Abstract
Background
So far, all clinical cases of new variant Creutzfeldt-Jakob disease (vCJD), thought to result from the Bovine Spongiform Encephalopathy (BSE) prion agent, have shown Methionine–Methionine (M/M) homozygosity at the M129V polymorphism of the PRNP gene. Although established, this relationship is still not understood. In both vCJD and experimental BSE models prion agents do reach the bloodstream, raising concerns regarding disease transmission through blood transfusion.
Methodology/Principal Findings
We investigated the impact of the M129V polymorphism on the expression and processing of the prion protein in human peripheral blood mononuclear cells (PBMCs) from three blood donor populations with Methionine-Methionine (M/M), Valine-Valine (V/V) and M/V genotypes. Using real-time PCR, ELISA and immunoblot assays we were unable to find differences in prion protein expression and processing relating to the M129V polymorphism.
Conclusions/Significance
These results suggest that in PBMCs, the M129V PrP polymorphism has no significant impact on PrP expression, processing and the apparent glycoform distribution. Prion propagation should be investigated further in other cell types or tissues.
Introduction
Transmissible spongiform encephalophathies (TSEs or prion diseases) are fatal neurodegenerative disorders, of which the main human form is Creutzfeldt–Jakob disease (CJD) [1], [2]. Importantly, a new variant of CJD (vCJD) has recently emerged [3], believed to result from oral exposure to Bovine Spongiform Encephalopathy (BSE). Although the exact nature of the infective agent remains controversial, many believe that infectivity is closely associated with the accumulation of a pathological misfolded protein, PrPSc (Scrapie Prion Protein), derived from the normal cellular protein PrPC [4]. PrPC and PrPSc isoforms share the same primary structure [5]: a single disulfide bond [6], two complex-type N–Linked oligosaccharide chains [7], [8], and a C-terminal glycosyl phosphatidylinositol (GPI) anchor [9]. The difference between the isoforms relates to their secondary structure: PrPC is rich in α-helices, while PrPSc has a higher content of β-pleated sheets [10]–[12]. During prion propagation, the increase in the level of infectivity is associated in most cases with the accumulation of the misfolded PrPSc protein, believed to convert PrPC through a pathological conformational rearrangement [13]. Although the mechanism explaining this transconformation remains unclear, two different models have been proposed: nucleation-polymerization and template-assisted conversion [14], [15]. Both involve the recruitment of normal prion protein PrPC. Biochemical studies in cultured cells demonstrate that conformational rearrangement of PrPC can occur in the absence of glycosylation [16] and that N-glycan chains can modulate the in vitro conversion of PrPC into PrPSc-like molecules [17]. In the normal brain, PrPC processing includes proteolysis of the full-length protein to generate a main N-terminal truncated protein named the C1 fragment. However, proteolysis can also generate an additional, longer C-terminus protein –the C2 fragment - that accumulates in prion-infected brain in particular. [18].The C2 fragment only accumulates as a resistant truncated protein in CJD-affected brains. This raises questions about whether this truncated form influences the possibility of developing the illness. Moreover, studies have described the accumulation of under-glycosylated, full-length and N-Terminal truncated PrP as a fingerprint for prion disease [19].
The PRNP gene, on the short arm of human chromosome 20, encodes the prion protein [20]. Some polymorphisms of the PRNP open reading frame (ORF) seem to be involved in susceptibility to different forms of CJD (iCJD, gCJD, sCJD, vCJD) [21]. This is illustrated by the fact that 100% of the clinical vCJD cases thus far have been Methionine –Methionine homozygous at the M129V polymorphism (129-M/M) [22], yet only 40% of the Caucasian population carry this genotype. Importantly, in classical CJD patients, PrPSc accumulates in neuronal, [23], [24] follicular dendritic cells, and muscle [25], but for vCJD patients, PrPSc also accumulates in lymphoid tissue, including the tonsils and appendix [26]–[29]. This distribution raised concerns about blood transfusion from the very beginning of the epidemic. Since then, studies by Houston F. et al [30] have demonstrated that prion transmission by blood transfusion occurs experimentally in sheep infected with BSE. In humans, three UK vCJD patients had previously been transfused with red cells donated by individuals who subsequently developed vCJD 1.5–3.5 years after donation. All three transfusion-recipient patients carried the 129-M/M genotype. A fourth recipient of red cells from a donor who later developed vCJD, died from unrelated causes. Post mortem investigation in this case found abnormal prion accumulation in the spleen and in one cervical lymph node, but not in the brain [31]–[33]. Recently, a person with hemophilia who died of a condition unrelated to vCJD, only showed evidence of infection with the abnormal prion protein in the spleen at post mortem. In these cases without clinical signs, both patients were heterozygous 129-M/V. This bears in mind the sub-clinical CJD cases with 129-V/V recently described in a retrospective study of appendix samples [34]. Polymorphism at position 129 could, therefore, not only influence susceptibility to prions, but also impact the incubation time or clinical onset of the disease. One possible mechanism to explain this relation would be the consequence of 129 polymorphism on PrPC expression and processing that are critical for prion propagation [4], [16], [17], [35]. Given the concerns regarding vCJD transmission through blood transfusion, studying PrPC in blood seems particularly relevant. Indeed, PrPC distribution determined by ELISA and flow cytometry, shows that plasma mononuclear cells and platelets express PrPC, although contradictory results were obtained for red cells and granulocytes [36], [37].
In order to investigate a possible relationship between the M129V polymorphism and susceptibility to vCJD and PrPC expression, we compared three blood donor populations of different genotypes: M/M, M/V, and V/V at position 129. We analyzed their expression of total PrPC (mRNA, protein) and PrPC isoforms (glycosylated and truncated proteins) in peripheral blood mononuclear cells (PBMCs).
Our results showed that in these cells, PrP expression and processing did not determine susceptibility to prion disease.
Materials and Methods
Blood Samples
After having obtained informed consent in writing from each blood donor, in compliance with the French law (code de la santé publique article L.1243-3), whole blood was taken in EDTA collection tubes. Given the well known distribution of codon M129V genotypes (41% M/M, 49% M/V and 10% V/V) in the French population, the study did not require the approval of the Bioethical Review Board. We genotyped a total of 119 donors in order to obtain a statistically representative V/V population.
Isolation of peripheral blood mononuclear cells (PBMCs)
In order to avoid bias due to physiological variation in the percentage of granulocytes (40 to 70%) in human blood, we examined only purified mononuclear cells. Indeed, preliminary studies performed on leukocyte populations (data not shown) indicate a lower PrPC mRNA expression in granulocytes than mononuclear cells. Consequently, we only purified mononuclear cells from blood by centrifugation through a Ficoll-Hypaque gradient (Pharmacia, 91 898 Orsay, France). In brief, whole blood diluted Vol./Vol. with phosphate-buffer saline (PBS) was carefully layered upon Ficoll and spun at 700 g. Mononuclear cells were harvested from the interface of the plasma-platelets layer and the Ficoll, washed in PBS and suspended in the same buffer. The cell suspension was analyzed in triplicate, on the ABX (Beckman Coulter, 95942 Roissy, France) hemocytometer for total cell number and mononuclear cell purity level. After elimination of PBS, aliquots of 106 PBS-cleared PBMCs were stored at −80°C.
RNA extraction
Total RNA was isolated (High Pure RNA isolation kit, Roche Diagnostics, 38240 Meylan France) from 106 PBMCs, according to the manufacturer's instructions. Briefly, cells were lysed by solubilizing them in lysis/binding buffer optimized for RNA extraction (4,5M guanidin hydrochloride, 50 mM Tris-HCl and 30% Triton X-100, pH 6.6). Homogenized lysate was then transferred to a filter tube for binding to the fiberglass filter. After centrifugation at 10 000 g for 15 sec, residual contaminated DNA was digested by DNase I (180 U/tube) applied directly to the fiberglass filter and incubated for 15 min. After washing three times - in order to eliminate enzymes and cellular impurities - purified RNA was eluted with 50 µl elution buffer (nuclease-free sterile water), and immediately stored at −80°C.
As the PRNP ORF sequence is located on only one exon, to exclude potential contamination by genomic sequences, we treated total RNA extracts with RNase-free DNase before amplification. Samples were then controlled for the absence of genomic sequence by performing PCR directly on the RNA extracts, thus avoiding the reverse transcription step.
cDNA synthesis
The first cDNA strand was synthesized by reverse transcription (R.T. at 42°C for 1 hour) from 5 µl of RNA (preheated for 7 min at 70°C) in the presence of hexanucleotides (7 µM), dNTPs (800 µM each), 60 U Rnasin Ribonuclease Inhibitor (Promega, 69260 Charbonnières France) and 300 U M-MLV-RT (Promega) in a final volume of 35 µl. cDNA was then purified with “High Pure PCR Product Purification Kit” (Roche Diagnostics) according to the manufacturer's instructions. The test principle was the same as previously described for RNA extraction; the only difference concerned the lysis buffer, which was adapted for DNA products. Finally, cDNA was eluted with 50 µl elution buffer, supplemented with salmon sperm DNA (10 ng/µl final), and kept at −20°C.
Real-time PCR
Standard curve preparation
First, RNA was produced by in vitro transcription with AmpliScribe T7, T3 and SP6 High Yield Transcription Kit (Epicentre/Tebu 78610 Le Perray-en-Yvelines France) according to the manufacturer's instructions. The ORF sequence (759 pb) of the human prion gene - inserted into a pcDNA3 plasmid - was used as the reaction template. Second, four one-in-ten serial dilutions were prepared from a stock RNA solution at 10−11 grams/µl (g/µl). This yielded a standard curve with concentrations ranging from 10−12 to 10−15 g/µl, corresponding to 2.33×106 and 2.33×103copies/reaction.
Primer Sequences
Primer Sequences selected for real-time PCR were those described by Dodelet et al. [38]. They ensure amplification of a fragment of 432 pb length, encompassing the M129V polymorphism within the human prion ORF.
Quantification
Quantification was carried out on a LightCycler by real-time PCR and SYBR Green I dye (Roche Diagnostics, Penzberg, Germany). Amplification assays were performed with 1 µl of cDNA in 20 µl final reaction mixture containing 2 µl LightCycler-FastStart DNA Master SYBR Green (Roche Diagnostics) and 0.6 µM of each primer (forward, nt 79 to 99 and reverse, nt 498 to 510). PCR conditions consisted of an initial denaturation at 95°C for 5 min, and 45 cycles at 95°C for 15 sec, 65°C for 5 sec, 72°C for 15 sec, 89°C for 5 sec. All samples were analyzed in duplicate. The amount of DNA was estimated after each PCR cycle by reading fluorescent dye incorporation (SYBR Green) at 530 nm in the PCR product. At the end of each run, a DNA melting step was performed and a fusion curve was recorded to control the homogeneity and specificity of the amplified DNA.
Analysis of the results
Readings were performed for each cycle after a fourth segment at 89°C, in order to exclude non-specific signal interference with the concentration calculation. Analysis was automatically performed by LightCycler software 4.0 as follows: after PCR completion, LightCycler software calculated the copy number of target molecules by plotting the logarithm of fluorescence versus cycle numbers and setting a baseline x-axis. From each sample, the baseline identified the cycle (crossing point, Ct) at which the log-linear signal could be distinguished from the background. Each run was analyzed by setting the noise band just above the background fluorescence. A value corresponding to F = 0.5 was systematically chosen for comparison between the runs. Regression of the x-axis crossing point of each standard with known concentrations defined the standard curve from which unknown samples were estimated. Specificity was achieved by plotting a melting curve graph based on the final PCR.
129 codon Genotyping
According to Teupser et al [39] the following probe pair spans the nucleic acid sequence responsible for the polymorphism: a detection probe complementary to the 129M allele and 3′-labeled with fluorescein, and the adjoining, anchoring probe 5′-labeled with Red 640. Genotyping assays were carried out in a reaction mixture containing 10 µl of amplicons and probes, at 2.5 picomoles each per reaction. Assay conditions consisted of initial denaturation at 95°C for 2 min, followed by hybridization at 42°C for 2 min and final denaturation by increasing the series to 75°C at a rate of 0.1°C with continuous reading at 640 nm. Analysis using LightCycler was performed by creating a melting profile that defined the melting temperature (Tm) of the hybrid, composed of the target DNA and the fluorescent probe pair. Tm depends on the degree of homology between the two strands, which distinguishes M129V polymorphism. At the end of the reading, LightCycler software 4 draws a melting curve, by plotting fluorescence (F) versus temperature (T), which is automatically converted to melting peaks (-dF/dT).
Preparation of protein lysate and PNGase F-deglycosylated protein
All samples were prepared from 106 PBMCs according to 2 protocols:
Protein lysate preparation only
Cells were dissociated and incubated on ice for 30 min in a lysis buffer containing 150 mM NaCl, 0.5% triton X-100, 0.5% sodium deoxycholate and 50 mM Tris-HCl pH 7.5, and supplemented with a protease inhibitor cocktail (Roche, 82372 Penzberg Germany). After centrifugation at 450 g for 2 min, the supernatant was harvested, added to 4× loading buffer (Invitrogen, Paisley, UK), heated at 95°C for 5 min and stored at −80°C.
Protein lysate preparation with PNGase treatment
Protein lysate preparation with PNGase treatment was carried out in order to reduce the heterogeneity of the PrPC bands resulting from protein-bound Asn-linked oligosaccharides.
Incubation for 30 min on ice was followed by protein reduction, performed by adding 1% 2-mercaptoethanol and incubating at 90°C for 5 min. After snap cooling on ice, 2 units of PNGase F (Roche Diagnostics) were added and digestion was carried out overnight at 37°C. Then, we precipitated the proteins with 3 volumes of cooled methanol for 2 hours at −20°C. Following centrifugation at 20,000 g for 10 min, the dry pellet was suspended in lysis buffer prior to denaturation with 4× loading buffer (Invitrogen, 95613 Cergy-Pontoise France) and heated at 95°C for 5 min. Finally, samples were stored at −80°C.
Total protein quantification
For all samples, we quantified the lysates for total protein by Bicinchoninic Acid Protein Assay (BCA assay, Sigma, 38297 Saint Quentin Fallavier France), before freezing, according to the manufacturer's instructions.
Determination of PrPC levels
“The Enzyme Immuno Assay (EIA) Kit for the determination of PrPC Protein” (SpiBio 91741 Massy France) was applied to 106 PBMCs, according to the manufacturer's instructions. Briefly, we lysed cells by solubilizing them in 65 µl of extraction buffer (10 mM Tris-HCl pH 4, 100 mM NaCl, 10 mM EDTA, 0.5% IGEPAL, and 1% Deoxycholic Acid). After spinning at 10,000 g for 5 min., the supernatant was denatured by adding 4 M Urea (final concentration, diluted in Tris-HCl 10 mM pH 7.4)) and heating at 100°C for 10 min. Then the EIA, based on a double-antibody sandwich principle, was performed. The micro-well plate was coated with a monoclonal antibody specific to the prion protein (aa 144–153). The detection antibody, conjugated with acetylcholinesterase (AchE), recognized the octo-repeat region located in the N-terminus part of the PrPC. Readings were performed at 414 nm.
SDS-PAGE and Immunoblotting
Antibodies
Four different mouse antibodies were used to span large regions of PrP: SAF32 (SpiBio, 91741 Massy, France) - which recognizes an octa-repeat region located in the N-terminus part of human PrP (human numbering 79–91), 8G8 (SpiBio) - directed against an epitope of human PrP residues 95–110, 3F4 (Signet/Proteogenix, 67412 Illkirch France) - specific for an epitope of human PrP residues 109–112, and an antibody provided from ascites fluid PRI 917 (J.Grassi, CEA Saclay, France) - spanning the COOH- terminus region of human PrPC (aa: 216–221). Normalization of the expected signals was achieved using the signal obtained with monoclonal anti -α- tubulin (Sigma).
SDS-PAGE and Immunoblotting
Protein samples (0.5 106 cells/lane i.e. 50 µg of total protein) were separated by 12% NUPAGE gel electrophoresis (Invitrogen) at 150 volts for 90 min. Proteins were then transferred to nitrocellulose membranes Hybond (Amersham biosciences, 91400 Orsay, France) for 45 min at 400 mA using the TE 22 transfer unit (Amersham biosciences). After blocking with 5% non-fat milk in PBS-0.05%Tween 20 buffer, blots were probed with the panel of selected antibodies in the dilution conditions recommended by the manufacturer. Immunoreactivity detection was carried out by a secondary anti-mouse IgG peroxydase-linked antibody, involved in an ECL reaction (ECL reagent, Amersham biosciences) and visualized on an ECL film (Amersham biosciences). Results were quantified by density readings using Sigmagel Analyzer Software (Sigma).
We prepared five protein samples for each genotype, both with and without PNGase F treatment. Samples were then analyzed using 12% SDS-PAGE separation. We normalized the samples, by migrating lysate obtained from 0.5×106 cells - a total protein concentration of 50 µg +/− 10%/lane - and by comparing the 50 kDa signals obtained by anti -α- tubulin antibody blotting (not shown).
Statistical analysis
Statistical analysis was performed by the test of mean comparison which obeys the Student-Fisher law (from Fisher and Yates, Statistical tables for biological, agricultural and medical research, Olivier and Boyd, Edinburgh). This means that the mean difference is not significant when the calculated absolute t value is less than 0.05. If the value is greater than 0.05, this represents a difference between populations.
Results and Discussion
Characterization of the peripheral blood mononuclear cells
One possible mechanism explaining the relationship between the M129V polymorphism and disease development, would be that the polymorphism influences PrPC expression and processing critical for prion propagation [4], [16], [17], [35]. We decided to test this hypothesis on peripheral blood mononuclear cells (PBMCs) for the following reasons: the controlled and ethical ease of access to PBMCs from blood donors allowed us to collect enough samples to perform a robust analysis. Collecting several hundred human samples is virtually impossible with other tissues. Secondly, as mentioned before, blood is an important issue in the context of vCJD and our results could be important to evaluate the relative risk of PrPSc transmission in different blood components. As PBMCs express the highest level of PrP compared to platelets and red blood cells the choice seemed evident even if no experimental data have showed yet that these cells could indeed propagate prions. In any case, it is not clear how the prion agent reaches the bloodstream. Theories propose local replication in specific cell types, like dendritic cells, or secretion from extra- vascular tissues [40], [41]. Finally, we hypothesized that if the M129V polymorphism exerted a more general impact on PrP expression, it could still be observed in the PBMCs.
Quality controls of the isolated cell population indicated systematic granulocyte contamination (10% to 15%), with an acceptable variation of 5% between samples. To determine the 129 polymorphism, we used melting curve analysis to show a specific melting peak for each genotype: 129-M/M homozygous genotype peaked at 60°C, V/V homozygous at 52°C, whereas M/V heterozygous gave 2 peaks at 52°C and 60°C. The genotype distribution of the donor population of the Languedoc region, France was 42% M/M, 42% M/V and 16% V/V, values that agree with the German and the Spanish populations (Table 1) [39], [42]. Of note, a previous study on the French population observed values of 44% M/M, 46,5% M/V and 9,5% V/V) [43]. However, this study used RFLP-PCR (Restriction Fragment Length Polymorphism-Polymerase Chain Reaction), a procedure dependent on digestion efficiency.
Table 1. M129V polymorphism distribution.
values calculated from 50, 50, and 19 samples respectively per genotype.
Languedoc region.
PrPC expression studies
Since PrPSc generation depends on PrPC expression [44], we examined PrPC expression in each of the three genotype populations. We first analyzed PrPC mRNA levels using real-time PCR amplification and SYBR Green detection for optimal sensitivity. Samples were quantified using a standard curve composed of 4 concentrations ranging from 2.33×106 to 2.33×103 copies/reaction. This agreed with the linear correlation between concentration and cycle number (correlation coefficient: 1 and slope at 3.47, indicating an acceptable PCR yield at 97%). Results from the 50 M/M, 49 M/V and 19 V/V blood samples showed no significant difference between the mRNA levels of the three populations (Table 2, observed t-value less than the 0.05 critical t-value). Nevertheless, the coefficient of variation (CV) indicated a maximum of 4 fold deviation within each group. In view of the numerous steps involved in this method and the normal variation (0.5–0.7 log10) in technologies based on the same number of steps [45], a deviation of around 0.6 log10 within each group, denoted low physiological variation. The absence of significant differences in mRNA expression between the three genotypes could indicate that transcription regulation does not relate specifically to genotype. On the other hand, gene expression profiling studies of scrapie-infected brain tissue have identified numerous genes that do modify mRNA expression. Likewise, expression of proteins encoded and involved in several pathways (e.g. inflammatory reaction, proteolysis, protease inhibition, cell grown, stress, immune responses…) [46]–[49] was modified. For these reasons, we needed to confirm our results at the protein level. We determined PrPC protein levels by using a commercial EIA kit, whose N- and C-terminal antibodies detect the full-length protein. As standard curves were unavailable, we compared absorbency values expressed as Absorbency Unit (AU) for each group. Results obtained from 10 blood samples from each group revealed no statistical difference (observed t-value less than the 0.05 critical t-values) in total PrPC levels between the groups (Table 2). Patients with CJD - and other neurodegenerative diseases - show increased PrPC expression in plasma compared to healthy control groups [50]. Our findings suggest that such over-expression in plasma does not result from PBMCs. Nor does it seem to be linked with M129V polymorphism. Moreover, our total PrPC analysis conflicted with those obtained in sheep, where PBMC surface PrPC expression did depend upon the genotype. Researchers found the highest levels of PrPC in scrapie-susceptible VRQ/VRQ sheep, and the lowest levels in scrapie-resistant ARR/ARR genotypes [51].
Table 2. PrPC expression and processing depending on the M129V polymorphism.
| PrPC mRNA c | PrPC d | PrPC glycosylationa | PrPC cleavage b | PrPC cleavage b | |
| (copies/2000 cells) | (Absorbance Unit) | diglyc./unglyc.e | F.L./C2f | C1/C2g | |
| Met/Met | 3.63E+05 | 1.81 | 8.86 (SAF 32); 3.56 (PRI 917); 1.72 (8G8) | 6.39 (8G8); 3.53 (3F4) | 2.99 (PRI 917) |
| Met/Val | 4.06E+05 | 2.02 | 7.25 (SAF 32); 3.65 (PRI 917); 1.83 (8G8) | 7.50 (8G8); 2.71 (3F4) | 2.20 (PRI 917) |
| Val/Val | 4.98E+05 | 2.15 | 8.87 (SAF 32); 3.56 (PRI 917); 2.09 (8G8) | 6.91 (8G8); 2.22 (3F4) | 2.18 (PRI 917) |
13%<VC<47%.
3%<VC<60%.
52%<VC<63%.
15%<VC<20%.
ratio diglycosylated PrPC/unglycosylated PrPC calculated by image analysis of the western blot assay performed with SAF 32, PRI 917 and 8G8 antibodies.
ratio full length unglycosylated PrPC/C2 fragment calculated by image analysis of the western blot assay performed with 8G8 and 3F4 antibodies.
ratio C1 fragment/C2 fragment calculated by image analysis of the western blot assay performed with PRI 917 antibody.
PrPC glycosylation studies
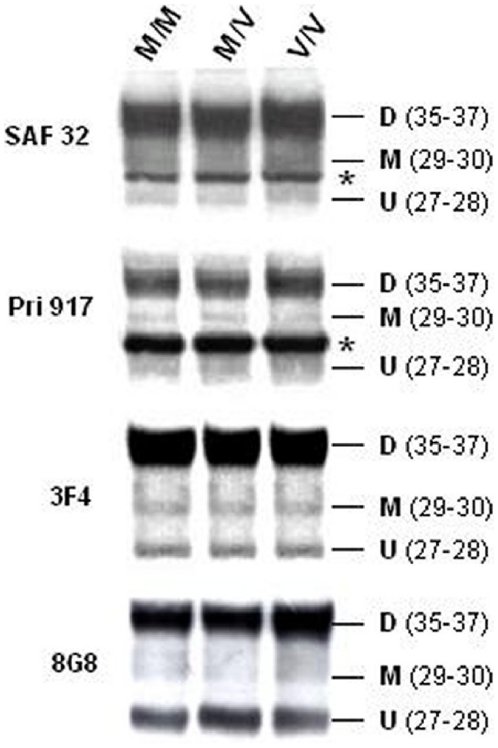
The PrP protein encounters various post-translational modifications, including truncation of signalling peptides, GPI anchoring, disulfide bond formation, N-glycosylation, and α- and β- physiological cleavage. PrPC is normally glycosylated at positions 182 and 198 and different paradigms show that glycans modulate the conversion of PrPC into PrPSc [16], [17]. For this reason, we investigated potential differences in PrPC glycosylation between the three genotypes by performing Western Blots (WB) before and after PrPC deglycosylation with PNGase. Before deglycosylation, both SAF32 (aa: 78–91) and 3F4 (aa: 109–112) revealed similar PrP patterns, composed of three bands (Fig. 1): a dominant 35–37 kDa band corresponding to the diglycosylated full-length PrPC, and two faint bands at 29–30 kDa and 27–28 kDa, corresponding to the monoglycosylated and unglycosylated PrPC isoforms respectively [17]. 8G8 antibody recognized two major bands- corresponding to diglycosylated and unglycosylated PrPC - and also a faint band corresponding to the monoglycosylated form (aa: 95–110) (Fig. 1). The C-terminal antibody Pri 917 (aa: 216–221) showed a major diglycosylated band, and two faint bands corresponding to monoglycosylated and unglycosylated ptoteins (Fig. 1).The α-tubuline signal intensity of the samples agrees with the normalization realized by using 0.5X106 cells per lane corresponding to 50 µg of total protein.
Figure 1. Analysis of PrPC glycosylation.
Immunoblots of PrPC glycoforms from the three PRNP codon 129 genotypes in PBMCs. Denatured lysates from 106 PBMCs were tested by Western Blot analysis using 4 antibodies to span the large PrP region: SAF32 (79–91), 8G8 (95–110), 3F4 (109–112) and PRI 917 (216–221). The 35–37 kDa band corresponds to the M.W. of the diglycosylated protein (D), while the 29–30 kDa and 27–28 kDa bands correspond to the M.W. of the monoglycosylated (M) and unglycosylated (U) proteins respectively. *: Non-specific signal.
In addition, we observed on blot Pri917 a prominent band between the unglycosylated and monoglycosylated bands. We identified this band as non-specific, firstly because the Pri 917 (aa: 216–221) antibody is not purified but an ascitic fluid extract, and secondly because this band is not present in the 3F4 (aa: 109–112) and 8G8 blots (aa: 95–110). Finally, we recently ran a blot on PBMCs using the 12F10 (aa: 142–160), antibody which identified both C1 and C2 fragments. No non-specific band was detected (results not shown)
We calculated the ratio between diglycosylated (35–37 kDa) and unglycosylated (27–28 kDa) isoforms for each sample, after quantifying SAF32 blot bands using the Sigmagel analyzer Software. We performed ratio calculations on the SAF32 blot, because we observed better resolution of low density bands with this antibody. Comparing the averages of the three ratios, from five samples of each genotype, showed no significant difference in expression (observed t-value less than the 0.05 critical t-value) between the groups (Table 2). These findings were confirmed by quantifying the Pri 917 (C-terminus region) and 8G8 blots (middle region among 3F4). Nevertheless, the absolute ratio values differed according to the antibody used (Table 2).
PrPC cleavage studies
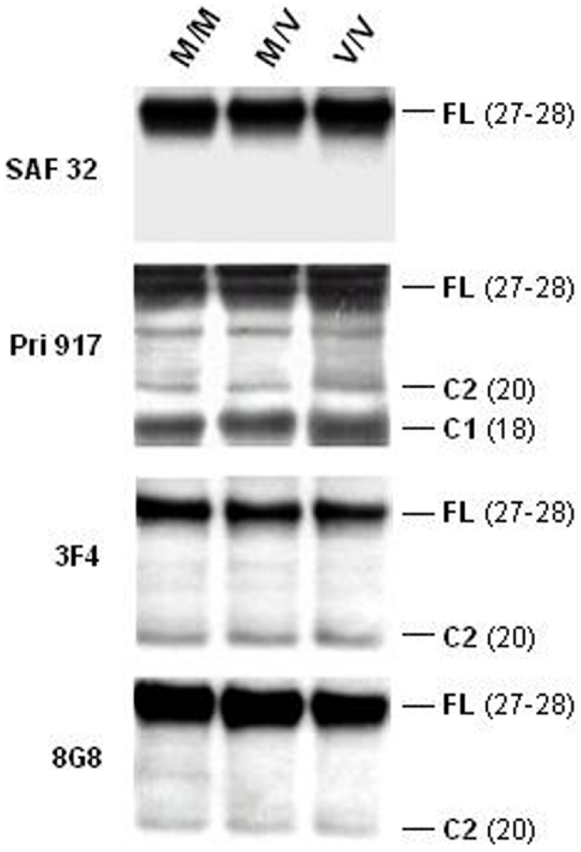
In the normal brain, PrPC proteolysis generates an N-terminal truncated protein - the C1 fragment - from α-cleavage at position 111/112 [18], [52]. However, β-cleavage at residue 90 can generate [52] an additional, longer C-terminal protein – the C2 fragment - that accumulates in prion-infected brain [4]. Importantly, C2 fragments correspond to protease-resistant and insoluble PrPSc. We therefore investigated potential differences in PrP cleavage that might affect PrPSc generation. Following deglycosylation, we noticed identical patterns with the different anti-PrP antibodies between the three genotypes. SAF32 (aa: 78–91) recognized a major band at 27–28 kDa, which corresponds to the estimated MW of the unglycosylated full-length PrPC [52]. As expected, with this N-terminal antibody, no truncated C-terminal forms could be identified (Fig. 2). Both 3F4 and 8G8 antibodies, whose epitopes are located between the α and β cleavage sites, demonstrated similar patterns: a major 27–28 kDa unglycosylated full-length protein band and a faint band near 20 kDa corresponding to the MW of the C2 NH2-terminal truncated fragment of PrPC protein(Fig. 2). This confirms previous results [52] As expected, only the C-terminal antibody Pri 917 (aa: 216–221) identified both the C1 and C2 truncated proteins [52], at approximately 18 kDa and 20 kDa respectively. It also identified the major, unglycosylated full-length protein (27–28 kDa) (Fig. 2).
Figure 2. PrPC cleavage analysis.
Immunoblots of truncated proteins from the three PRNP codon 129 genotypes in PBMCs. After PNGase treatment, denatured lysates from 106 PBMCs were tested by Western Blot analysis using 4 antibodies to span the large PrP region: SAF32 (79–91), 8G8 (95–110), 3F4 (109–112) and PRI 917 (216–221). The 27–28 kDa band corresponds to the M.W. of the unglycosylated Full-Length (F.L.) protein, while the bands near 20 kDa and 18 kDa correspond to the C2 and C1 fragments, respectively.
We calculated the ratio of the unglycosylated full-length PrPC (27–28 kDa) to the C2 NH2-terminal truncated protein for each genotype. Even though the absolute ratios from the 8G8 and 3F4 blots were slightly different, we found no significant difference between the M/M, M/V and V/V populations (five samples per genotype) (observed t-value less than 0.05) for C2 expression (Table 2). As Pri 917 antibody recognizes both C1 and C2 fragments, we also compared the C1/C2 ratios. Values for the M/M, M/V and V/V populations (five samples per genotype) showed no significant difference in expression (t-value less than 0.05) with a good coefficient of variation (Table 2).
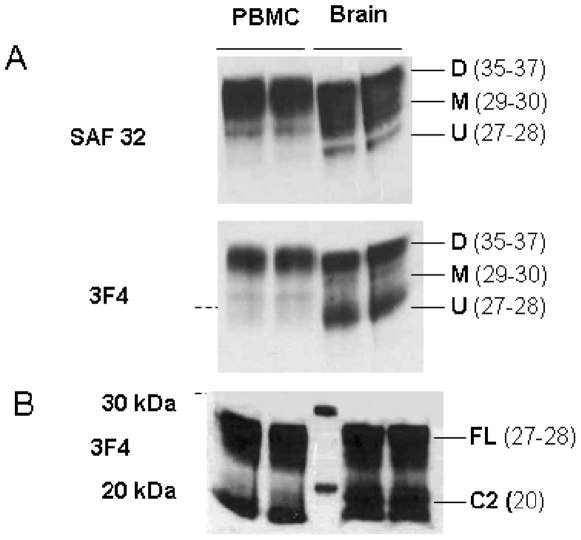
Comparative study with human brain tissue (Fig. 3)
Figure 3. Comparative biochemical analysis between brain and PBMCs.
A) Immunoblots of PrPC glycoforms from PBMCs and brain, Denatured lysates from 106 PBMCs and 10% brain were tested by Western Blot analysis using 2 antibodies: SAF32 (79–91), 3F4 (109–112). The 35–37 kDa band corresponds to the M.W. of the diglycosylated protein (D), while the 29–30 kDa and 27–28 kDa bands correspond to the M.W. of the monoglycosylated (M) and the unglycosylated (U) proteins, respectively. B) Immunoblots of truncated proteins from PBMCs and brain, After PNGase treatment, denatured lysates from 106 PBMCs and brain were tested by Western Blot analysis using 3F4 (109–112). The 27–28 kDa bands correspond to the M.W. of the unglycosylated Full-Length (F.L.) protein while the band near 20 kDa corresponds to the C2 fragment.
We observed two differences when examining normal human brain with the same antibodies (SAF32 and 3F4). First, brain tissue showed higher levels of monoglycosylated and unglycosylated isoforms (Blots SAF 32 and 3F4, respectively), than PBMCs (fig. 3A). Secondly, brain tissue showed the C2 fragment after deglycosylation more strongly than PBMCs (fig. 3B). As the antibodies recognize the same epitope, we cannot explain variations in expression by differences in antibody affinity. These findings agree with previous studies [18], [52]. Our results suggest that both the C2 fragment and under- glycosylated PrPC are under-expressed in PBMCs in comparison to brain tissue. This may result from different physiological functions or protein processing and may influence prion propagation in these tissues. We suggest that low levels of under-glycosylated isoforms and C2 fragments in PBMCs, compared to normal human brain, explains the lower blood levels of PrPSc measured when contamination does occur [53], [54].
So, the molecular mechanism explaining the relationship between the PrP M129V genotype and disease development remains elusive. Although studies have identified some specific amino-acid interactions for the methionine genotype in vitro [55], NMR analysis indicated no PrPC instability related to M129V polymorphism that could explain disease susceptibility [56]. On the other hand, a recent study using molecular dynamic technology demonstrated a higher stability in methionine variants than valine variants [57]. In addition, a comparison of the misfolding pathway, leading to the formation of oligomeric isoforms rich in β-sheets, revealed that methionine 129 human prion protein oligomerized more rapidly than the valine 129 variant [56]. In patients with iatrogenic CJD from contaminated human growth hormone, M129V polymorphism also influences outcome. In this case, homozygous M/M and V/V patients had a shorter incubation period than the heterozygous patients [58], [59].
Taken together, our data suggest that in PBMCs, M129V PrP polymorphism has no significant effect on either PrP mRNA or protein expression. Neither processing nor apparent PrP glycoform distribution seem affected by this M129V polymorphism, which suggests that we should investigate its relationship with prion propagation elsewhere.
Acknowledgments
The authors thank Janine Viladomat for her excellent technical assistance and Julia Roche and Chantal Fournier-Wirth for reviewing the manuscript.
Footnotes
Competing Interests: The authors have declared that no competing interests exist.
Funding: This work was supported by the grant of Etablissement Français du Sang (EFS2003-18). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Pocchiari M. Prions and related neurological diseases. Mol Aspects Med. 1994;15:195–291. doi: 10.1016/0098-2997(94)90042-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Prusiner SB. Genetic and infectious prion diseases. Arch Neurol. 1993;50:1129–53. doi: 10.1001/archneur.1993.00540110011002. [DOI] [PubMed] [Google Scholar]
- 3.Will RG, Ironside JW, Zeidler M, Cousens SN, Estibeiro K, et al. A new variant of Creutzfeldt-Jakob disease in the UK. Lancet. 1996;347:921–5. doi: 10.1016/s0140-6736(96)91412-9. [DOI] [PubMed] [Google Scholar]
- 4.Prusiner SB. Prions. Proc Natl Acad Sci U S A. 1998;95:13363–83. doi: 10.1073/pnas.95.23.13363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stahl N, Baldwin MA, Teplow DB, Hood L, Gibson BW, et al. Structural studies of the scrapie prion protein using mass spectrometry and amino acid sequencing. Biochemistry. 1993;32:1991–2002. doi: 10.1021/bi00059a016. [DOI] [PubMed] [Google Scholar]
- 6.Turk E, Teplow DB, Hood LE, Prusiner SB. Purification and properties of the cellular and scrapie hamster prion proteins. Eur J Biochem. 1988;176:21–30. doi: 10.1111/j.1432-1033.1988.tb14246.x. [DOI] [PubMed] [Google Scholar]
- 7.Endo T, Groth D, Prusiner SB, Kobata A. Diversity of oligosaccharide structures linked to asparagines of the scrapie prion protein. Biochemistry. 1989;28:8380–8. doi: 10.1021/bi00447a017. [DOI] [PubMed] [Google Scholar]
- 8.Haraguchi T, Fisher S, Olofsson S, Endo T, Groth D, et al. Asparagine-linked glycosylation of the scrapie and cellular prion proteins. Arch Biochem Biophys. 1989;274:1–13. doi: 10.1016/0003-9861(89)90409-8. [DOI] [PubMed] [Google Scholar]
- 9.Stahl N, Baldwin MA, Hecker R, Pan KM, Burlingame AL, et al. Glycosylinositol phospholipid anchors of the scrapie and cellular prion proteins contain sialic acid. Biochemistry. 1992;31:5043–53. doi: 10.1021/bi00136a600. [DOI] [PubMed] [Google Scholar]
- 10.Caughey BW, Dong A, Bhat KS, Ernst D, Hayes SF, et al. Secondary structure analysis of the scrapie-associated protein PrP 27-30 in water by infrared spectroscopy. Biochemistry. 1991;30:7672–80. doi: 10.1021/bi00245a003. [DOI] [PubMed] [Google Scholar]
- 11.Pan KM, Baldwin M, Nguyen J, Gasset M, Serban A, et al. Conversion of alpha-helices into beta-sheets features in the formation of the scrapie prion proteins. Proc Natl Acad Sci U S A. 1993;90:10962–6. doi: 10.1073/pnas.90.23.10962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Safar J, Roller PP, Gajdusek DC, Gibbs CJ., Jr Conformational transitions, dissociation, and unfolding of scrapie amyloid (prion) protein. J Biol Chem. 1993;268:20276–84. [PubMed] [Google Scholar]
- 13.Moroncini G, Kanu N, Solforosi L, Abalos G, Telling GC, et al. Motif-grafted antibodies containing the replicative interface of cellular PrP are specific for PrPSc. Proc Natl Acad Sci U S A. 2004;101:10404–9. doi: 10.1073/pnas.0403522101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cohen FE, Prusiner SB. Pathologic conformations of prion proteins. Annu Rev Biochem. 1998;67:793–819. doi: 10.1146/annurev.biochem.67.1.793. [DOI] [PubMed] [Google Scholar]
- 15.Caughey B, Raymond GJ, Callahan MA, Wong C, Baron GS, et al. Interactions and conversions of prion protein isoforms. Adv Protein Chem. 2001;57:139–69. doi: 10.1016/s0065-3233(01)57021-7. [DOI] [PubMed] [Google Scholar]
- 16.Taraboulos A, Rogers M, Borchelt DR, McKinley MP, Scott M, et al. Acquisition of protease resistance by prion proteins in scrapie-infected cells does not require asparagine-linked glycosylation. Proc Natl Acad Sci U S A. 1990;87:8262–6. doi: 10.1073/pnas.87.21.8262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lehmann S, Harris DA. Blockade of glycosylation promotes acquisition of scrapie-like properties by the prion protein in cultured cells. J Biol Chem. 1997;272:21479–87. doi: 10.1074/jbc.272.34.21479. [DOI] [PubMed] [Google Scholar]
- 18.Chen SG, Teplow DB, Parchi P, Teller JK, Gambetti P, et al. Truncated forms of the human prion protein in normal brain and in prion diseases. J Biol Chem. 1995;270:19173–80. doi: 10.1074/jbc.270.32.19173. [DOI] [PubMed] [Google Scholar]
- 19.Pan T, Wong P, Chang B, Li C, Li R, et al. Biochemical fingerprints of prion infection: accumulations of aberrant full-length and N-terminally truncated PrP species are common features in mouse prion disease. J Virol. 2005;79:934–43. doi: 10.1128/JVI.79.2.934-943.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee IY, Westaway D, Smit AF, Wang K, Seto J, et al. Complete genomic sequence and analysis of the prion protein gene region from three mammalian species. Genome Res. 1998;8:1022–37. doi: 10.1101/gr.8.10.1022. [DOI] [PubMed] [Google Scholar]
- 21.Alperovitch A, Zerr I, Pocchiari M, Mitrova E, de Pedro Cuesta J, et al. Codon 129 prion protein genotype and sporadic Creutzfeldt-Jakob disease. Lancet. 1999;353:1673–4. doi: 10.1016/s0140-6736(99)01342-2. [DOI] [PubMed] [Google Scholar]
- 22.No author. Genetic epidemiology of Creutzfeldt-Jakob disease in Europe. 2001;157:633–7. [PubMed] [Google Scholar]
- 23.Kretzschmar HA, Prusiner SB, Stowring LE, DeArmond SJ. Scrapie prion proteins are synthesized in neurons. Am J Pathol. 1986;122:1–5. [PMC free article] [PubMed] [Google Scholar]
- 24.Moser M, Colello RJ, Pott U, Oesch B. Developmental expression of the prion protein gene in glial cells. Neuron. 1995;14:509–17. doi: 10.1016/0896-6273(95)90307-0. [DOI] [PubMed] [Google Scholar]
- 25.Peden AH, Ritchie DL, Head MW, Ironside JW. Detection and localization of PrPSc in the skeletal muscle of patients with variant, iatrogenic, and sporadic forms of Creutzfeldt-Jakob disease. Am J Pathol. 2006;168:927–35. doi: 10.2353/ajpath.2006.050788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hill AF, Butterworth RJ, Joiner S, Jackson G, Rossor MN, et al. Investigation of variant Creutzfeldt-Jakob disease and other human prion diseases with tonsil biopsy samples. Lancet. 1999;353:183–9. doi: 10.1016/s0140-6736(98)12075-5. [DOI] [PubMed] [Google Scholar]
- 27.Hilton DA, Fathers E, Edwards P, Ironside JW, Zajicek J. Prion immunoreactivity in appendix before clinical onset of variant Creutzfeldt-Jakob disease. Lancet. 1998;352:703–4. doi: 10.1016/S0140-6736(98)24035-9. [DOI] [PubMed] [Google Scholar]
- 28.Klein MA, Frigg R, Flechsig E, Raeber AJ, Kalinke U, et al. A crucial role for B cells in neuroinvasive scrapie. Nature. 1997;390:687–90. doi: 10.1038/37789. [DOI] [PubMed] [Google Scholar]
- 29.Klein MA, Frigg R, Raeber AJ, Flechsig E, Hegyi I, et al. PrP expression in B lymphocytes is not required for prion neuroinvasion. Nat Med. 1998;4:1429–33. doi: 10.1038/4022. [DOI] [PubMed] [Google Scholar]
- 30.Houston F, Foster JD, Chong A, Hunter N, Bostock CJ. Transmission of BSE by blood transfusion in sheep. Lancet. 2000;356:999–1000. doi: 10.1016/s0140-6736(00)02719-7. [DOI] [PubMed] [Google Scholar]
- 31.Pincock S. Patient's death from vCJD may be linked to blood transfusion. Committee to discuss need for further precautions to prevent possible vCJD transmission through blood. Lancet. 2004;363:43. doi: 10.1016/s0140-6736(03)15247-6. [DOI] [PubMed] [Google Scholar]
- 32.Pincock S. Government confirms second case of vCJD transmitted by blood transfusion. Bmj. 2004;329:251. doi: 10.1136/bmj.329.7460.251-a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Turner ML, Ludlam CA. An update on the assessment and management of the risk of transmission of variant Creutzfeldt-Jakob disease by blood and plasma products. Br J Haematol. 2009;144:14–23. doi: 10.1111/j.1365-2141.2008.07376.x. [DOI] [PubMed] [Google Scholar]
- 34.Ironside JW, Bishop MT, Connolly K, Hegazy D, Lowrie S, et al. Variant Creutzfeldt-Jakob disease: prion protein genotype analysis of positive appendix tissue samples from a retrospective prevalence study. Bmj. 2006;332:1186–8. doi: 10.1136/bmj.38804.511644.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harris DA, Huber MT, van Dijken P, Shyng SL, Chait BT, et al. Processing of a cellular prion protein: identification of N- and C-terminal cleavage sites. Biochemistry. 1993;32:1009–16. doi: 10.1021/bi00055a003. [DOI] [PubMed] [Google Scholar]
- 36.Barclay GR, Houston EF, Halliday SI, Farquhar CF, Turner ML. Comparative analysis of normal prion protein expression on human, rodent, and ruminant blood cells by using a panel of prion antibodies. Transfusion. 2002;42:517–26. doi: 10.1046/j.1537-2995.2002.00095.x. [DOI] [PubMed] [Google Scholar]
- 37.Barclay GR, Hope J, Birkett CR, Turner ML. Distribution of cell-associated prion protein in normal adult blood determined by flow cytometry. Br J Haematol. 1999;107:804–14. doi: 10.1046/j.1365-2141.1999.01789.x. [DOI] [PubMed] [Google Scholar]
- 38.Dodelet VC, Cashman NR. Prion protein expression in human leukocyte differentiation. Blood. 1998;91:1556–61. [PubMed] [Google Scholar]
- 39.Teupser D, Heino N, Wilfert W, Thiery J. Rapid detection of the prion protein M129V polymorphism with the LightCycler. J Neurosci Methods. 2002;115:93–6. doi: 10.1016/s0165-0270(02)00008-0. [DOI] [PubMed] [Google Scholar]
- 40.Simak J, Holada K, D'Agnillo F, Janota J, Vostal JG. Cellular prion protein is expressed on endothelial cells and is released during apoptosis on membrane microparticles found in human plasma. Transfusion. 2002;42:334–42. doi: 10.1046/j.1537-2995.2002.00072.x. [DOI] [PubMed] [Google Scholar]
- 41.Starke R, Drummond O, MacGregor I, Biggerstaff J, Gale R, et al. The expression of prion protein by endothelial cells: a source of the plasma form of prion protein? Br J Haematol. 2002;119:863–73. doi: 10.1046/j.1365-2141.2002.03847.x. [DOI] [PubMed] [Google Scholar]
- 42.Vega A, Ruiz-Ponte C, Carracedo A, Barros F. Rapid genotyping of the M129V polymorphism of prion protein using real-time fluorescent PCR. Clin Chem. 2001;47:1874–5. [PubMed] [Google Scholar]
- 43.Lucotte G, Mercier G. The population distribution of the Met allele at the PRNP129 polymorphism (a high risk factor for Creutzfeldt-Jakob disease) in various regions of France and in West Europe. Infect Genet Evol. 2005;5:141–4. doi: 10.1016/j.meegid.2004.08.002. [DOI] [PubMed] [Google Scholar]
- 44.Prusiner SB, Groth D, Serban A, Koehler R, Foster D, et al. Ablation of the prion protein (PrP) gene in mice prevents scrapie and facilitates production of anti-PrP antibodies. Proc Natl Acad Sci U S A. 1993;90:10608–12. doi: 10.1073/pnas.90.22.10608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee SC, Antony A, Lee N, Leibow J, Yang JQ, et al. Improved version 2.0 qualitative and quantitative AMPLICOR reverse transcription-PCR tests for hepatitis C virus RNA: calibration to international units, enhanced genotype reactivity, and performance characteristics. J Clin Microbiol. 2000;38:4171–9. doi: 10.1128/jcm.38.11.4171-4179.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Xiang W, Windl O, Wunsch G, Dugas M, Kohlmann A, et al. Identification of differentially expressed genes in scrapie-infected mouse brains by using global gene expression technology. J Virol. 2004;78:11051–60. doi: 10.1128/JVI.78.20.11051-11060.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Riemer C, Neidhold S, Burwinkel M, Schwarz A, Schultz J, et al. Gene expression profiling of scrapie-infected brain tissue. Biochem Biophys Res Commun. 2004;323:556–64. doi: 10.1016/j.bbrc.2004.08.124. [DOI] [PubMed] [Google Scholar]
- 48.Baker CA, Manuelidis L. Unique inflammatory RNA profiles of microglia in Creutzfeldt-Jakob disease. Proc Natl Acad Sci U S A. 2003;100:675–9. doi: 10.1073/pnas.0237313100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Baker CA, Lu ZY, Manuelidis L. Early induction of interferon-responsive mRNAs in Creutzfeldt-Jakob disease. J Neurovirol. 2004;10:29–40. doi: 10.1080/13550280490261761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Volkel D, Zimmermann K, Zerr I, Bodemer M, Lindner T, et al. Immunochemical determination of cellular prion protein in plasma from healthy subjects and patients with sporadic CJD or other neurologic diseases. Transfusion. 2001;41:441–8. doi: 10.1046/j.1537-2995.2001.41040441.x. [DOI] [PubMed] [Google Scholar]
- 51.Halliday S, Houston F, Hunter N. Expression of PrPC on cellular components of sheep blood. J Gen Virol. 2005;86:1571–9. doi: 10.1099/vir.0.80561-0. [DOI] [PubMed] [Google Scholar]
- 52.Mange A, Beranger F, Peoc'h K, Onodera T, Frobert Y, et al. Alpha- and beta- cleavages of the amino-terminus of the cellular prion protein. Biol Cell. 2004;96:125–32. doi: 10.1016/j.biolcel.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Grassi J, Maillet S, Simon S, Morel N. Progress and limits of TSE diagnostic tools. Vet Res. 2008;39:33. doi: 10.1051/vetres:2008009. [DOI] [PubMed] [Google Scholar]
- 54.Gonzalez-Romero D, Barria MA, Leon P, Morales R, Soto C. Detection of infectious prions in urine. FEBS Lett. 2008;582:3161–6. doi: 10.1016/j.febslet.2008.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Alonso DO, DeArmond SJ, Cohen FE, Daggett V. Mapping the early steps in the pH-induced conformational conversion of the prion protein. Proc Natl Acad Sci U S A. 2001;98:2985–9. doi: 10.1073/pnas.061555898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hosszu LL, Jackson GS, Trevitt CR, Jones S, Batchelor M, et al. The residue 129 polymorphism in human prion protein does not confer susceptibility to Creutzfeldt-Jakob disease by altering the structure or global stability of PrPC. J Biol Chem. 2004;279:28515–21. doi: 10.1074/jbc.M313762200. [DOI] [PubMed] [Google Scholar]
- 57.Shamsir MS, Dalby AR. One gene, two diseases and three conformations: molecular dynamics simulations of mutants of human prion protein at room temperature and elevated temperatures. Proteins. 2005;59:275–90. doi: 10.1002/prot.20401. [DOI] [PubMed] [Google Scholar]
- 58.Brandel JP, Preece M, Brown P, Croes E, Laplanche JL, et al. Distribution of codon 129 genotype in human growth hormone-treated CJD patients in France and the UK. Lancet. 2003;362:128–30. doi: 10.1016/S0140-6736(03)13867-6. [DOI] [PubMed] [Google Scholar]
- 59.Huillard d'Aignaux J, Costagliola D, Maccario J, Billette de Villemeur T, Brandel JP, et al. Incubation period of Creutzfeldt-Jakob disease in human growth hormone recipients in France. Neurology. 1999;53:1197–201. doi: 10.1212/wnl.53.6.1197. [DOI] [PubMed] [Google Scholar]