Abstract
Background
Y-SNP haplogroup G (hgG), defined by Y-SNP marker M201, is relatively uncommon in the United States general population, with only 8 additional sub-markers characterized. Many of the previously described eight sub-markers are either very rare (2–4%) or do not distinguish between major populations within this hg. In fact, prior to the current study, only 2% of our reference Caucasian population belonged to hgG and all of these individuals were in sub-haplogroup G2a, defined by P15. Additional Y-SNPs are needed in order to differentiate between individuals within this haplogroup.
Principal Findings
In this work we have investigated whether we could differentiate between a population of 63 hgG individuals using previously uncharacterized Y-SNPs. We have designed assays to test these individuals using all known hgG SNPs (n = 9) and an additional 16 unreported/undefined Y-SNPS. Using a combination of DNA sequence and genetic genealogy databases, we have uncovered a total of 15 new hgG SNPs that had been previously reported but not phylogenetically characterized. Ten of the new Y-SNPs are phylogenetically equivalent to M201, one is equivalent to P15 and, interestingly, four create new, separate haplogroups. Three of the latter are more common than many of the previously defined Y-SNPs. Y-STR data from these individuals show that DYS385*12 is present in (70%) of G2a3b1-U13 individuals while only 4% of non-G2a3b1-U13 individuals posses the DYS385*12 allele.
Conclusions
This study uncovered several previously undefined Y-SNPs by using data from several database sources. The new Y-SNPs revealed in this paper will be of importance to those with research interests in population biology and human evolution.
Introduction
Single nucleotide polymorphisms (SNPs) are the smallest and most abundant type of human DNA polymorphisms [1]. SNPs have been extensively used in the study of human evolutionary and migratory patterns [2] and are increasingly being used in genome-wide association studies [3]. Y-SNPs, in particular, are of interest due to their paternal inheritance, lack of recombination, abundance, and low mutation rate and are currently being investigated for characterizing male population structure and ethnogeographic origin in forensic science [4]–[10]. These unique polymorphisms within the non-recombining region (NRY) of the Y-chromosome (mainly SNPs) have created population specific paternal lineages (commonly called haplogroups) that have persisted throughout human history. Large scale parsimonious phylogenetic trees representing world wide Y chromosomal variation have been constructed and comprise the major haplogroups A-T [11]–[13]. The rules for naming haplogroups have been designed to adjust for new SNPs that are continuously being identified and characterized to be added to the tree and potentially reshaping it as in the most recently published y-chromosomal haplogroup tree [13]. Although some are rare (e.g. [14]), some can still be useful for individual identification especially if found at higher frequencies in certain defined populations. Many of these polymorphisms have proven highly informative in tracing human prehistoric migrations and generating new hypotheses on human colonization and migrations [15].
This study recruited 54 hgG men from the pool available in Ysearch.org (http://www.ysearch.org) in addition to 9 that we possessed in-house. We have uncovered a total of 15 new hgG SNPs, four of which create new sub-haplogroups with the hgG clade. Additionally, we have also discovered that the Y-STR DYS385*12 is present in (70%) of the new hg G2a3b1-U13 individuals and only 4% of non-G2a3b1-U13 individuals.
Results
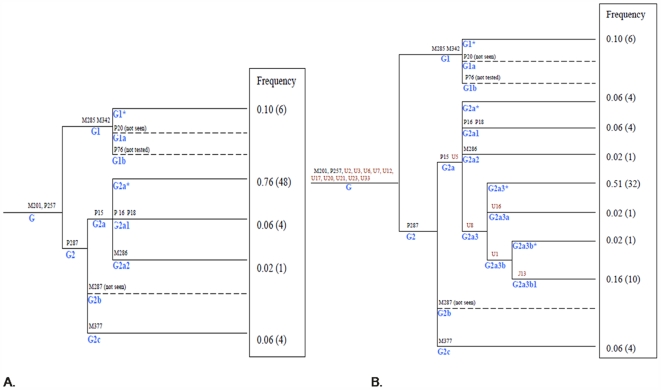
Phylogenetic trees were constructed to show the evolutionary relationships between the previously characterized hgG SNPs and 15 newly characterized hgG SNPs (Figure 1A and 1B). The population frequencies of Haplogroup G individuals in sub hgG groups without (Figure 1A) and with the newly characterized Y-SNPs are also shown (Figure 1B). Eleven of the new Y-SNPs were phylogenetically indistinct from the current G-M201 and G2a-P15 markers. However, four newly characterized SNPs (U8, U16, U1, U13) permitted the definition of four new sub-clades of hg G (G2a3*, G2a3a, G2a3b*, G2a3b1) which, in our population sample, increased the number of observable hg G genotypes by 80% (from five to nine). To ascertain the extent to which the new markers are useful for differentiating hgG individuals, the probability of discrimination (DP) obtained by typing individuals with and without the four new informative markers was calculated [16]. The DP was increased 72%, from 0.40 to 0.69, in our Caucasian sample set.
Figure 1. Phylogenetic Trees Indicating the Frequencies of Haplogroup G Individuals with (B) and without (A) the Newly Characterized Y-SNPs.
The haplogroup names are assigned base on the most recent haplogroup tree by Karafet et al. [13].
Examination of the allele distribution of DYS385 showed that the allele DYS385*12 was over-represented in the G2a3b1-U13 samples (7/10 samples possess the 12 allele). Only two copies of the DYS385*12 allele were found in the other 53 non-G2a3b1-U13 samples (4%). Thus the DYS385*12 is highly predictive (P = 0.70) of a G2a3b1-U13 individual if the individual is known to belong to hgG. Such Y-STR information can sometimes facilitate a speedier haplogroup assignment and differentiation than would be available from a strict hierarchy-based SNP analysis.
In summary, we have characterized 15 new hgG SNPs that had been previously reported but not phylogenetically defined. Ten of the new Y-SNPs are phylogenetically indistinguishable from G-M201, one is equivalent to G2a-P15 but four create new, separate hg G sub-haplogroups. Three of the latter are more common than many of the previously defined hgG Y-SNPs.
Discussion
Y-SNP haplogroup G (hgG), defined by the Y-SNP marker G-M201, is relatively uncommon in the European American population of the United States. This haplogroup is thought to have originated in the Caucasus region of Eurasia, especially in the North Ossetians [17] and specifically, the Digora population with an average frequency of 74% [18]. Also approximately 11% of individuals in Anatolia [19] and 17% in Northern Sardenia [20] belong to hgG. In a study on Y-chromosomes in the Caucasus, it was found that the hgG genotype frequency ranges from 21%–74% in seven different populations of the north Caucasus region while it is only found within 3 populations in south Caucasus region (at frequencies of 11%, 18%, and 31%) [18]. It has been proposed that the peoples from these regions originated from West Asia rather than Europe due the high frequencies of the G, J2* and F* haplogroups [17].
Prior to this study, only 8 sub-markers had been described within haplogroup G[11], [19], [21] with the most common haplogroup being G2a*, defined by P15. Many of the previously described eight sub-markers are either very rare or do not distinguish between major populations within this haplogroup. In fact, prior to the current study, only 2% of our reference European American population was within hgG and all of the individuals were in sub-haplogroup G2a defined by the P15 polymorphism [10]. In this work we have investigated whether we could differentiate between a population of 63 hgG individuals using previously uncharacterized Y-SNPs as well as their associated 19 marker Y-STR haplotypes. Here we describe the characterization of new hgG sub-markers, four of which can further differentiate between sub-populations within this hg.
The subjects were recruited from a selection of over 500 haplogroup G men available in public genetic-genealogy databases in the fall of 2006. This was especially valuable in the case of a somewhat rare European haplogroup like haplogroup G (about 2–4% of the general population) [10], [19], [22], [23]. The selection of hgG men from already SNP typed populations provided a tremendous savings in time and cost for this project. To have found this large of a sample of HgG men without the resource of the genetic genealogy community's results would have necessitated the SNP typing of approximately 1500 men. This study exemplifies the success obtainable by productive collaboration between genetics researchers and the genetic genealogy community.
Materials and Methods
This study was conducted according to the principles expressed in the Declaration of Helsinki. The study was approved by the Institutional Review Board of the University of Central Florida. All participants provided written informed consent for the collection of samples and subsequent analysis.
Candidate SNP Identification
DNA sequence traces from the NCBI Trace Archives were used to identify SNPs of an individual from haplogroup G, in silico. Most of the earliest mapping of the Y chromosome was done using the BAC library from the California Institute of Technology called CTC. This included the contigs: AC005942.2 CTC-298B15, AC002992.1 CTC-203M13, AC004617.2 CTC-264M20, AC002531.1 CTC-486O8, AC004474.1 CTC-475I1, AC006565.4 CTC-484O7, AC005820.1 CTC-494G17, and AC078938.3 CTC-480L15. The Y-SNP marker M201 in contig AC004474 was seen to be derived rather than ancestral in the reference sequence. This means that the man used in the CTC library belonged to YCC haplogroup G. The assumption was made that the contents of the NCBI Trace Archives were not likely to contain the re-sequencing of any other haplogroup G men (since G is seen in only about 3% of the males of European descent). Candidate YSNPs were chosen on the basis of appearing in only the above reference contigs and none of the traces in the Trace Archives. Those candidate SNPs were typed and characterized in a panel of 63 haplogroup G men. Additionally, all previously defined hgG Y-SNPs were compiled from various publications and a 25 member candidate list of SNPs suspected to be polymorphic inside haplogroup G was developed for assay development and subsequent population studies.
Biological Sample Donors
Buccal swabs were obtained from a total of 74 individuals including 63 human males belonging to Y-SNP hgG, one human female, one male chimp, and one human individual from each of the following hgs: A, B, C, E3a, F, H, I, J, and R1b. Fifty-four of the individuals were recruited from the list of individuals whose 37 locus Y-STR haplotypes and Y-SNP haplogroups are available in http://www.ysearch.org. All DNA samples were obtained with the individual's informed consent in accordance with the University of Central Florida's Institutional Review Board.
Genomic DNA isolation
A small piece of the buccal swab was cut and placed in a Spin-ease tube (Gibco-BRL, Grand Island, NY) and incubated overnight at 56°C in 400 µL DNA extraction buffer (100 mM NaCl, 10 mM Tris-HCl, pH 8.0, 25 mM EDTA, 0.5% SDS, and 0.1 mg/ml Proteinase K). The cut material was then removed from the tube and placed in a Spin-ease basket and the basket was then placed back in the original tube and centrifuged (Eppendorf Centrifuge 5415D) at 13,200 rpm for 5 min. The extract was then purified and isolated using 25∶24∶1 phenol/chloroform/isoamyl achohol (Fisher Scientific, Norcross, GA) followed by filtration using Microcon® 100 centrifugal filter devices (Millipore, Bedford, MA) according to the manufacturer's instructions. Samples were brought to a final volume of 50 µL in TE−4 (10 mM Tris-HCl and 0.1 mM EDTA, pH 7.5) and stored at 4°C until analysis. DNA was quantified by gel electrophoresis in a 1% Agarose gel. Samples were visualized using the ΩMega 12ic™ Gel documentation system (Ultra-Lum, Claremont, CA). Quantification was accomplished by a comparison of the fluorescence intensity of the unknown bands to a set of known quantity standards that were run simultaneously.
SNP Primer Design and PCR
Assays were developed to amplify regions flanking the SNP for use with pyrosequencing technology. Extracted female DNA was also tested to ensure male specificity. All SNPs were tested against individuals from hgs A, B, C, E, F, H, I, J, and R for haplogroup determination and ancestral vs. derived states. A male chimp was also tested to facilitate determination of the ancestral vs. derived states. PCR primers were designed using a combination of Primer3[24] and SNP Primer Design Pyrosequencing AB v.1.0.1.software. The 50 µL PCR reaction contained: 1 ng DNA, 0.2 µM each primer, 125 µM dNTPs, 1X PCR Buffer II (10 mM Tris-HCl, pH 8.3, 50 mM KCl), 2.0 mM MgCl2, 10 µg non-acetylated BSA (Sigma, St. Louis, MO) and 1.5 units of AmpliTaq® Gold Polymerase (Applied Biosystems, Foster City, CA). Cycling conditions were: (1) 95°C for 10 min, (2) 40 cycles: 95°C for 15 s, 54°C for 30 s, 72°C for 15 s, and (3) final extension at 72°C for 5 min, unless otherwise noted.
Table 1 provides details of the SNP loci used and the PCR primers used.
Table 1. List of Y-SNPS, their associated database references, and location of the SNP within the product for the indicated primers.
| SNP Name | rs # or AC#: SNP† | Product Size (bp) | Position (bp) | Forward (5′-3′) | Reverse (5′-3′) |
| M201 | AC004474.1: G>T | 310 | 199 | CATGGGTAATTCGGTTGTTACC | GCCCTTTGGTGGCATAGTA |
| P15 | AC007876.2: C>T | 75 | 34 | GAATAGAGCCAATGCTTGAGGT | TATGGGAATCACTTTTGCAACT |
| P16 | AC068541.7: A>T | 84 | 34 | GTCGTTTATTTGGTGCCTGAA | CTATGACCTCAGCAGAATGGA |
| P18 | AC016698.3: C>T | 141 | 80 | GGTTGGGATTGTGACTCCTCT | CTGGGCAAATTTACCTGTCTC |
| P20 | AC016698.3: C>del | 141 | 30 | GGTTGGGATTGTGACTCCTCT | CTGGGCAAATTTACCTGTCTC |
| M285 | AC007678.3: C>G | 188 | 63 | GAGCCGTTGTCCCTGTGTTT | TGCAGGCATCAGCTAGATTGT |
| M287 | AC007678.3: A>T | 188 | 93 | GAGCCGTTGTCCCTGTGTTT | TGCAGGCATCAGCTAGATTGT |
| M286 | AC007678.3: G>A | 188 | 123 | GAGCCGTTGTCCCTGTGTTT | TGCAGGCATCAGCTAGATTGT |
| M342 | AC007876.2: C>T | 103 | 40 | AACAGGGCGTAACAAATAGGT | TGGCTCTATTCATGTGAGGAA |
| M377 | AC004474.1: A>G | 195 | 93 | TCGGTTGTTACCTTGAGCATT | CGAAAAACCTCAGTTGATACTGG |
| U1 | rs9785956: A>G | 189 | 118 | TTTCTGCTCCAAATCTGCTG | CACCTGTAATCGGGAGGCTA |
| U2 | rs9786712: G>A | 150 | 123 | AGCTCATCTTCACGGGTGTG | ACAGGGCAAAGGAATCGTTA |
| U3 | rs11799152: A>G | 153 | 45 | CTTGAACCCAAGACGAGGAG | CAACAGTGGATCCCACATCA |
| U6 | rs2740980: G>A | 71 | 43 | CCTCAGTCTCTCCGATTCCTT | CTTTCATCTCCAACCCCCATC |
| U5 | rs2178500: T>G | 64 | 24 | CTATCACCCAGAGACCCCTCA | GAATCGGGTCCCATAACAAT |
| U7 | rs7067251: G>C | 222 | 124 | GATCCCACAGAGTGCTCAGG | CATGGGGTAAGAGAATGGGTA |
| U8 | rs7892988: T>C | 70 | 45 | TATAACCAAAAATGGCACGAT | GGATTAAGGTTGCCATCAGG |
| U12 | rs9786640: A>C | 162 | 43 | CTATGAGCATTTGGGGGCTTA | CCTTGTATCCTCCCTCCCTTT |
| U13 | rs9786706: C>T | 81 | 48 | GTGGTAACAGCTCCTGGTGAG | TGCTGCTTTGGTTAACTGTCC |
| U16 | rs35474563: C>T | 107 | 30 | CTCATTTGACTTCCCGCTGT | CTAGGACGCAGACGTCTTACC |
| U17 | rs34742138: C>T | 175 | 150 | CAGTTGTGCATCAACCATTCA | AACCTAGGTACTTCTTCCACTTCTC |
| U21 | AC005820.1: C>T | 98 | 53 | CTGGCACCTCCTCTCACTTC | TCAAGGAGCTTCCACTCACG |
| U20 * | rs16648: AG>del | 119 | 61 | AGACAAAGTCGGGGTTTTGA | GATCTGCCTCTTTCCCAAAAT |
| U23 * | rs9786460: G>A | 181 | 58 | GGTGGGAGAATTACCTGAACC | AGCTTCCTGAGTTCCCTTTTT |
| U33 * | rs1125978: C>G | 94 | 68 | CCCAATGTCCCTCTTTCCTT | ACCCACTTTGACCCAATCTG |
SNP is listed as: ancestral>derived.
U20 and U33 were amplified at 56°C annealing temperature and U23 was amplified at 57°C for only 35 cycles.
SNP/STR Genotyping and Phylogenetic Analysis
SNP genotyping was performed by pyrosequencing on a PSQ™ 96 MA instrument according to the manufacturer’s recommendations (Biotage, Uppsala, Sweden, http://www.biotage.com). SNP genotype data were collected and the phylogenetic relationships were depicted in a phylogenetic tree showing the corresponding frequencies for each haplogroup observed. Haplogroups were assigned based on the most recent comprehensive Y chromosomal haplogroup tree published by Karafet et al. [13]. All STR genotyping (19 locus haplotypes) was performed on a Macintosh-based ABI Prism 310 capillary electrophoresis system using two validated multiplex systems, MP I and MP II, as previously described [25], [26]. For two individuals whose STR haplotypes were not determined as just described, some Y STR data were available from http://www.ysearch.org. The probability of discrimination (DP)[16] was calculated as: DP = 1−∑pi 2, where pi is the observed frequency of the derived allele at each of the sub hg G haplogroups.
Acknowledgments
Points of view in this manuscript are those of the authors and do not necessarily represent the official position of the US Department of Justice. The authors would like to thank the 54 donors from http://www.ysearch.org who gladly offered their DNA samples.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported under Award Numbers 1998-IJ-CX-K003 and 2005-MU-MU-K044 from the Office of Justice Programs, National Institute of Justice, Department of Justice. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Brookes AJ. The essence of SNPs. Gene. 1999;234:177–186. doi: 10.1016/s0378-1119(99)00219-x. [DOI] [PubMed] [Google Scholar]
- 2.Shastry BS. SNP alleles in human disease and evolution. J Hum Genet. 2002;47:561–566. doi: 10.1007/s100380200086. [DOI] [PubMed] [Google Scholar]
- 3.Syvanen AC. Toward genome-wide SNP genotyping. Nat Genet. 2005;37(Suppl):S5–10. doi: 10.1038/ng1558. [DOI] [PubMed] [Google Scholar]
- 4.Brion M, Sanchez JJ, Balogh K, Thacker C, Blanco-Verea A, et al. Introduction of an single nucleodite polymorphism-based “Major Y-chromosome haplogroup typing kit” suitable for predicting the geographical origin of male lineages. Electrophoresis. 2005;26:4411–4420. doi: 10.1002/elps.200500293. [DOI] [PubMed] [Google Scholar]
- 5.Hammer MF, Chamberlain VF, Kearney VF, Stover D, Zhang G, et al. Population structure of Y chromosome SNP haplogroups in the United States and forensic implications for constructing Y chromosome STR databases. Forensic Sci Int. 2005 doi: 10.1016/j.forsciint.2005.11.013. [DOI] [PubMed] [Google Scholar]
- 6.Jobling MA. Y-chromosomal SNP haplotype diversity in forensic analysis. Forensic Sci Int. 2001;118:158–162. doi: 10.1016/s0379-0738(01)00385-1. [DOI] [PubMed] [Google Scholar]
- 7.Kidd KK, Pakstis AJ, Speed WC, Grigorenko EL, Kajuna SL, et al. Developing a SNP panel for forensic identification of individuals. Forensic Sci Int. 2005 doi: 10.1016/j.forsciint.2005.11.017. [DOI] [PubMed] [Google Scholar]
- 8.Sanchez JJ, Borsting C, Hallenberg C, Buchard A, Hernandez A, et al. Multiplex PCR and minisequencing of SNPs–a model with 35 Y chromosome SNPs. Forensic Sci Int. 2003;137:74–84. doi: 10.1016/s0379-0738(03)00299-8. [DOI] [PubMed] [Google Scholar]
- 9.Vallone PM, Butler JM. Y-SNP typing of U.S. African American and Caucasian samples using allele-specific hybridization and primer extension. J Forensic Sci. 2004;49:723–732. [PubMed] [Google Scholar]
- 10.Sims LM, Garvey D, Ballantyne J. Sub-populations within the major European and African derived haplogroups R1b3 and E3a are differentiated by previously phylogenetically undefined Y-SNPs. Hum Mutat. 2007;28:97. doi: 10.1002/humu.9469. [DOI] [PubMed] [Google Scholar]
- 11.YCC. A nomenclature system for the tree of human Y-chromosomal binary haplogroups. Genome Res. 2002;12:339–348. doi: 10.1101/gr.217602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Jobling MA, Tyler-Smith C. The human Y chromosome: an evolutionary marker comes of age. Nat Rev Genet. 2003;4:598–612. doi: 10.1038/nrg1124. [DOI] [PubMed] [Google Scholar]
- 13.Karafet TM, Mendez FL, Meilerman MB, Underhill PA, Zegura SL, et al. New binary polymorphisms reshape and increase resolution of the human Y chromosomal haplogroup tree. Genome Res. 2008;18:830–838. doi: 10.1101/gr.7172008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sims LM, Ballantyne J. A Rare Y Chromosome Missense Mutation in Exon 25 of Human USP9Y Revealed by Pyrosequencing. Biochem Genet. 2008 doi: 10.1007/s10528-007-9139-1. [DOI] [PubMed] [Google Scholar]
- 15.Rosser ZH, Zerjal T, Hurles ME, Adojaan M, Alavantic D, et al. Y-chromosomal diversity in Europe is clinal and influenced primarily by geography, rather than by language. Am J Hum Genet. 2000;67:1526–1543. doi: 10.1086/316890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jones DA. Blood samples: probability of discrimination. J Forensic Sci Soc. 1972;12:355–359. doi: 10.1016/s0015-7368(72)70695-7. [DOI] [PubMed] [Google Scholar]
- 17.Nasidze I, Quinque D, Dupanloup I, Rychkov S, Naumova O, et al. Genetic evidence concerning the origins of South and North Ossetians. Ann Hum Genet. 2004;68:588–599. doi: 10.1046/j.1529-8817.2004.00131.x. [DOI] [PubMed] [Google Scholar]
- 18.Nasidze I, Ling EY, Quinque D, Dupanloup I, Cordaux R, et al. Mitochondrial DNA and Y-chromosome variation in the caucasus. Ann Hum Genet. 2004;68:205–221. doi: 10.1046/j.1529-8817.2004.00092.x. [DOI] [PubMed] [Google Scholar]
- 19.Cinnioglu C, King R, Kivisild T, Kalfoglu E, Atasoy S, et al. Excavating Y-chromosome haplotype strata in Anatolia. Hum Genet. 2004;114:127–148. doi: 10.1007/s00439-003-1031-4. [DOI] [PubMed] [Google Scholar]
- 20.Zei G, Lisa A, Fiorani O, Magri C, Quintana-Murci L, et al. From surnames to the history of Y chromosomes: the Sardinian population as a paradigm. Eur J Hum Genet. 2003;11:802–807. doi: 10.1038/sj.ejhg.5201040. [DOI] [PubMed] [Google Scholar]
- 21.Repping S, van Daalen SK, Brown LG, Korver CM, Lange J, et al. High mutation rates have driven extensive structural polymorphism among human Y chromosomes. Nat Genet. 2006;38:463–467. doi: 10.1038/ng1754. [DOI] [PubMed] [Google Scholar]
- 22.Flores C, Maca-Meyer N, Larruga JM, Cabrera VM, Karadsheh N, et al. Isolates in a corridor of migrations: a high-resolution analysis of Y-chromosome variation in Jordan. J Hum Genet. 2005;50:435–441. doi: 10.1007/s10038-005-0274-4. [DOI] [PubMed] [Google Scholar]
- 23.Marjanovic D, Fornarino S, Montagna S, Primorac D, Hadziselimovic R, et al. The peopling of modern Bosnia-Herzegovina: Y-chromosome haplogroups in the three main ethnic groups. Ann Hum Genet. 2005;69:757–763. doi: 10.1111/j.1529-8817.2005.00190.x. [DOI] [PubMed] [Google Scholar]
- 24.Rozen S, Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 25.Hall A, Ballantyne J. The development of an 18-locus Y-STR system for forensic casework. Anal Bioanal Chem. 2003;376:1234–1246. doi: 10.1007/s00216-003-2039-2. [DOI] [PubMed] [Google Scholar]
- 26.Daniels DL, Hall AM, Ballantyne J. SWGDAM developmental validation of a 19-locus Y-STR system for forensic casework. J Forensic Sci. 2004;49:668–683. [PubMed] [Google Scholar]
- 27.Bandelt HJ, Forster P, Rohl A. Median-joining networks for inferring intraspecific phylogenies. Mol Biol Evol. 1999;16:37–48. doi: 10.1093/oxfordjournals.molbev.a026036. [DOI] [PubMed] [Google Scholar]