Abstract
The [PSI+] prion may enhance evolvability by revealing previously cryptic genetic variation, but it is unclear whether such evolvability properties could be favored by natural selection. Sex inhibits the evolution of other putative evolvability mechanisms, such as mutator alleles. This paper explores whether sex also prevents natural selection from favoring modifier alleles that facilitate [PSI+] formation. Sex may permit the spread of “cheater” alleles that acquire the benefits of [PSI+] through mating without incurring the cost of producing [PSI+] at times when it is not adaptive. Using recent quantitative estimates of the frequency of sex in Saccharomyces paradoxus, we calculate that natural selection for evolvability can drive the evolution of the [PSI+] system, so long as yeast populations occasionally require complex adaptations involving synergistic epistasis between two loci. If adaptations are always simple and require substitution at only a single locus, then the [PSI+] system is not favored by natural selection. Obligate sex might inhibit the evolution of [PSI+]-like systems in other species.
Author Summary
Can evolvability evolve? One obvious way to evolve faster is via mutator alleles that increase the mutation rate. Unfortunately, recombination will rapidly separate a mutator allele from the advantageous alleles that it creates. Mutators, therefore, gain very little benefit from promoting adaptations and are thought not to evolve in sexual organisms. Here we find that the [PSI+] prion, unlike mutator alleles, will evolve to promote evolvability in sexual yeast species. Together with previous laboratory studies of [PSI+]–mediated adaptation, and with bioinformatic studies consistent with [PSI+]–mediated adaptation in the wild, our theoretical results firmly establish [PSI+] as a model system for the evolution of evolvability. We also shed light on the importance of complex adaptations involving multiple genes. Adaptations involving multiple simultaneous genes drive the evolution of evolvability in this system. This work is an important proof of principle, showing that evolvability can sometimes evolve under realistic conditions.
Introduction
[PSI+] is the aggregated prion form of the protein Sup35 [1]. [PSI+] aggregates appear spontaneously at a low rate [2]. Once established, [PSI+] causes normal Sup35 proteins to misfold to form more [PSI+] [3]. This self-catalytic conversion allows for transgenerational inheritance [4].
The normal, non-prion form of Sup35 is involved in stop codon recognition during gene translation [5],[6]. The depletion of normal Sup35 through its incorporation into prion aggregates leads to readthrough errors at stop codons [7]. This phenotypically reveals previously cryptic genetic variation beyond stop codons [8]–[11]. Revealed variation can sometimes lead to faster growth [10] and adaptation [12] under stressful lab conditions. These observations have controversially suggested a role for [PSI+] in promoting evolvability [13]–[17]. [PSI+] may tap into stocks of variation at times of stress when they are most likely to be needed [18].
[PSI+] induces only low levels of any given adaptive readthrough product. A simple point mutation at the stop codon will produce much higher levels. Let the ancestral allele at locus i with an intact stop codon be designated  (appended gene product - sensu Masel and Bergman [15]) and the derived, adaptive allele with its stop codon destroyed by mutation be designated
(appended gene product - sensu Masel and Bergman [15]) and the derived, adaptive allele with its stop codon destroyed by mutation be designated  (Figure 1). [PSI+] promotes evolvability by acting as a stopgap mechanism. [PSI+] spontaneously appears far more often than stop codon mutations ([2],[19],[20]; see parameter estimates below). This provides partial but nevertheless rapid and therefore easily accessible adaptation. [PSI+] buys time for the lineage to expand, providing more opportunities for more precise adaptation later through genetic assimilation via the appearance of the stop codon mutant agp
+
[9],[14].
(Figure 1). [PSI+] promotes evolvability by acting as a stopgap mechanism. [PSI+] spontaneously appears far more often than stop codon mutations ([2],[19],[20]; see parameter estimates below). This provides partial but nevertheless rapid and therefore easily accessible adaptation. [PSI+] buys time for the lineage to expand, providing more opportunities for more precise adaptation later through genetic assimilation via the appearance of the stop codon mutant agp
+
[9],[14].
Figure 1. Different mechanisms of readthrough translation.
Either the presence of [PSI+] (B) or an agp + point mutation (C) can lead to readthrough of the wild-type stop codon (A).
[PSI+] may provide a convenient model system for the more general study of evolvability via evolutionary capacitors [21]. Evolutionary capacitors are molecular mechanisms that act as switches to control the storage and release of cryptic genetic variation. Cryptic stocks of variation are likely to be pre-enriched for potential adaptations, making this mechanism of evolvability more potent than a reliance on new mutation [22].
Despite these experimental and theoretical results, a role for [PSI+] in evolvability has remained controversial. In particular, although data directly show that [PSI+] can sometimes promote rapid growth and adaptation in novel environments [10],[12], this does not imply that indirect selection for evolvability explains the emergence and evolutionary conservation of the [PSI+] system.
Theoretical results support the evolution of the evolvability properties of the [PSI+] system, but all such studies to date have neglected sex [15],[21],[23],[24]. This is of concern, since the evolution of another putative evolvability mechanism, namely mutator alleles, is dramatically inhibited by sex [25]–[27]. This is because recombination rapidly breaks up linkage between mutator alleles and the adaptations they generate, severely limiting the mutator's ability to hitchhike on the success of those adaptations. This argument does not, however, apply in an identical form to the [PSI+] system, since revealed variants remain dependent on continued [PSI+] expression, preventing their adaptive separation by recombination until genetic assimilation has occurred [15]. When linkage equilibrium evolves only slowly, evolvability may be favored by natural selection [28].
Here we examine for the first time the effect of realistic rates of Saccharomyces sex on the evolution of the evolvability properties of the [PSI+] system. Consider a modifier locus prf (prion-forming - sensu Masel and Bergman [15]) that affects whether [PSI+] is formed. Examples of modifiers of [PSI+] formation in nature include the [PIN +] prion [29], chaperone molecules [30]–[32] and changes in the Sup35 sequence [33],[34]. In our analysis, prf is an abstract modifier in the tradition of theoretical population genetics, rather than a specific, empirically identified modifier. Let the prf0 allele completely suppress de novo [PSI+] formation and the prf+ allele allow for it. We track allele frequencies at the prf locus in order to infer whether the [PSI+] system is favored by natural selection. Both alleles allow propagation of [PSI+], once present.
Usually, [PSI+] is deleterious, and so the prf+ allele incurs small ongoing costs by generating [PSI+] lineages. But on rare occasions, [PSI+] and hence prf+ may be adaptive. The prf0 allele avoids the costs, but is still partially able to usurp the benefits by acquiring the cytoplasmically inherited [PSI+] element through sex with a [PSI+] strain. prf0 can therefore be thought of as a “cheater” allele. When outcrossed sex is rare, however, as it is in Saccharomyces [35], prf0 will on average acquire [PSI+] only after a potentially significant delay, during which a prf+ lineage may have already hitchhiked to high frequency in association with [PSI+]-facilitated adaptation. Here we determine whether prf+ is able to outcompete prf0, implying that the [PSI+] system is favored by natural selection on evolvability, given empirically estimated [35] rates of sex in Saccharomyces.
An interesting aspect of evolutionary capacitors in general, and the [PSI+] system in particular, is the fact that variants at many loci are exposed simultaneously. It has long been speculated that certain adaptations might involve multiple simultaneous changes, and that a temporary period of relaxed selection would allow multiple mutations to accumulate, providing greater diversity as the raw material for evolution [10],[36]. Of course, a potential problem with this idea is that cryptic genetic variation may also contain an accumulation of highly deleterious mutations. This may thwart adaptation, since revealing a stock of variation that includes both highly deleterious and mildly adaptive mutations will on balance likely be deleterious. However, capacitors such as [PSI+] tap into stocks of cryptic genetic variation that had remained subject to low levels of selection while in the cryptic state [22]. This low level of “pre-selection” is sufficient to weed out strongly deleterious alleles, while allowing mutations of small effect to accumulate [22]. One consequence of this pre-selection is that when variation is finally released through a capacitor, adaptations involving multiple simultaneous changes occur far more readily than they would without a capacitance mechanism [22].
Here we consider the evolution of the [PSI+] system via the prf modifier locus in the presence of sex, a fluctuating environment in which [PSI+] occasionally promotes adaptation, and both with and without complex adaptations involving multiple loci. We find that in the presence of realistic frequencies of Saccharomyces sex, complex adaptations are both necessary and sufficient for natural selection on evolvability to drive the evolution of the [PSI+] system.
Materials and Methods
Overview
The simulated diploid Saccharomyces population experiences a fluctuating environment. All environments where [PSI+] is deleterious we label “1” and the environments where [PSI+] generates an adaptation we label “2”. The probability of switching from environment 1 to 2 is Ω12 per generation, and the probability of switching from environment 2 to 1 is Ω21 per generation. We explore environmental switching rates between 10−7 and 10−3 per generation. The population starts in environment 1, with prf+ and prf0 allele frequencies of 0.5, and evolves for 5/Ω12 generations. This process is replicated to determine the proportion of runs for which the prf+ frequency increases. Model parameters are listed in Table 1, including default values when not otherwise specified.
Table 1. Parameters in the Model.
| Parameter | Definition | Value and supporting references |
| δpsi− | Probability of stop codon readthrough in [psi−] individuals | 0.003 [7] |
| δPSI+ | Probability of stop codon readthrough in [PSI+] individuals | 0.01 [7] |
| μ | Per stop codon mutation rate (forwards and backwards) per replication | 1.3×10−9 [19] |
| m | Probability of [PSI+] formation per diploid offspring | 10−5 [2, Tuite MF, pers comm] |
| m′ | Probability of [PSI+] loss per diploid cell division | 10−5 [38] |
| psex | Probability an offspring is formed sexually | 0.001 [35], also 0.01, 0.1 |
| pauto | Given sex, probability of automixis (within tetrad mating) | 0.94 [35] |
| pamphi | Given sex, probability of amphimixis (random mating) | 0.01 [35] |
| phaplo | Given sex, probability of haplo-selfing (mother-daughter mating) | 0.05 [35] |
| Ne | Effective population size | 5×106 [19],[20],[35] |
| ε | Frequency of [PSI +] in wild prf+ populations, used to infer selection against [PSI +] in environment 1 | 0.01 and lower [39],[40] |
| α, s 1 | parameters inferred, primarily from ε and m, that determine the strength of selection against [PSI +] in environment 1 | |
| s 2 | Strength of selection for [PSI +]Agpwt in environment 2 | 0.1-0.001 |
| h | Dominance of prf+ | {0,1} |
| Ω12 | Per generation switching probability from environment 1 to 2 | 10−7–10−5 |
| Ω21 | Per generation switching probability from environment 2 to 1 | 10−5–10−3 |
| k−/k+ | Equilibrium constant of adaptive dimerization in expression concentration units (see Text S2) | 10−3 |
| k1 | Rate constant associated with adaptive readthrough dimer function, per unit concentration (see Text S2) | 50 |
In environment 2, [PSI+] can mediate adaptation by expressing novel gene product(s) at either one locus (simple adaptation; i = 1) or two loci (complex adaptation; i∈{1,2}). Our “agp” notation and our parameter estimates are based on the assumption that adaptation comes from addition to a protein C-terminal through stop codon readthrough. The same formalism can, however, still be applied if the variation revealed by [PSI+] is instead mediated via nonstop mRNA decay [11], via +1 frameshifting at shifty-stop sites [37], or via variation in genes regulated downstream through any of these mechanisms [37]. Each switch to environment 2 is considered unique and involves a new set of agp loci, whose frequencies are initialized at this time. After switching back to environment 1, this set of agp loci is no longer tracked.
If only a single agp
+ allele is required for adaptation, and it is already present or very soon appears in the population, then adaptation will proceed via this more direct route rather than via [PSI+], yielding no benefit to a prf+ allele. If, however, two different readthrough products are simultaneously involved in a complex adaptation, then it becomes exceedingly unlikely that both  alleles will initially be present in the same individual. In this case [PSI+] will have an advantage, since it will cause simultaneous readthrough at both loci, reaping synergistic benefits and promoting complex adaptations. Competing paths of direct vs. [PSI+] mediated adaptation are shown in Figure 2 for the two locus case.
alleles will initially be present in the same individual. In this case [PSI+] will have an advantage, since it will cause simultaneous readthrough at both loci, reaping synergistic benefits and promoting complex adaptations. Competing paths of direct vs. [PSI+] mediated adaptation are shown in Figure 2 for the two locus case.
Figure 2. Alternative pathways, with and without [PSI+], leading to the same readthrough adaptation.
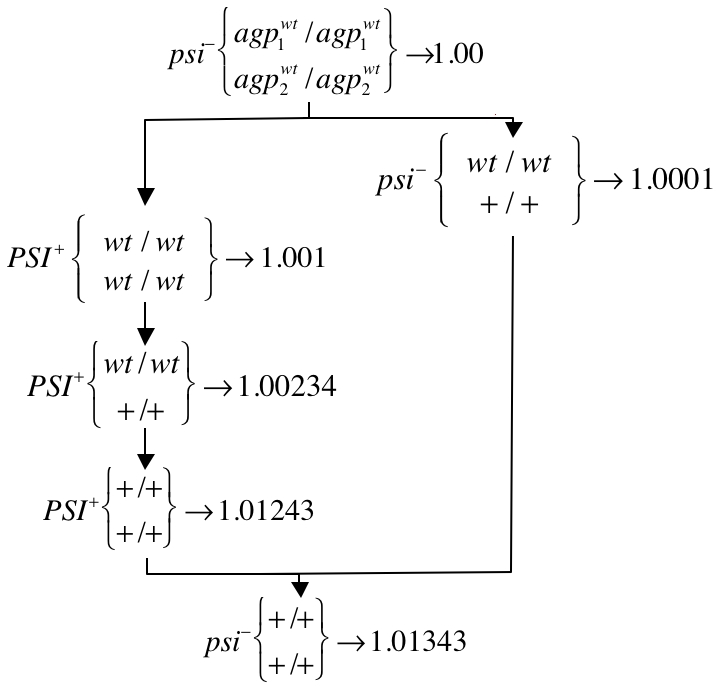
Adaptation in environment 2 can proceed either via: (1) [PSI+] appearance followed by point mutation at agp loci (genetic assimilation) and finally reversion to [psi −] (left) and (2) direct adaptation at agp loci without involvement of [PSI+] (right). The fitness of an individual is given to the right of its genotype, calculated using s 2 = 0.001. Only homozygous states are shown because inbreeding quickly leads to homozygosity. The genetic assimilation pathway typically occurs more often because [PSI+] individuals appear far more often than [psi−] individuals who carry the “+” allele at both agp loci.
Individual Genetic and Cytoplasmic States
We track individual genotypes at the agp and prf loci. Haploids must have different alleles (α vs. a) at the mating-type mat locus in order to conjugate, and so we also track the mat locus for its potential effect on inbreeding. We do not model mutation at the mat and prf loci, except implicitly through the possibility of mother-daughter haplo-selfing (see Reproduction below). There is free recombination between all loci. Individuals therefore have either three or four genetic loci, depending on whether we are modeling simple or complex adaptations with one or two agp loci, respectively. Each of the three to four loci has two alleles, plus there are also two possible cytoplasmic states ([PSI+] versus [psi−]).
Mutation Rate at agp Locus
The point mutation rate in Saccharomyces is around 5×10−10 per base pair per cell division [19]. We approximate the frequencies of the 3 stop codons TAA, TAG and TGA as equal and all mutational substitution types as equally likely. All point mutations at the first position destroy the stop codon. So do all but two at the second position (namely those between TAA and TGA) out of 3 possible substitutions at each of the 3 stop codons. Similarly, of the 9 possible substitutions at the third position, only those between TAA and TAG preserve the stop codon. The total rate of stop codon destruction by point mutation is therefore estimated as (1+7/9+7/9)×5×10−10 = 1.3×10−9 per cell division.
Although mutations that precisely reverse stop codon loss are rarer than this, compensatory mutations can also create alternative stop codons nearby, leading to a functionally equivalent gene product. We therefore assume symmetric mutation rates at the agp loci. The back mutation rate is primarily important only for setting agp allele frequencies at mutation-selection-drift equilibrium in environment 1 (see Text S1).
[PSI+] Appearance and Loss
We explore both the case where the prf+ allele is completely dominant (h = 1), and the case where it is completely recessive (h = 0) relative to the prf0 allele. prf+ individuals form [PSI+] with probability m = 10−5 per generation [2, Tuite MF pers. comm.]. [PSI+] is lost with probability m′ per generation during cell division. Empirical work shows that m′<0.0002 [38]; here we follow the common assumption that m′≈m.
Note that m increases by as much as 60-fold in response to environmental stress [18]. This responsiveness increases the ability of prf+ to promote evolvability. Here we make the conservative assumption that m does not depend on the degree of adaptation to the current environment.
Fitness
In both environments, readthrough products at any locus induced either by [PSI+] or by point mutations in stop codons are likely to incur a fitness cost. This cost could be related to gain or loss of function, and hence specific to the gene in question, or it could be a more general metabolic cost. Here, in order to develop a general, parameterized model, we assume a metabolic cost.
In environment 2, the metabolic cost of readthrough is ameliorated because of the adaptive effects of a substrate-dimer reaction involving readthrough products at one or two loci. Readthrough probabilities are δpsi− = 0.003 and δPSI+ = 0.01 in [psi−] and [PSI+] cells respectively [7]. Let Ei be the level of readthrough at locus i. Ei is equal to δj for  genotypes, (1+δpsi−)/2 for
genotypes, (1+δpsi−)/2 for  genotypes and 1 for
genotypes and 1 for  genotypes, where j∈{psi−, PSI+}. The unit concentrations for all equations below is now given relative to a typical expression level of a gene defined as E = 1.
genotypes, where j∈{psi−, PSI+}. The unit concentrations for all equations below is now given relative to a typical expression level of a gene defined as E = 1.
The fitness of an individual in environment 1 is
| (1) |
where L is either 1 or 2, depending on whether simple or complex adaptation is assumed, and βd is a constant that weights the metabolic cost of readthrough at the potentially adaptive loci relative to the metabolic cost of readthrough across the whole genome. Since there are ∼5000 genes in Saccharomyces, we assume that βd = 1/5000. Equation 1 yields a fitness of 1 in the absence of readthrough, decaying exponentially towards zero as levels of readthrough increase. The parameter α controls the strength of selection against readthrough.
In environment 2, an individual's fitness depends both on the metabolic cost above, and on a benefit accruing from readthrough at agp loci. We assume that the readthrough Agp+ gene product has adaptive function when in the form of a dimer. For simple adaptations, this is an Agp+ homodimer. For complex adaptations, this is an  heterodimer. These dimeric scenarios allow us to capture synergistic epistasis in a realistic way that allows direct comparison between one-locus and two-locus models. Fitness in environment 2 is given by
heterodimer. These dimeric scenarios allow us to capture synergistic epistasis in a realistic way that allows direct comparison between one-locus and two-locus models. Fitness in environment 2 is given by
| (2) |
where βb is a parameter controlling the magnitude of the adaptive effects and t 1/2 is the half-life of a substrate acted on by a catalytic Agp+ dimer. The first term represents the metabolic cost of readthrough, and is identical to fitness in environment 1. βb is set such that the relative fitness of [PSI+] homozygous agp + individuals is 1+s 2 where s 2 = 0.001, 0.01 or 0.1. Fixing s 2 in this way allows appropriate comparisons between the 1-locus and 2-locus models. t 1/2 captures how the strength of adaptation depends on the extent of readthrough at each of the L loci. The biochemical model for calculating t 1/2 depends on the Agp+ dimer concentration and is presented in the Text S2 and Figure S1.
Masel and Griswold [39] estimate the strength of selection against [PSI+]. This estimate depends on the frequency of [PSI+] as a rare polymorphism in wild, [PSI+]-competent Saccharomyces populations. Following expression of a Sup35-GFP fusion protein, a few cells from wild populations show aggregates almost immediately [40]. This suggests the pre-existence of [PSI+] cells containing Sup35 aggregates at a frequency of ε = 1%. If some of these aggregates are false positives, then the true value of ε could be lower. Assuming populations are in epimutation-selection balance, the strength of selection against [PSI+] is [39]
 |
(3) |
where psex is the probability an offspring is formed sexually. Given that an individual is formed sexually, pauto is the probability it is formed via automixis and pamphi is the probability it is formed via amphimixis (see below). Although Equation 3 is complex, its inference of the strength of selection against [PSI+] depends largely just on the observed [PSI+] frequency ε and the rate of [PSI+] appearance m [39].
Given selection s1 against [PSI+] in environment 1, α is given by
 |
(4a) |
 |
(4b) |
Equation 4 is derived from Equation 1 by equating 1−s 1 to the fitness of [PSI+] individuals relative to psi− individuals. We useε to calculate s 1 and hence α, and α to run our simulations. Since there is uncertainty in the estimate of the equilibrium frequency ε of [PSI+] when deleterious, we explore cases when ε is 0.01%, 0.1% and 1%.
Simulated Evolution
We analyze evolutionary competition between prf+ and prf0 alleles by initializing a population in environment 1 with a 0.5 frequency of each allele, and simulating evolution for 5/Ω12 generations to determine how often prf+ increases in frequency.
We use mutation-selection-drift balance theory to initialize [PSI+] frequencies, and also to initialize agp frequencies at the moment when the switch to environment 2 occurs (see Text S1 for details). We assume initial linkage equilibrium between all loci. Although epimutation tends to associate [PSI+] with prf+, we neglect this association during initialization since it is not tractable, and since in any case it establishes itself very rapidly on the timescale of our simulations. Reduced covariance between prf+ and [PSI+] prior to a change from environment 1 to 2 inhibits the maintenance of prf+ (pers. obs.) and so the approximation of linkage equilibrium is conservative relative to inferring whether prf+ can be maintained.
Given genotype and epigenotype frequencies in one generation, we calculate the effects first of reproduction and epimutation (described below), then of mutation at the agp locus, and finally of selection (according to fitnesses described above) to calculate expected genotype frequencies in the next generation. We then sample realized genotype frequencies from expected genotype frequencies using the multinomial distribution to capture genetic drift in a finite population of size Ne. The effective population size Ne in Saccharomyces can be estimated as θ(1+F)/(4μ) where θ is the pairwise sequence divergence estimated as 0.0032–0.0038 [35], the inbreeding coefficient F = 0.98 [35], and the per-base pair per replication point mutation rate μ is around 3.3×10−10 [20] to 5×10−10 [19]. This yields Ne≈3×106–6×106. We use Ne = 5×106.
Reproduction and Epimutation
Saccharomyces is generally diploid, and normally reproduces asexually, with only around psex = 0.1% of offspring formed via sex [35]. We ignore the haploid stage of the life cycle in our calculations of both mutation and selection, thus assuming that there is no fitness cost to sex in terms of a delay in forming the next generation of diploid offspring. We calculate a combination of sexual and asexual diploid offspring produced instantaneously in each generation.
Given sex, only around pamphi = 1% of offspring are generated through amphimictic random mating in the population [35]. pauto = 94% of sexual offspring are formed by automictic within-tetrad mating, while phaplo = 5% of sexual offspring are formed when the products of a haploid mother-daughter mitotic division mate with one another following mating-type switching [35]. In our simulations we explore the effect of varying the overall probability of sex psex, but hold the relative proportions of amphimixis pamphi, automixis pauto and haplo-selfing phaplo constant at the values estimated by Tsai et al. [35]. Amphimictic and automictic mating are only allowed to occur between cells of opposite mating type specified at the mat locus. All sexual reproduction involves independent segregation at each genetic locus.
Propagation of [PSI+] state is slightly more complex. Both sexual and asexual reproduction consist of cell division followed by cell growth. During cell division, [PSI+] is lost with probability m′. During subsequent cell growth, [PSI+] appears spontaneously in prf+ cells with probability m. When reproduction is sexual, both contributing individuals first have the opportunity to lose [PSI+] during meiosis with probability m′. The new diploid individual is then [psi−] only if both parent cells are [psi−]. This allows prf0 lineages to capture the benefits of [PSI+]. During diploid cell growth following mating, [PSI+] has a single opportunity to appear with probability m in prf+ cells.
Simulation from Initially Rare prf+ Mutants
Some simulations were initialized with only a single prf+ mutant rather than with a 50% allele frequency. This single mutant appears in a random genetic background, and in environment 1 rather than environment 2 with probability Ω21/(Ω12+Ω21). When prf+ appeared in environment 1, simulations were carried out in the same way as for an initial 0.5 frequency described above. For single mutants, simulations continued forward in time until prf+ went either extinct or fixed in the population, rather than observing whether its frequency was greater or less than 0.5 after a certain number of generations. Fixation probability was then compared to the neutral expectation of 1/N.
When prf+ appeared in environment 2, simulations began at the time of the previous environmental switch from 1 to 2. Both the time tprf+ of the appearance of the prf+ allele by mutation and the time t21 of switching back to environment 1 were preset as follows. First, tprf+ and t21 were drawn from geometric distributions with mean 1/μprf+ and 1/Ω21 respectively where μprf+ is the probability a prf+ allele arises per generation and was set to an arbitrarily low value. Then while tprf+>t21, we reset t21 to equal t21−tprf+. It is important to note that it is possible for the population to adapt to environment 2 prior to the arrival of the prf+ allele. If the population adapts prior to the arrival of the prf+ allele, prf+ will be unconditionally deleterious.
Confidence Intervals
95% confidence intervals in the figures are calculated using the approximate method suggested by Agresti and Coull [41].
Results
The simulated yeast population experiences a fluctuating environment between times when [PSI+] is deleterious and times when it is adaptive. A sample run showing prf+ fixation is shown for illustrative purposes in Figure 3. prf+ very slowly declines in frequency until the switch from environment 1 to 2 occurs. A [PSI+] selective sweep then immediately begins, with prf+ hitchhiking to high frequency. Later, first one and then both rare agp+ mutants appear. Once recombination has combined the two, a [psi−] revertent sweeps through the population. By this stage prf+ has already become fixed.
Figure 3. An example of two-locus adaptation mediated by [PSI+].
ε = 0.01, Ω12 = Ω21 = 10−5, s 2 = 0.001, h = 1.
prf+ Is Expected to Be Maintained in Yeast Even with Sex, Provided Adaptation Is Complex
We see in Figure 4 that with psex = 10−3, as estimated for S. paradoxus [35], prf+ is favored given complex but not simple adaptations. This inference does not depend on the extreme rarity of yeast sex: with complex adaptations, prf+ would still be maintained even if the probability of sex were raised an order of magnitude. Once sex becomes as frequent as 0.1, prf+ is maintained only if selection on [PSI+]-mediated adaptations is strong. From these results, it seems unlikely that a [PSI+]-like evolvability system could be favored by natural selection in an obligately sexual species under the conditions considered here.
Figure 4. prf+ is maintained in the two-locus but not the one-locus model.
The y-axis gives the probability that the frequency of prf+ after 5×105 generations is greater than its starting frequency of 0.5. The strength of selection s2 for adaptation in environment 2 affects the cutoff frequency of sex. ε = 0.01, Ω12 = Ω21 = 10−5. prf+ is maintained in the two-locus model unless sex is very frequent.
For most of our simulations, we assume prf+ is dominant (h = 1). When prf+ is completely recessive (h = 0), sex provides even less of a barrier to the evolution of evolvability (Figure 4).
Uncertainty in the Strength of Selection against [PSI+]
Inference of the strength of selection against [PSI+] in Equation 3 depends on the estimate ε = 1% of the mean frequency of [PSI+] in prf+ populations at mutation-selection-drift equilibrium (see [39] for details). This estimate may contain false positives and instead be an upper bound. In Figure S2, we see that uncertainty in ε is not important, since lower values of ε, implying stronger selection against [PSI+] in environment 1, lead us to the same conclusions.
Environmental Switching Rates Ω12 and Ω21
If environment 2 is too short-lived for selective sweeps to be completed, then capacitance cannot evolve (Figure 5). This agrees with previous work using a different modeling approach [42].
Figure 5. If environment 2 is too short-lived (high Ω21), then prf+ is not maintained.
This is because there is insufficient time for the selective sweeps shown in Figure 3 to be completed. ε = 0.01, Ω12 = 10−5, h = 1.
Opportunities for adaptation must also arise at a minimum frequency for capacitance to evolve (Figure 6). Previous work in an asexual model found that a capacitor must be useful at a minimum frequency of Ω12>1/Ne per generation in order to be favored by natural selection [23]. With realistic levels of Saccharomyces sex (i.e., psex = 0.001), we see in Figure 6 that prf+ increases in frequency when Ω12>2×10−6, corresponding instead to Ω12 Ne>10. This still corresponds to an exceptionally mild and plausible absolute requirement on the rate of environmental change.
Figure 6. Very rare opportunities for adaptation (low Ω12) cause prf+ to be lost.
ε = 0.01, Ω21 = 10−5.
Fixation of Initially Rare prf + Mutants
A prf+ frequency of 0.5 is a very artificial starting condition, and was chosen for computational efficiency. To test the sensitivity of our results to this starting condition, we also did an invasion analysis starting with a single new prf+ mutant (Figures 7 and 8). The neutral expectation of fixation with probability 1/N is shown by a dashed line. In agreement with results using a 0.5 starting condition (Figure 3), we find that prf+ will fix with a probability greater than the neutral expectation, provided that sex is not too common and selection is not too weak (Figure 7). prf+ fixes more often than the neutral expectation when Ω12≥10−7 (Figure 8), favoring evolvability at even lower levels of Ω12 than with a 0.5 starting condition (Figure 6), in agreement with previous work in an asexual model that Ω12>1/Ne per generation is the necessary and sufficient condition for prf+ to be favored by natural selection [23]. Our more comprehensive calculations above that began with a prf+ frequency of 0.5 seem to be mildly conservative with respect to the evolution of evolvability.
Figure 7. Fixation probabilities starting from a single copy.
prf+ fixes more often than the neutral expectation (10−7), except when selection is weak (s 2≤0.001) and sex is common psex≥0.1. All parameters are equal to values in Figure 3, with the additional parameter μprf+ equal to 10−9. For psex = 0.001 and psex = 0.01, results are based on 106 replicates. For psex = 0.1 and s2 = 0.001, the number of replicates is 3.5×107. Otherwise the number of replicates is 6×106.
Figure 8. Fixation probabilities starting from a single copy.
prf+ allele fixes above neutral expectations (10−7) when the transition probability to environment 2 is greater than 10−8. All parameters are equal to values in Figure 6, with the additional parameter μprf+ = 10−9. Results are based on 106 replicates (Ω12 = 10−5), 5×106 replicates (Ω12 = 10−6), 2×107 replicates (Ω12 = 10−7) and 2×107 replicates (Ω12 = 10−8).
Discussion
When realistic levels of yeast sex are accounted for, indirect selection for evolvability can still favor the evolution of the [PSI+] system. This is only true, however, if adaptation involves at least two loci with synergistic epistatic effects on fitness. Otherwise, with an effective population size as large as that of yeast, all single-locus mutants are readily accessible through mutation. [PSI+] is a stopgap adaptation that incurs costs as well as benefits, and is never preferred to direct adaptation. However, simultaneous direct adaptation at multiple loci is extremely rare, and modifiers of [PSI+] hitchhike to high frequency by virtue of facilitating it.
Evolutionary capacitors, by exposing multiple variants simultaneously, have long been believed to facilitate complex adaptations involving multiple sites [10],[22],[36]. Here we find that the converse is also true: complex adaptations facilitate the evolution of capacitors. This illustrates the intricate relationship between the two.
Sex strongly inhibits the evolution of mutator genes, but here we find that its effect on modifiers of capacitance is much weaker. Nevertheless, were yeast to undergo obligate sex, this would be sufficient to disrupt the evolution of [PSI+] under a model of 2-locus adaptation. Our model is specific to the parameters of the [PSI+] system in Saccharomyces, and the evolution of other putative capacitors in the presence of sex still remains to be determined.
Supporting Information
Fitness contours in environment two as a function of  and
and  . The approximation
. The approximation  was used to calculate optimal expression. This approximation is reasonably accurate because δj, j∈{psi−,[PSI+]} is small 0<δj≤0.01.
was used to calculate optimal expression. This approximation is reasonably accurate because δj, j∈{psi−,[PSI+]} is small 0<δj≤0.01.
(4.09 MB TIF)
Values of ε lower than the upper bound ε = 0.01 do not change our results. Lower values of ε imply stronger selection against [PSI+] in environment one. This could inhibit [PSI+]-mediated adaptation, but the effect is negligible unless selection for [PSI+] in environment two is very weak. Ω12 = Ω21 = 10−5, h = 1.
(8.80 MB TIF)
Initial allele frequencies.
(0.03 MB PDF)
Adaptive gene product activity.
(0.03 MB PDF)
Acknowledgments
We thank Lilach Hadany for helpful discussions.
Footnotes
The authors have declared that no competing interests exist.
Work was supported by National Institute of Health grant GM-076041. JM is a Pew Scholar in the Biomedical Sciences. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Wickner RB, Masison DC, Edskes HK. [PSI] and [URE3] as yeast prions. Yeast. 1995;11:1671–1685. doi: 10.1002/yea.320111609. [DOI] [PubMed] [Google Scholar]
- 2.Lund PM, Cox BS. Reversion analysis of [psi −] mutations in Saccharomyces cerevisiae. Genet Res. 1981;37:173–182. doi: 10.1017/s0016672300020140. [DOI] [PubMed] [Google Scholar]
- 3.Paushkin SV, Kushnirov VV, Smirnov VN, Ter-Avanesyan MD. Propagation of the yeast prion-like [PSI+] determinant is mediated by oligomerization of the SUP35-encoded polypeptide chain release factor. EMBO J. 1996;15:3127–3134. [PMC free article] [PubMed] [Google Scholar]
- 4.Serio TR, Lindquist SL. [PSI+]: An epigenetic modulator of translation termination efficiency. Annu Rev Cell Dev Biol. 1999;15:661–703. doi: 10.1146/annurev.cellbio.15.1.661. [DOI] [PubMed] [Google Scholar]
- 5.Stansfield I, Jones KM, Kushnirov VV, Dagkesamanskaya AR, Poznyakovski AI, et al. The products of the sup45 (eRF1) and sup35 genes interact to mediate translation termination in Saccharomyces cerevisiae. EMBO J. 1995;14:4365–4373. doi: 10.1002/j.1460-2075.1995.tb00111.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Zhouravleva G, Frolova L, Legoff X, Leguellec R, Inge-Vechtomov S, et al. Termination of translation in eukaryotes is governed by two interacting polypeptide-chain release factors, eRF1 and eRF3. EMBO J. 1995;14:4065–4072. doi: 10.1002/j.1460-2075.1995.tb00078.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Firoozan M, Grant CM, Duarte JAB, Tuite MF. Quantitation of readthrough of termination codons in yeast using a novel gene fusion assay. Yeast. 1991;7:173–183. doi: 10.1002/yea.320070211. [DOI] [PubMed] [Google Scholar]
- 8.Eaglestone SS, Cox BS, Tuite MF. Translation termination efficiency can be regulated in Saccharomyces cerevisiae by environmental stress through a prion-mediated mechanism. EMBO J. 1999;18:1974–1981. doi: 10.1093/emboj/18.7.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.True HL, Berlin I, Lindquist SL. Epigenetic regulation of translation reveals hidden genetic variation to produce complex traits. Nature. 2004;431:184–187. doi: 10.1038/nature02885. [DOI] [PubMed] [Google Scholar]
- 10.True HL, Lindquist SL. A yeast prion provides a mechanism for genetic variation and phenotypic diversity. Nature. 2000;407:477–483. doi: 10.1038/35035005. [DOI] [PubMed] [Google Scholar]
- 11.Wilson MA, Meaux S, Parker R, van Hoof A. Genetic interactions between [PSI+] and nonstop mRNA decay affect phenotypic variation. Proc Natl Acad Sci USA. 2005;102:10244–10249. doi: 10.1073/pnas.0504557102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Joseph SB, Kirkpatrick M. Effects of the [PSI+] prion on rates of adaptation in yeast. J Evol Biol. 2008;21:773–780. doi: 10.1111/j.1420-9101.2008.01515.x. [DOI] [PubMed] [Google Scholar]
- 13.Brookfield JFY. Evolution: The evolvability enigma. Curr Biol. 2001;11:R106–R108. doi: 10.1016/s0960-9822(01)00041-0. [DOI] [PubMed] [Google Scholar]
- 14.Giacomelli MG, Hancock AS, Masel J. The conversion of 3′ UTRs into coding regions. Mol Biol Evol. 2007;24:457–464. doi: 10.1093/molbev/msl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Masel J, Bergman A. The evolution of the evolvability properties of the yeast prion [PSI+]. Evolution. 2003;57:1498–1512. doi: 10.1111/j.0014-3820.2003.tb00358.x. [DOI] [PubMed] [Google Scholar]
- 16.Pal C. Yeast prions and evolvability. Trends Genet. 2001;17:167–169. doi: 10.1016/s0168-9525(01)02235-1. [DOI] [PubMed] [Google Scholar]
- 17.Partridge L, Barton NH. Evolving evolvability. Nature. 2000;407:457–458. doi: 10.1038/35035173. [DOI] [PubMed] [Google Scholar]
- 18.Tyedmers J, Madariaga ML, Lindquist S. Prion switching in response to environmental stress. PLoS Biol. 2008;6:e294. doi: 10.1371/journal.pbio.0060294. doi/10.1371/journal.pbio.0060294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lang GI, Murray AW. Estimating the per-base-pair mutation rate in the yeast Saccharomyces cerevisiae. Genetics. 2008;178:67–82. doi: 10.1534/genetics.107.071506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lynch M, Sung W, Morris K, Coffey N, Landry CR, et al. A genome-wide view of the spectrum of spontaneous mutations in yeast. Proc Natl Acad Sci USA. 2008;105:9272–9277. doi: 10.1073/pnas.0803466105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Masel J. Evolutionary capacitance may be favored by natural selection. Genetics. 2005;170:1359–1371. doi: 10.1534/genetics.105.040493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masel J. Cryptic genetic variation is enriched for potential adaptations. Genetics. 2006;172:1985–1991. doi: 10.1534/genetics.105.051649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.King O, Masel J. The evolution of bet-hedging adaptations to rare scenarios. Theor Popul Biol. 2007;72:560–575. doi: 10.1016/j.tpb.2007.08.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lancaster AK, Masel J. The evolution of reversible switches in the presence of irreversible mimics. Evolution in press. 2009 doi: 10.1111/j.1558-5646.2009.00729.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pigliucci M. Is evolvability evolvable? Nat Rev Genet. 2008;9:75–82. doi: 10.1038/nrg2278. [DOI] [PubMed] [Google Scholar]
- 26.Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. BioEssays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Sniegowski PD, Murphy HA. Evolvability. Curr Biol. 2006;16:R831–R834. doi: 10.1016/j.cub.2006.08.080. [DOI] [PubMed] [Google Scholar]
- 28.Wagner GP, Bürger R. On the evolution of dominance modifiers II: a non-equilibrium approach to the evolution of genetic systems. J Theor Biol. 1985;113:475–500. doi: 10.1016/s0022-5193(85)80034-5. [DOI] [PubMed] [Google Scholar]
- 29.Derkatch IL, Bradley ME, Zhou P, Chernoff YO, Liebman SW. Genetic and environmental factors affecting the de novo appearance of the [PSI+] prion in Saccharomyces cerevisiae. Genetics. 1997;147:507–519. doi: 10.1093/genetics/147.2.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chernoff YO, Newnam GP, Kumar J, Allen K, Zink AD. Evidence for a protein mutator in yeast: Role of the Hsp70-related chaperone Ssb in formation, stability, and toxicity of the [PSI] prion. Mol Cell Biol. 1999;19:8103–8112. doi: 10.1128/mcb.19.12.8103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kushnirov VV, Kryndushkin DS, Boguta M, Smirnov VN, Ter-Avanesyan MD. Chaperones that cure yeast artificial [PSI+] and their prion- specific effects. Curr Biol. 2000;10:1443–1446. doi: 10.1016/s0960-9822(00)00802-2. [DOI] [PubMed] [Google Scholar]
- 32.Sharma D, Masison DC. Functionally redundant isoforms of a yeast Hsp70 chaperone subfamily have different antiprion effects. Genetics. 2008;179:1301–1311. doi: 10.1534/genetics.108.089458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Parham SN, Resende CG, Tuite MF. Oligopeptide repeats in the yeast protein Sup35p stabilize intermolecular prion interactions. EMBO J. 2001;20:2111–2119. doi: 10.1093/emboj/20.9.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Resende CG, Outeiro TF, Sands L, Lindquist S, Tuite MF. Prion protein gene polymorphisms in Saccharomyces cerevisiae. Mol Microbiol. 2003;49:1005–1017. doi: 10.1046/j.1365-2958.2003.03608.x. [DOI] [PubMed] [Google Scholar]
- 35.Tsai IJ, Bensasson D, Burt A, Koufopanou V. Population genomics of the wild yeast Saccharomyces paradoxus: Quantifying the life cycle. Proc Natl Acad Sci USA. 2008;105:4957–4962. doi: 10.1073/pnas.0707314105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Koch AL. Enzyme evolution. I. Importance of untranslatable intermediates. Genetics. 1972;72:297–316. doi: 10.1093/genetics/72.2.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Namy O, Galopier A, Martini C, Matsufuji S, Fabret C, et al. Epigenetic control of polyamines by the prion [PSI+]. Nat Cell Biol. 2008;10:1069–1075. doi: 10.1038/ncb1766. [DOI] [PubMed] [Google Scholar]
- 38.Tank EMH, Harris DA, Desai AA, True HL. Prion protein repeat expansion results in increased aggregation and reveals phenotypic variability. Mol Cell Biol. 2007;27:5445–5455. doi: 10.1128/MCB.02127-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Masel J, Griswold CK. The strength of selection against the yeast prion [PSI+]. Genetics. 2009;181:1057–1063. doi: 10.1534/genetics.108.100297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nakayashiki T, Kurtzman CP, Edskes HK, Wickner RB. Yeast prions [URE3] and [PSI+] are diseases. Proc Natl Acad Sci USA. 2005;102:10575–10580. doi: 10.1073/pnas.0504882102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Agresti A, Coull BA. Approximate is better than “exact” for interval estimation of binomial proportions. Amer Statistician. 1998;52:119–126. [Google Scholar]
- 42.Kim Y. Rate of adaptive peak shifts with partial genetic robustness. Evolution. 2007;61:1847–1856. doi: 10.1111/j.1558-5646.2007.00166.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fitness contours in environment two as a function of  and
and  . The approximation
. The approximation  was used to calculate optimal expression. This approximation is reasonably accurate because δj, j∈{psi−,[PSI+]} is small 0<δj≤0.01.
was used to calculate optimal expression. This approximation is reasonably accurate because δj, j∈{psi−,[PSI+]} is small 0<δj≤0.01.
(4.09 MB TIF)
Values of ε lower than the upper bound ε = 0.01 do not change our results. Lower values of ε imply stronger selection against [PSI+] in environment one. This could inhibit [PSI+]-mediated adaptation, but the effect is negligible unless selection for [PSI+] in environment two is very weak. Ω12 = Ω21 = 10−5, h = 1.
(8.80 MB TIF)
Initial allele frequencies.
(0.03 MB PDF)
Adaptive gene product activity.
(0.03 MB PDF)









