsense mutations create premature stop codons (e.g. in a C>T mutation: CAG, which codes for a glutamine, becomes TAG or UAG, which stops translation at that point, resulting in a truncated unstable protein). Splicing mutations come in many variations (Fig. 2), the underlying common denomination being that a critical sequence of nucleotides is disrupted by the mutation so that its chemical entropy is no longer the optimal site for splicing of pre-mRNA. This creates aberrant or cryptic splice sites that translate to aberrant proteins. Frameshift mutations most commonly result from the insertion or deletion of one or two nucleotides, which destroy the reading frame for any subsequent downstream codons. Missense mutations are typically single-nucleotide changes that either alter the amino acid in translated proteins (nonsynonymous) or do not alter the amino acids (synonymous or ‘silent’). The potential disease-causing consequence of a missense change is often difficult to evaluate [12••]. Thus, it can be anticipated that many reported ‘missense mutations’ will later be re-evaluated as nondeleterious DNA variants or single-nucleotide polymorphisms (SNPs), which occur as frequently as one in every 100–1000 nucleotides in the human genome [13]. It should also be emphasized that mutation-based therapy requires that only one allele carries the targeted mutation. Thus, if 17% of ataxia–telangiectasia (A–T) patients carry a nonsense mutation, approximately 34% would be amenable to such read-through chemicals if all patients carried two different mutations. One can even envision patients receiving two different medicines, one for each mutation.
Figure 1. Distribution of mutations in primary immunodeficiencies.
The percentage of each type of mutation is presented for individual PIDs. The gene responsible for the primary immunodeficiency disease and the total number of reported mutations are listed at the top and bottom of each pie-chart, respectively. ATM, ataxia–telangiectasia; WASP, Wiskott–Aldrich syndrome; BTK (Burton’s tyrosine kinase), X-Linked agammaglobulinemia; ADA, adenosine deaminase – severe combined immunodeficiency (SCID); CD40L, X-linked hyper-IgM; and CYBB, X-linked chronic granulomatous disease. PID, primary immunodeficiency.
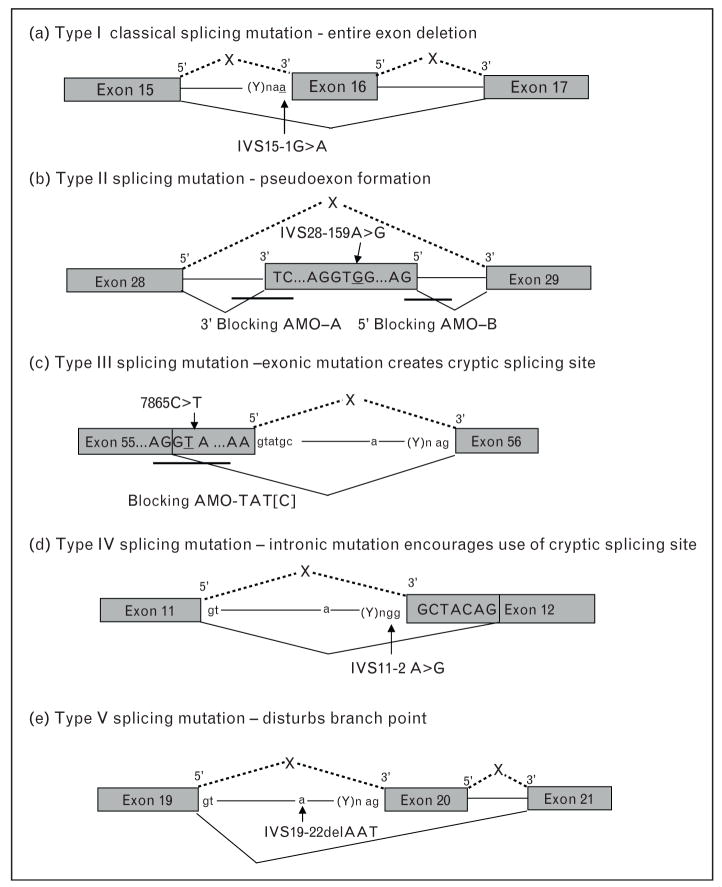
Figure 2. Types of splicing mutations and correction with antisense morpholino oligonucleotides.
(a) Type I refers to the classical splicing mutation that causes the deletion of an entire exon during pre-mRNA splicing. (b) Type II splicing mutations occur in the mid-intron and result in the insertion of a pseudoexon at the cDNA level. Blocking antisense morpholino oligonucleotides have been used to restore normal splicing. (c) Type III mutations lie within the coding region, resulting in the partial deletion of an exon. An antisense morpholino oligonucleotide has been used to block the mutation-generated 5′ splicing site. (d) Type IV mutations lie within the intron and lead to partial deletion of an exon. (e) Type V mutations disturb the lariat branch point and result in deletion of an exon.
Nonsense, splicing, and frameshift mutations usually lead to loss of or unstable protein, whereas missense mutations produce nonfunctional or partially functional proteins. In-frame insertion or deletion mutations also give rise to proteins; thus, we combined the in-frame mutations with the missense subgroup in Fig. 1. Common groups of mutations prompted the hypothesis of common group-specific molecular pathogenesis. It also follows that therapeutic strategies developed against a common mutation group will prove effective against similar mutations, irrespective of the gene. This overreaching concept of mutation-targeted therapy has been shown effective by read-through compounds that correct nonsense mutations in many disease models. A similar argument can be made for antisense oligonucleotides that correct splicing mutations. With the availability of mutation databases for most PIDs [14], we can summarize here the distribution of mutation types for some of the more common PIDs (Fig. 1). Herein lies the future of mutation-based therapy for PIDs.
Correction of nonsense mutations by read-through compounds
Approximately 1800 inherited human diseases are caused by nonsense mutations [15]. Suppressing (i.e. reading through) the resulting premature termination codons with chemicals, thereby allowing translation to continue to the true end of the transcript, is a promising approach for correcting nonsense mutations.
Proof of concept for such a therapeutic approach in a series of genetic diseases has already been demonstrated for aminoglycosides, the widely used bacterial antibiotics [16•]. Aminoglycosides bind to the decoding site of the 16S ribosomal RNA, inducing a local conformational change that allows translation through what would otherwise be read as a premature termination codon. After the aminoglycosides paromomycin and G418 were first shown to partially restore the full-length protein of nonsense mutation in mammalian cells [17], the aminoglyco-side gentamicin was tested in clinical trials for cystic fibrosis, Duchenne muscular dystrophy, and hemophilia [18–20]. Aminoglycosides have also been employed ex vivo to achieve read-through expression of full-length ATM protein in A–T [21]. These studies demonstrated that true stop codons were not affected by the read-through compounds. Studies are now underway to correct nonsense mutations in other forms of PIDs. However, the high concentration of aminoglycosides required for effective read-through is often toxic to cells, restricting their uses in clinical trials. Furthermore, most antibiotics do not cross the blood–brain barrier efficiently and would be of limited use for treating central nervous system diseases. Some attempts have been made to redesign aminoglycosides so as to reduce the toxicity while retaining the read-through activity. A newly designed compound based on paromomycin seems to have this property [22].
PTC124 is another recently identified nonsense-suppression read-through compound [23••], that is not an amino-glycoside antibiotic. PTC124 promotes suppression of human and mouse nonsense alleles of the dystrophin genes and restores ex vivo dystrophin expression. Systemic (sometimes in combination with oral and intraperitoneal) delivery of PTC124 into mouse models for Duchnne muscular dystrophy and cystic fibrosis showed restoration of protein and function in vivo without any obvious toxicity [23••,24]. Of particular interest was the in vivo demonstration of specificity for PTC124 read-through activity to the premature stop codons rather than to the true termination codons. The latter is apparently read in the context of other factors besides simply the TAG, TGA, or TAA stop codon sequences. PTC124 demonstrates a new, nontoxic, small-molecule drug that promotes selective and specific read-through of disease-causing premature stop codons. However, it should be noted that more than 800 000 chemicals are needed to be screened, and then chemically modified, in the selection of PTC124 for the clinic [23••].
In order to discover additional read-through chemicals, as well as structure–activity relationship (SAR) groups, our laboratory has developed a high throughput screen assay based on the Protein Truncation Test (PTT) [25]. We have identified more than 50 nonaminoglycoside chemicals with read-through activity. Although many of these have low EC50 (half-maximal effective concentration) scores, others are more promising, and several potential SAR groups are evident (Du et al., manuscript submitted). This will allow rapid expansion of the discovery aspect of read-through chemicals and hopefully will lead to a better understanding of the critical mechanisms underlying suppression/read-through.
Nonsense mutations activate a RNA surveillance system, known as nonsense mediated decay (NMD), to destroy PTC-containing transcripts in order to prevent the synthesis of truncated proteins that might produce dominant negative effects and waste cellular resources. NMD has been reported to be a major mechanism of nonsense transcript elimination and governs the patient’s response to read-through compounds [26••]. Because NMD also degrades the physiologically occurring PTC-containing mRNAs such as those resulted from alternative splicing, disruption of NMD might result in undesired side effects [27]. Interestingly, PTC124 and aminoglycosides have minimal effects on NMD [23••]. In theory, developing a method to specifically disrupt NMD of a disease-causing mRNA without influencing normally degraded mRNA might greatly improve the read-through efficiency of targeted nonsense mutation by read-through compounds.
Correction of splicing mutations with antisense oligonucleotides
A significant fraction (15–30%) of disease-causing mutations in mammalian genes affect pre-mRNA splicing, and antisense oligonucleotides have been successfully used to correct splicing mutations [28]. Basically, antisense oligonucleotides target pre-mRNA in a sequence-specific manner, sterically blocking the targeted splice sites and redirecting the splicing machinery to a more appropriate nearby splice site [28].
Splicing mutations can be grouped into at least five types (Fig. 2, using ATM data as an example) [29]. Type I is the classical splicing mutation resulting in deletion of an entire exon. Type II, III, and IV are nonclassical splicing mutations that result in pseudoexon inclusion or partial exon deletion. Type V involves the branch point and although such mutations exist ([30,31], unpublished), they have not been targeted for therapy. Our laboratory has used antisense morpholino oligonucelotides (AMOs) to redirect and restore normal splicing in the ATM gene for type II and type III mutations [32•]. AMOs have also been used to successfully modulate RNA splicing for many other genetic diseases including cystic fibrosis, β-thalassemia, and Duchnne muscular dystrophy [28]. As splicing mutations are a prevalent and important class of mutation in PIDs (Fig. 1), antisense oligonucleotides should be applicable for most PIDs.
To be effective, antisense oligonucleotides also need to be delivered into cell nuclei in relative high concentrations. The traditional delivery is with the aid of physical and chemical techniques, such as using the proprietary Endo-porter and cationic lipids. Significant improvements are being made in the delivery efficiency, such as the recent use of Arginine-rich, cell-penetrating peptides (CPP) or nine-arginine tags fused to rabies virus glycoprotein peptides (RVG-9R) to deliver antisense oligonucleotides directly to mammalian cells, including primary leukocytes and neurons [33,34••]. In our experiments, the correction of intranuclear ATM protein levels with CPP-mediated AMOs sometimes exceeds 50% of control levels (unpublished) – this would exceed the ATM levels of A–T carriers [35] who remain largely asymptomatic. Also encouraging, RVG-9R mediated oli-gonucleotide delivery can cross the blood–brain barrier [34••]. This has important implications for PIDs with central nervous system involvement, as exemplified by A–T, Nijmegen Breakage syndrome, SCID-Cernunnos, and DNA ligase IV deficiency [36].
Potential approaches for correction of frameshift mutations
Frameshifts resulting from nucleotide insertions or deletions account for a large fraction of mutations in commonly occurring PIDs such as A–T, Wiskott–Aldrich syndrome, and X-linked agammaglobulinemia (Fig. 1). To our knowledge, pharmacogenetic agents capable of correcting frameshift mutations at the DNA or RNA (genomic or transcriptional) level have not yet been identified. Meanwhile some promising approaches can be considered.
Gene therapy can mitigate a frameshift mutation in the interleukin 2 receptor common gamma chain (IL2Rγ), responsible for X-linked severe combined immunodeficiency (SCID) [37]. In this approach, an engineered zinc-finger nuclease fusion protein (ZFN) specifically binds and cleaves the two DNA regions flanking the insertion and enhances ex vivo homologous recombination to a potentially therapeutic level (18%), without any selection steps [37–39].
Antisense oligonucelotide-mediated exon skipping may be another potential approach for correcting frameshift mutations. The principle of this approach is to use anti-sense oligonucleotides to skip a mutation-containing exon and generate an in-frame deletion and a functional protein. This has been achieved for Duchnne muscular dystrophy using an antisense oligonucleotide PRO051 [40•]. Treated patients improved from a severe phenotype to a much milder phenotype. The antisense PRO051 produces a shortened dystrophin with partial function [40•]. This approach has two requisites: the exon skipping retains in-frame codon reading and the resultant partially truncated protein retains function. Most likely, this approach will be applicable primarily to highly repetitive genes, such as those coding for structure proteins.
Revertant mosaicism, a naturally occurring event, can also correct a frameshift by reverse mutations (i.e. revertants) or by second-site mutations that lead to the neutralization of mutant phenotype. Revertant phenotypes have been studied in several PIDs, such as X-linked SCID, Wiskott–Aldrich syndrome, Fanconi anemias, and Bloom syndrome [41–44]. The possible mechanisms responsible for reversion include intragenic recombination, mitotic gene conversion, second-site compensatory mutation, DNA slippage, and site-specific reversion [45]. Understanding these mechanisms might provide insights for developing new high throughput screening assays to discover chemicals that would encourage such events.
Potential approaches for correcting missense and in-frame mutations
Missense and in-frame mutations produce proteins with null, partial, or abnormal functions. These mutant proteins exert their deleterious effects through multiple mechanisms such as dominant negative or gain-of-function. As discussed above, however, more work remains to be done with regard to the proper diagnosis of missense mutations, as opposed to nondeleterious polymorphism [12••].
Depending on the functions of missense or in-frame mutated proteins, different approaches can be developed for treatment. For example, a phenylalanine 508 deletion (ΔF508) in cystic fibrosis patients produces a cystic fibrosis transmembrane conductance regulator (CFTR) protein with defects in folding, stability, and channel gating. Small-molecule compounds, identified by high-throughput screening, correct the channel activity of ΔF508–CFTR mutated protein [46]. Another example of gain-of-function for a missense mutation is gain of glycosylation, in which a glycan is added to the mutant amino acid. Up to 4% of known disease-causing missense mutations are predicted to give rise to gain of glycosylation [47]. The first gain-of-glycosylation mutation to be thoroughly characterized was the T168N mutation in the IFNGR2 gene, a gene causing primary immunodeficiency [48]. The identification of the large fraction of gain-of-glycosylation mutations also suggests a therapeutic approach to target these mutations, using tunicamycin, catanospermine, glycosylation inhibitors, or glycosidases [47]. New chemicals targeting glycosylation need to be further identified for specificity and nontoxicity based on the mechanisms involved [49].
Chimeric RNA/DNA oligonucleotides (chimeraplasts) may provide an alternative approach to missense mutation treatment. A well designed chimeraplast pairs perfectly and anneals to the mutated region except for a base pair mismatch. This mismatch activates the mismatch repair system, which reverses the missense mutation. The ability of chimeraplasts to correct point mutations has been demonstrated in a mouse Duchnne muscular dystrophy model for nonsense and frameshift mutations [50].
Conclusion
PTC124 was an encouraging breakthrough development for the discovery and design of read-through compounds for mitigating nonsense mutations. We envision that other promising chemicals with improved pharmacokinetics will be developed. These compounds will increase the therapeutic options for patients with nonsense mutations in PID genes. With the progress in delineating the mechanisms of NMD, it may be possible to reduce the destruction of disease-causing transcripts if, at the same time, the efficiency of translational read-through of these transcripts can be significantly improved.
Antisense oligonucleotide-mediated approaches that redirect splicing can also be applied to exon skipping, both to avoid unwanted exons and to control alternative splicing. New advances in the delivery and stability of antisense oligonucleotides can be expected [51], especially with regard to crossing the blood–brain barrier. Some antisense oligonucleotides are already in clinical trials and will soon be ready for testing in PIDs.
Acknowledgments
Work related to this article is supported by NINDS, the A–T Medical Research Foundation (Los Angeles, USA), the A–T Ease Foundation (New York, USA), the APRAT Foundation (Chermont-Ferrand, France), and the A–T Charity (U.K.).
References and recommended reading
Papers of particular interest, published within the annual period of review, have been highlighted as:
•of special interest
••of outstanding interest
Additional references related to this topic can also be found in the Current World Literature section in this issue (pp. 592–593).
- 1.Fischer A. Human primary immunodeficiency diseases. Immunity. 2007;27:835–845. doi: 10.1016/j.immuni.2007.11.012. [DOI] [PubMed] [Google Scholar]
- 2.Marodi L, Notarangelo LD. Immunological and genetic bases of new primary immunodeficiencies. Nat Rev Immunol. 2007;7:851–861. doi: 10.1038/nri2195. [DOI] [PubMed] [Google Scholar]
- 3•.Geha RS, Notarangelo LD, Casanova JL, et al. Primary immunodeficiency diseases: an update from the International Union of Immunological Societies Primary Immunodeficiency Diseases Classification Committee. J Allergy Clin Immunol. 2007;120:776–794. doi: 10.1016/j.jaci.2007.08.053. A comprehensive update on the classification of primary immunodeficiencies and the responsible genes. This is the official classification of the world body I.U.I.S. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ballow M. Safety of IGIV therapy and infusion-related adverse events. Immunol Res. 2007;38:122–132. doi: 10.1007/s12026-007-0003-5. [DOI] [PubMed] [Google Scholar]
- 5.Gatti RA, Meuwissen HJ, Allen HD, et al. Immunological reconstitution of sex-linked lymphopenic immunological deficiency. Lancet. 1968;2:1366–1369. doi: 10.1016/s0140-6736(68)92673-1. [DOI] [PubMed] [Google Scholar]
- 6.Notarangelo LD, Forino C, Mazzolari E. Stem cell transplantation in primary immunodeficiencies. Curr Opin Allergy Clin Immunol. 2006;6:443–448. doi: 10.1097/01.all.0000246616.47708.2f. [DOI] [PubMed] [Google Scholar]
- 7.Dvorak CC, Cowan MJ. Hematopoietic stem cell transplantation for primary immunodeficiency disease. Bone Marrow Transplant. 2008;41:119–126. doi: 10.1038/sj.bmt.1705890. [DOI] [PubMed] [Google Scholar]
- 8.Chinen J, Puck JM. Perspectives of gene therapy for primary immunodeficiencies. Curr Opin Allergy Clin Immunol. 2004;4:523–527. doi: 10.1097/00130832-200412000-00008. [DOI] [PubMed] [Google Scholar]
- 9.Aiuti A, Bachoud-Levi AC, Blesch A, et al. Progress and prospects: gene therapy clinical trials (part 2) Gene Ther. 2007;14:1555–1563. doi: 10.1038/sj.gt.3303033. [DOI] [PubMed] [Google Scholar]
- 10.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 11.Zhang XB, Beard BC, Trobridge GD, et al. High incidence of leukemia in large animals after stem cell gene therapy with a HOXB4-expressing retroviral vector. J Clin Invest. 2008;118:1502–1510. doi: 10.1172/JCI34371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12••.Mitui M, Nahas S, Du LT, et al. Functional and computational assessment of missense variants in the ATM gene: mutations with increased cancer risk. Hum Mut. doi: 10.1002/humu.20805. Epub ahead of print This is the first attempt to evaluate heterozygous nonsynonymous DNA changes by stably transfecting each allele into a single host cell so that functional consequences can be evaluated in detail and compared to clinical phenotypes. [DOI] [Google Scholar]
- 13.Orro A, Guffanti G, Salvi E, et al. SNPLims: a data management system for genome wide association studies. BMC Bioinformatics. 2008;9 (Suppl 2):S13. doi: 10.1186/1471-2105-9-S2-S13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Knerr V, Grimbacher B. Primary immunodeficiency registries. Curr Opin Allergy Clin Immunol. 2007;7:475–480. doi: 10.1097/ACI.0b013e3282f2162c. [DOI] [PubMed] [Google Scholar]
- 15.Kellermayer R. Translational readthrough induction of pathogenic nonsense mutations. Eur J Med Genet. 2006;49:445–450. doi: 10.1016/j.ejmg.2006.04.003. [DOI] [PubMed] [Google Scholar]
- 16•.Zingman LV, Park S, Olson TM, et al. Aminoglycoside-induced translational read-through in disease: overcoming nonsense mutations by pharmacogenetic therapy. Clin Pharmacol Ther. 2007;81:99–103. doi: 10.1038/sj.clpt.6100012. This is an excellent overview of genetic disease with nonsense mutations treated by aminoglycosides in vitro and in vivo. [DOI] [PubMed] [Google Scholar]
- 17.Burke JF, Mogg AE. Suppression of a nonsense mutation in mammalian cells in vivo by the aminoglycoside antibiotics G-418 and paromomycin. Nucleic Acids Res. 1985;13:6265–6272. doi: 10.1093/nar/13.17.6265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wilschanski M, Yahav Y, Yaacov Y, et al. Gentamicin-induced correction of CFTR function in patients with cystic fibrosis and CFTR stop mutations. N Engl J Med. 2003;349:1433–1441. doi: 10.1056/NEJMoa022170. [DOI] [PubMed] [Google Scholar]
- 19.Politano L, Nigro G, Nigro V, et al. Gentamicin administration in Duchenne patients with premature stop codon. Preliminary results Acta Myol. 2003;22:15–21. [PubMed] [Google Scholar]
- 20.James PD, Raut S, Rivard GE, et al. Aminoglycoside suppression of nonsense mutations in severe hemophilia. Blood. 2005;106:3043–3048. doi: 10.1182/blood-2005-03-1307. [DOI] [PubMed] [Google Scholar]
- 21.Lai CH, Chun HH, Nahas SA, et al. Correction of ATM gene function by aminoglycoside-induced read-through of premature termination codons. Proc Natl Acad Sci U S A. 2004;101:15676–15681. doi: 10.1073/pnas.0405155101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nudelman I, Rebibo-Sabbah A, Shallom-Shezifi D, et al. Redesign of aminoglycosides for treatment of human genetic diseases caused by premature stop mutations. Bioorg Med Chem Lett. 2006;16:6310–6315. doi: 10.1016/j.bmcl.2006.09.013. [DOI] [PubMed] [Google Scholar]
- 23••.Welch EM, Barton ER, Zhuo J, et al. PTC124 targets genetic disorders caused by nonsense mutations. Nature. 2007;447:87–91. doi: 10.1038/nature05756. This important report identified and characterized a novel translational read-through compound PTC124 that can suppress nonsense mutations of Duchnne muscular dystrophy ex vivo in human and murine. PTC124 demonstrates a new, non-toxic, and bioavailable small-molecule drug for nonsense mutations, which has many implications for other genetic diseases. [DOI] [PubMed] [Google Scholar]
- 24.Du M, Liu X, Welch EM, et al. PTC124 is an orally bioavailable compound that promotes suppression of the human CFTR-G542X nonsense allele in a CF mouse model. Proc Natl Acad Sci U S A. 2008;105:2064–2069. doi: 10.1073/pnas.0711795105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Du L, Lai CH, Concannon P, Gatti RA. Rapid screen for truncating ATM mutations by PTT-ELISA. Mutat Res. 2008;640:139–144. doi: 10.1016/j.mrfmmm.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 26••.Linde L, Boelz S, Nissim-Rafinia M, et al. Nonsense-mediated mRNA decay affects nonsense transcript levels and governs response of cystic fibrosis patients to gentamicin. J Clin Invest. 2007;117:683–692. doi: 10.1172/JCI28523. This study showed that nonsense-mediated decay was a major molecular mechanism to control patients’ response to gentamicin and suggested that manipulation of NMD might be useful clinically. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Holbrook JA, Neu-Yilik G, Hentze MW, Kulozik AE. Nonsense-mediated decay approaches the clinic. Nat Genet. 2004;36:801–808. doi: 10.1038/ng1403. [DOI] [PubMed] [Google Scholar]
- 28.Vacek M, Sazani P, Kole R. Antisense-mediated redirection of mRNA splicing. Cell Mol Life Sci. 2003;60:825–833. doi: 10.1007/s00018-003-3042-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Eng L, Coutinho G, Nahas S, et al. Nonclassical splicing mutations in the coding and noncoding regions of the ATM gene: maximum entropy estimates of splice junction strengths. Hum Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 30.Burrows NP, Nicholls AC, Richards AJ, et al. A point mutation in an intronic branch site results in aberrant splicing of COL5A1 and in Ehlers-Danlos syndrome type II in two British families. Am J Hum Genet. 1998;63:390–398. doi: 10.1086/301948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Di Leo E, Panico F, Tarugi P, et al. A point mutation in the lariat branch point of intron 6 of NPC1 as the cause of abnormal premRNA splicing in Niemann-Pick type C disease. Hum Mutat. 2004;24:440. doi: 10.1002/humu.9287. [DOI] [PubMed] [Google Scholar]
- 32•.Du L, Pollard JM, Gatti RA. Correction of prototypic ATM splicing mutations and aberrant ATM function with antisense morpholino oligonucleotides. Proc Natl Acad Sci U S A. 2007;104:6007–6012. doi: 10.1073/pnas.0608616104. This study confirmed the ability of antisense oligonucleotides to correct splicing mutations in primary immunodeficiency diseases. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marshall NB, Oda SK, London CA, et al. Arginine-rich cell-penetrating peptides facilitate delivery of antisense oligomers into murine leukocytes and alter premRNA splicing. J Immunol Methods. 2007;325:114–126. doi: 10.1016/j.jim.2007.06.009. [DOI] [PubMed] [Google Scholar]
- 34••.Kumar P, Wu H, McBride JL, et al. Transvascular delivery of small interfering RNA to the central nervous system. Nature. 2007;448:39–43. doi: 10.1038/nature05901. This important study shows small RNA can be delivered across the blood–brain barrier to central nervous system with a Rabies virus glycoprotein peptide. This has broad implications to treatment of neurodegenerative genetic diseases. [DOI] [PubMed] [Google Scholar]
- 35.Chun HH, Sun X, Nahas SA, et al. Improved diagnostic testing for ataxia-telangiectasia by immunoblotting of nuclear lysates for ATM protein expression. Mol Genet Metab. 2003;80:437–443. doi: 10.1016/j.ymgme.2003.09.008. [DOI] [PubMed] [Google Scholar]
- 36.Gatti RA, Boder E, Good RA. Immunodeficiency, radiosensitivity, and the XCIND syndrome. Immunol Res. 2007;38:87–101. doi: 10.1007/s12026-007-0018-y. [DOI] [PubMed] [Google Scholar]
- 37.Urnov FD, Miller JC, Lee YL, et al. Highly efficient endogenous human gene correction using designed zinc-finger nucleases. Nature. 2005;435:646–651. doi: 10.1038/nature03556. [DOI] [PubMed] [Google Scholar]
- 38.Miller JC, Holmes MC, Wang J, et al. An improved zinc-finger nuclease architecture for highly specific genome editing. Nat Biotechnol. 2007;25:778–785. doi: 10.1038/nbt1319. [DOI] [PubMed] [Google Scholar]
- 39.Lombardo A, Genovese P, Beausejour CM, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25:1298–1306. doi: 10.1038/nbt1353. [DOI] [PubMed] [Google Scholar]
- 40•.van Deutekom JC, Janson AA, Ginjaar IB, et al. Local dystrophin restoration with antisense oligonucleotide PRO051. N Engl J Med. 2007;357:2677–2686. doi: 10.1056/NEJMoa073108. This clinical trial illustrated antisense oligonucleotide-mediated exon skipping for correcting frameshift mutations. [DOI] [PubMed] [Google Scholar]
- 41.Ellis NA, Ciocci S, German J. Back mutation can produce phenotype reversion in Bloom syndrome somatic cells. Hum Genet. 2001;108:167–173. doi: 10.1007/s004390000447. [DOI] [PubMed] [Google Scholar]
- 42.Wada T, Konno A, Schurman SH, et al. Second-site mutation in the Wiskott-Aldrich syndrome (WAS) protein gene causes somatic mosaicism in two WAS siblings. J Clin Invest. 2003;111:1389–1397. doi: 10.1172/JCI15485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wada T, Toma T, Okamoto H, et al. Oligoclonal expansion of T lymphocytes with multiple second-site mutations leads to Omenn syndrome in a patient with RAG1-deficient severe combined immunodeficiency. Blood. 2005;106:2099–2101. doi: 10.1182/blood-2005-03-0936. [DOI] [PubMed] [Google Scholar]
- 44.Kalb R, Neveling K, Hoehn H, et al. Hypomorphic mutations in the gene encoding a key Fanconi anemia protein, FANCD2, sustain a significant group of FA-D2 patients with severe phenotype. Am J Hum Genet. 2007;80:895–910. doi: 10.1086/517616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hirschhorn R. In vivo reversion to normal of inherited mutations in humans. J Med Genet. 2003;40:721–728. doi: 10.1136/jmg.40.10.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pedemonte N, Lukacs GL, Du K, et al. Small-molecule correctors of defective DeltaF508-CFTR cellular processing identified by high-throughput screening. J Clin Invest. 2005;115:2564–2571. doi: 10.1172/JCI24898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vogt G, Vogt B, Chuzhanova N, et al. Gain-of-glycosylation mutations. Curr Opin Genet Dev. 2007;17:245–251. doi: 10.1016/j.gde.2007.04.008. [DOI] [PubMed] [Google Scholar]
- 48.Vogt G, Chapgier A, Yang K, et al. Gains of glycosylation comprise an unexpectedly large group of pathogenic mutations. Nat Genet. 2005;37:692–700. doi: 10.1038/ng1581. [DOI] [PubMed] [Google Scholar]
- 49.Freeze HH. Novel perspectives on glycosylation and human disease. Curr Mol Med. 2007;7:387. doi: 10.2174/156652407780831566. [DOI] [PubMed] [Google Scholar]
- 50.Bertoni C. Clinical approaches in the treatment of Duchenne muscular dystrophy (DMD) using oligonucleotides. Front Biosci. 2008;13:517–527. doi: 10.2741/2697. [DOI] [PubMed] [Google Scholar]
- 51.Roberts J, Palma E, Sazani P, et al. Efficient and persistent splice switching by systemically delivered LNA oligonucleotides in mice. Mol Ther. 2006;14:471–475. doi: 10.1016/j.ymthe.2006.05.017. [DOI] [PubMed] [Google Scholar]