Abstract
Human T-cell leukemia virus type-I (HTLV-I) is the etiologic agent of adult T-cell leukemia (ATL), an aggressive lymphoproliferative disease. MicroRNAs (miRNAs) are differentially expressed during hematopoiesis and lineage commitment of hematopoietic stem cell progenitors (HSCPs). Here, we report aberrant expression of hematopoietic-specific miR-223, miR-181a, miR-150, miR-142.3p, and miR-155 in HTLV-I–infected cells in vitro and uncultured ex vivo ATL cells. Our results suggest that HTLV-I–infected cells have an unbalanced expression of miRNA that favors T-cell differentiation. We also found altered expression of miRNA previously recognized as innate immunity regulators: miR-155, miR-125a, miR-132, and miR-146. Strikingly, our data also revealed significant differences between ex vivo ATL tumor cells and in vitro HTLV-I cell lines. Specifically, miR-150 and miR-223 were up-regulated in ATL patients but consistently down-regulated in HTLV-I cell lines, suggesting that ATL cells and in vitro–established cells are derived from distinct cellular populations.
Introduction
miRNAs play critical roles in tumorigenesis by acting on oncogenes and/or tumor suppressor genes.1,2 In addition, studies strongly suggest that miRNAs may play important roles in normal or malignant hematopoiesis, including the differentiation of hematopoietic stem cell progenitors (HSCPs), their self-renewal, and the function of immune cells.3–6 Although human T-cell leukemia virus type-I (HTLV-I) infects and replicates in a broad spectrum of cells in vitro, the in vivo tropism is restricted. The typical adult T-cell leukemia (ATL) cell is usually an activated, mature CD4+ T-cell. However, HTLV-I has also been shown to infect and replicate in CD34+ hematopoietic stem cells (HSCs),7 and recent studies suggest that the Tax protein can alter hematopoiesis and differentiation of CD34+ cells.8 In addition to lymphocytosis, granulocytosis and monocytosis are frequently observed in ATL patients,9 and the number of naive T lymphocytes is markedly suppressed in patients with acute ATL.10 It is also well documented that ATL patients suffer severe immune dysfunctions.
Methods
Patient characteristics11 are provided in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). All samples were collected after informed consent was obtained in accordance with the Declaration of Helsinki and in agreement with regulations for the protection of human subjects and after internal IRB approval from the University of Kansas Medical Center. The MicroRNA Bioarrays and stem loop quantitative real-time reverse-transcription–polymerase chain reaction (RT-PCR) are described in Document S1.
Results and discussion
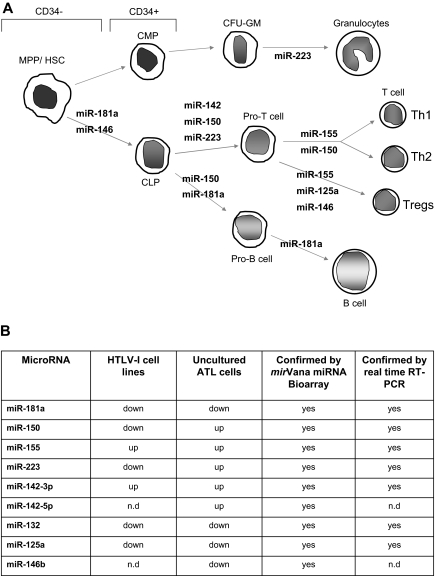
We analyzed the miRNA profile from 7 ATL patients' samples and 3 HTLV-I–negative donors using the mirVana miRNA Bioarrays (Ambion, Austin, TX). Several of the miRNAs whose expression were found to be deregulated in ATL cells are involved in hematopoiesis, differentiation, and innate immunity (Figure 1A,B). In fact, in silico analysis with several miRNA targeting programs identified similar targets (Table S2). We found that all of the HTLV-I deregulated miRNAs in our study had the potential to target genes involved with hematopoiesis and/or the immune response.
Figure 1.
Deregulation of miRNA involved in hematopoiesis and innate immunity in HTLV-I–infected cells. (A) miRNA involved in the different steps of hematopoiesis and showing deregulated expression in HTLV-I–transformed cells in vitro and ATL tumor cells ex vivo. MPP indicates myeloid pluripotent progenitors, HSC, hematopoietic stem cells; CMP, common myeloid progenitors; CLP, common lymphocyte progenitors; and CFU-GM, colony-forming units granulocyte-macrophage. (B) Table representation of mature miRNA deregulated in HTLV-I–infected cells in vitro and ex vivo identified by mirVana miRNA Bioarrays platform and real-time RT-PCR. The miRNA enriched fraction was obtained by passing total RNA through a flashPAGE Fractionator apparatus (Ambion, Austin, TX). The 3′ ends of the RNA molecules were tailed and labeled using the mirVana miRNA Labeling Kit (Ambion) and analyzed as described in “Methods.” nd indicates not determined.
A role for cancer-initiating stem cells has not been investigated in HTLV-I, despite being accepted for many types of leukemia.12 However, several lines of evidence demonstrate the possibility of cancer-initiating stem cells during HTLV-I infection: (1) HTLV-I can infect HSCPs and proviral sequences are maintained during differentiation in vitro and in vivo.7 (2) Reconstitution of T lymphopoiesis with HTLV-I–infected CD34+ cells in severe combined immunodeficient (SCID) mice suggests that HTLV-I perturbs hematopoiesis.7 (3) Tax induces cell-cycle withdrawal of HSCPs, which may prevent proliferation and exhaustion of HTLV-I–infected leukemia stem cells.8
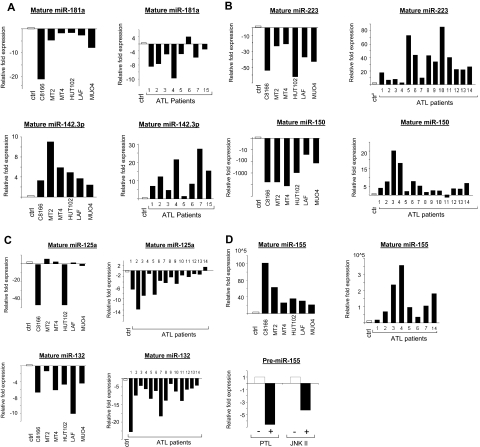
To confirm the results obtained through our microarrays, real-time RT-PCRs on mature miRNA were performed. The results revealed a down-regulation of miR-181a in HTLV-I cell lines and ex vivo ATL tumor cells (Figure 2A). miR-181a favors differentiation toward the B-cell lineage in HSCPs,13 and, when expressed in the double-positive stage of thymocytes, miR-181a results in a decrease in peripheral blood T cells.14 Decreased expression of miR-181a also results in hyporesponsiveness to TCR signaling and a decrease in sensitivity to antigens.15 The decrease in miR-181a expression is extremely relevant to HTLV-I biology, as HTLV-I has evolved several strategies to dampen TCR signaling and T-cell activation to persist in the host.
Figure 2.
Real-time quantification of mature miRNA in HTLV-I cell lines and ex vivo ATL samples. (A-D) TaqMan real-time RT-PCR from HTLV-I–transformed cells (C8166, MT2, MT4, and HUT102) and immortalized cells (LAF and MU04) in vitro and ATL tumor cells ex vivo. The TaqMan probes that were used ensure accurate discrimination between miRNAs that may differ by a single nucleotide. Mature miRNAs were analyzed because maturation of pri-miRNA is subject to posttranscriptional regulations. The threshold cycle (CT) is defined as the fractional cycle number at which the fluorescence passes the fixed threshold. TaqMan CT values are used to measure the fold change. ATL samples and HTLV-I cell lines were compared with peripheral blood mononuclear cells (PBMCs) and purified CD4 T cells as controls. miR-24 was used as the internal control, because of its constitutive expression among ATL samples, control T cells, and HTLV-I cell lines. These experiments were performed by Asuragen (Austin, TX). All primers and probes were tested and validated by Asuragen. Statistical analysis is provided in Document S1. For panel D, MT4 cells were treated overnight with either 6 μM parthenolide (PTL) or 90 μM JNK II. Control cells were treated with DMSO. SYBR green real-time PCR was performed for pre-miR-155 expression (F: 5′-CTGTTAATGCTAATCGTGATAG-3′ and R: 5′-AATGCTAATATGTAGGAGTCAG-3′) with GAPDH (F: 5′-GAAGGTGAAGGTCGGAGTC-3′ and R: 5′-GAAGATGGTGATGGGATTTC-3′) as an internal control. All experiments were performed in triplicate.
In contrast to miR-181a, it has been shown that expression of miR-142 and miR-223 alters differentiation toward T lymphopoiesis. Ectopic expression of miR-223 and miR-142 in HSCPs induced a 30% to 50% increase in the proportion of T cells, but not B-lineage or myeloid cells. Interestingly, our studies revealed increased expression of miR-142.3p (−5p) and miR-223 in all ATL cells tested (Figure 2A,B). miR-223 has also been shown to influence granulocytic differentiation.16 Up to 20% of acute ATL patients have marked granulocytosis, which could be due to the elevated levels of miR-223.
Whereas miR-142.3p was regulated in a similar way in HTLV-I cell lines and ex vivo ATL cells, miR-223 was consistently up-regulated in ATL cells but down-regulated in all in vitro HTLV-I cell lines (Figure 2B). This unexpected pattern was also found for miR-150 (Figure 2B), which is strongly up-regulated during the differentiation of T and B cells. Ectopic expression of miR-150 in HSCPs has been shown to block the transition from the pro-B to pre-B-cell stage, thereby significantly reducing the amount of mature B cells.17 miR-150 also moderately enhances T lymphopoiesis. Our results clearly identify 2 microRNAs, miR-150 and miR-223, that distinguish ATL tumor cells and in vitro HTLV-I–transformed cell lines and raise the possibility that these cells derive from a different cellular origin. Previous reports suggest that miR-223 is restricted to the bone marrow, HSCPs, and myeloid, erythroid, and lymphoid progenitor cells. Thus, a fascinating possibility would be that the target cell for HTLV-I in vivo is different and represents a less differentiated CD34+ or CD34− common lymphocyte progenitor (CLP) rather than a mature, differentiated CD4+ T cell. Undeniably, more studies in this area are needed.
Our microarray and RT-PCR data also demonstrated a decrease in miR-146b and miR-125a in ATL cells (Figures 1B,2C). miR-146a/b and miR-125 play roles in innate immunity and act as terminal transducers of TLR4 signaling by targeting NF-κB activation by TLR4 and NF-κB induction of TNFα, respectively.18 miR-146a/b has also been proposed to target TRAF6 and IRAK1. In addition to its role in innate immunity, expression of miR-125a is increased in regulatory T cells (T-regs).19 The decrease in miR-146b and miR-125a in ATL cells may, therefore, lead to increased inflammation and decreased T-reg functions, characteristics observed in HTLV-I–infected patients.20 We also found that miR-132, another miRNA involved in innate immunity, was consistently down-regulated in in vitro and ex vivo ATL cells (Figure 2C).
Studies suggest that deregulated expression of miR-155 can have deleterious effects on normal immune function,21 and that high levels of miR-155 can lead to leukemia/lymphoma.22 We found elevated expression of miR-155 in HTLV-I–infected cells in vitro and ex vivo (Figure 2D). This makes sense, as miR-155 induction involves both NF-κB and JNK, both of which are activated by HTLV-I. In fact, treatment of MT4 cells with NF-κB and JNK inhibitors caused a decrease in miR-155 expression (Figure 2D). Among the numerous functions of miR-155, is its role in dendritic and T-cell interactions. This may be important for persistence of HTLV-I, because dendritic cells can transfer HTLV-I infectious virus to CD4+ cells.23 miR-155 also regulates T-cell lineage fate by promoting T-helper type 1 (Th1) versus type 2 (Th2) differentiation.24 Because an efficient immune response to helminthic parasites, such as Strongyloides, involves a dominant Th2-type response and the incidence of ATL disease increases in patients carrying Strongyloides,25 miR-155 may indirectly contribute to the pathogenesis of ATL.
In summary, we found that miRNAs involved in normal hematopoiesis and immune responses are profoundly deregulated in ATL tumor cells ex vivo. Our findings also suggest that in vivo ATL cells and in vitro–established HTLV-I cell lines are derived from different cell populations. We propose that, in vivo, ATL cells are derived from infected HSCPs, a hypothesis that challenges the current notion that mature CD4+ differentiated T lymphocytes are the target for HTLV-I–associated leukemia. Reduced expression of miR-181a and miR-146b may prevent myeloid pluripotent progenitor's differentiation and preserve stemness, a phenotype recently observed in vitro in CD34+ infected cells by HTLV-I or Tax. However, if HTLV-I–infected progenitors initiate differentiation, the miRNA program may favor T-cell lineage and T-cell leukemia. Potential existence of leukemia-initiating stem cells controlled by miRNA in HTLV-I–infected individuals offers an exciting hypothesis for future studies.
Supplementary Material
Acknowledgments
This work was supported by awards R01CA106258 and R01CA115398 from the National Cancer Institute (Bethesda, MD). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Cancer Institute or the National Institutes of Health.
Footnotes
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Authorship
Contribution: M.B. prepared RNA samples, performed real-time PCR, analyzed data, and prepared the figures; Y.L. and O.H. provided patient samples; and C.N. was responsible for design of the study, interpretation of the data, and writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Christophe Nicot, Director, Center for Viral Oncology, Associate Professor, Department of Pathology and Laboratory Medicine, University of Kansas Medical Center, 3901 Rainbow Blvd, Kansas City, KS 66160; e-mail: cnicot@kumc.edu.
References
- 1.Calin GA, Croce CM. MicroRNA signatures in human cancers. Nat Rev Cancer. 2006;6:857–866. doi: 10.1038/nrc1997. [DOI] [PubMed] [Google Scholar]
- 2.Lu J, Getz G, Miska EA, et al. MicroRNA expression profiles classify human cancers. Nature. 2005;435:834–838. doi: 10.1038/nature03702. [DOI] [PubMed] [Google Scholar]
- 3.Kluiver J, Kroesen BJ, Poppema S, van den BA. The role of microRNAs in normal hematopoiesis and hematopoietic malignancies. Leukemia. 2006;20:1931–1936. doi: 10.1038/sj.leu.2404387. [DOI] [PubMed] [Google Scholar]
- 4.García P, Frampton J. Hematopoietic lineage commitment: miRNAs add specificity to a widely expressed transcription factor. Dev Cell. 2008;14:815–816. doi: 10.1016/j.devcel.2008.05.011. [DOI] [PubMed] [Google Scholar]
- 5.Garzon R, Croce CM. MicroRNAs in normal and malignant hematopoiesis. Curr Opin Hematol. 2008;15:352–358. doi: 10.1097/MOH.0b013e328303e15d. [DOI] [PubMed] [Google Scholar]
- 6.Chen CZ, Lodish HF. MicroRNAs as regulators of mammalian hematopoiesis. Semin Immunol. 2005;17:155–165. doi: 10.1016/j.smim.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 7.Feuer G, Fraser JK, Zack JA, et al. Human T-cell leukemia virus infection of human hematopoietic progenitor cells: maintenance of virus infection during differentiation in vitro and in vivo. J Virol. 1996;70:4038–4044. doi: 10.1128/jvi.70.6.4038-4044.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tripp A, Banerjee P, Sieburg M, et al. Induction of cell cycle arrest by human T-cell lymphotropic virus type 1 Tax in hematopoietic progenitor (CD34+) cells: modulation of p21cip1/waf1 and p27kip1 expression. J Virol. 2005;79:14069–14078. doi: 10.1128/JVI.79.22.14069-14078.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nimer SD, Gasson JC, Hu K, et al. Activation of the GM-CSF promoter by HTLV-I and -II tax proteins. Oncogene. 1989;4:671–676. [PubMed] [Google Scholar]
- 10.Tokioka T, Shimamoto Y, Motoyoshi K, Yamaguchi M. Clinical significance of monocytosis and human monocytic colony-stimulating factor in patients with adult T-cell leukaemia/lymphoma. Haematologia (Budap) 1994;26:1–269. [PubMed] [Google Scholar]
- 11.Datta A, Bellon M, Sinha-Datta U, et al. Persistent inhibition of telomerase reprograms adult T-cell leukemia to p53-dependent senescence. Blood. 2006;108:1021–1029. doi: 10.1182/blood-2006-01-0067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huntly BJ, Gilliland DG. Leukaemia stem cells and the evolution of cancer-stem-cell research. Nat Rev Cancer. 2005;5:311–321. doi: 10.1038/nrc1592. [DOI] [PubMed] [Google Scholar]
- 13.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 14.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–589. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Li QJ, Chau J, Ebert PJ, et al. miR-181a is an intrinsic modulator of T cell sensitivity and selection. Cell. 2007;129:147–161. doi: 10.1016/j.cell.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 16.Fazi F, Rosa A, Fatica A, et al. A minicircuitry comprised of microRNA-223 and transcription factors NFI-A and C/EBPalpha regulates human granulopoiesis. Cell. 2005;123:819–831. doi: 10.1016/j.cell.2005.09.023. [DOI] [PubMed] [Google Scholar]
- 17.Xiao C, Calado DP, Galler G, et al. MiR-150 controls B cell differentiation by targeting the transcription factor c-Myb. Cell. 2007;131:146–159. doi: 10.1016/j.cell.2007.07.021. [DOI] [PubMed] [Google Scholar]
- 18.Taganov KD, Boldin MP, Chang KJ, Baltimore D. NF-kappaB-dependent induction of microRNA miR-146, an inhibitor targeted to signaling proteins of innate immune responses. Proc Natl Acad Sci U S A. 2006;103:12481–12486. doi: 10.1073/pnas.0605298103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou X, Jeker LT, Fife BT, et al. Selective miRNA disruption in T reg cells leads to uncontrolled autoimmunity. J Exp Med. 2008;205:1983–1991. doi: 10.1084/jem.20080707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yamano Y, Takenouchi N, Li HC, et al. Virus-induced dysfunction of CD4+CD25+ T cells in patients with HTLV-I-associated neuroimmunological disease. J Clin Invest. 2005;115:1361–1368. doi: 10.1172/JCI23913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baltimore D, Boldin MP, O'Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–845. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 22.Costinean S, Zanesi N, Pekarsky Y, et al. Pre-B cell proliferation and lymphoblastic leukemia/high-grade lymphoma in E(mu)-miR155 transgenic mice. Proc Natl Acad Sci U S A. 2006;103:7024–7029. doi: 10.1073/pnas.0602266103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jones KS, Petrow-Sadowski C, Huang YK, Bertolette DC, Ruscetti FW. Cell-free HTLV-1 infects dendritic cells leading to transmission and transformation of CD4(+) T cells. Nat Med. 2008;14:429–436. doi: 10.1038/nm1745. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez A, Vigorito E, Clare S, et al. Requirement of bic/microRNA-155 for normal immune function. Science. 2007;316:608–611. doi: 10.1126/science.1139253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gabet AS, Mortreux F, Talarmin A, et al. High circulating proviral load with oligoclonal expansion of HTLV-1 bearing T cells in HTLV-1 carriers with strongyloidiasis. Oncogene. 2000;19:4954–4960. doi: 10.1038/sj.onc.1203870. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.