Abstract
The 20S proteasome has been shown to be largely responsible for the degradation of oxidatively modified proteins in the cytoplasm. Nuclear proteins are also subject to oxidation, and the nucleus of mammalian cells contains proteasome. In human beings, tumor cells frequently are subjected to oxidation as a consequence of antitumor chemotherapy, and K562 human myelogenous leukemia cells have a higher nuclear proteasome activity than do nonmalignant cells. Adaptation to oxidative stress appears to be one element in the development of long-term resistance to many chemotherapeutic drugs and the mechanisms of inducible tumor resistance to oxidation are of obvious importance. After hydrogen peroxide treatment of K562 cells, degradation of the model proteasome peptide substrate suc-LLVY-MCA and degradation of oxidized histones in nuclei increases significantly within minutes. Both increased proteolytic susceptibility of the histone substrates (caused by modification by oxidation) and activation of the proteasome enzyme complex occur independently during oxidative stress. This rapid up-regulation of 20S proteasome activity is accompanied by, and depends on, poly-ADP ribosylation of the proteasome, as shown by inhibitor experiments, 14C-ADP ribose incorporation assays, immunoblotting, in vitro reconstitution experiments, and immunoprecipitation of (activated) proteasome with anti-poly-ADP ribose polymerase antibodies. The poly-ADP ribosylation-mediated activated nuclear 20S proteasome is able to remove oxidatively damaged histones more efficiently and therefore is proposed as an oxidant-stimulatable defense or repair system of the nucleus in K562 leukemia cells.
Free radicals are involved in the cytotoxic action of many antitumor drugs, both in vitro and in vivo (1–4). Radical production may be caused by intratumor drug metabolism or redox cycling reactions (4, 5). Ultimately, the desirable DNA damage that chemotherapy causes in tumor cells is both the consequence of direct reaction of cytotoxic drugs with DNA, e.g., alkylating drugs (6), and strand breaks or nucleobase oxidation, caused by radical-producing redox cycling in close proximity to the chromatin, e.g., anthracyclines (1, 2, 5). Thus the antioxidant defense and repair capacity of tumor cells may reduce the efficiency of antitumor chemotherapy and may be a major cause of chemotherapy resistance (7). Although the repair of oxidative DNA damage has been widely studied in both normal and transformed cells, repair or removal mechanisms for oxidatively damaged histones are unknown. Transcription, replication, and DNA repair require a constant cyclic detachment and reassembly of histones with DNA (8), and this important function of “histone shuttling” may be impaired in oxidatively damaged histones. We hypothesized that selective degradation of oxidatively damaged histones might be a necessary cellular function for maintaining chromatin integrity. If true, a detailed understanding of such mechanisms would be important in antitumor chemotherapy and radiation therapy.
The degradation of oxidatively modified proteins now is a well-accepted physiological function of the proteasomal system in the cytoplasm (9–17), where ATP- and ubiquitin-independent proteolysis by the 20S proteasome seems to be the major pathway (12–14). The 20S proteasome is a 700-kDa, soluble proteinase complex that is found in the cytosol and the nucleus of mammalian cells (18). Tumor cells seem to have much higher proteasome activity than do nonmalignant cells, as recently demonstrated for K562 human myelogenous leukemia cells (19), and much of the “extra” proteasome appears to be localized in the nucleus. We previously have reported the rapid turnover of oxidatively modified proteins, by proteasome, in intact K562 cells but most of this degradation was assumed to be cytoplasmic (13).
In the present studies we have investigated the selective proteolytic degradation of oxidatively damaged histones by K562 cells and isolated nuclei during conditions of oxidative stress. Our work has identified a nuclear proteasome-activating system in tumor cells, which leads to a stimulatable and highly selective degradation of damaged histones in the nucleus of K562 human hematopoietic cells.
MATERIALS AND METHODS
Tissue Culture.
Cell line propagation. K 562 cells (human chronic myelogenous leukemia cells, ATCC CCL 243) were cultured in RPMI medium 1640 supplemented with 10% FCS. Cells initially were seeded at a density of 0.4 × 106 cells/ml and then grown for 3 days to a density of 5 × 106 cells/ml.
Metabolic labeling of endogenous cell proteins.
Endogenous proteins were metabolically labeled with [3H]leucine for five population doublings by addition of 50 μCi of [3H]leucine to each T75 culture flask containing K562 cells in complete medium for 4–5 days. After this period the unincorporated radioactivity was washed out, and the cells were either treated with hydrogen peroxide or used as controls. After the indicated time periods the histones were analyzed by electrophoresis. The bands reflecting individual histones were cut out of the gels and analyzed for radioactivity.
Cell treatment.
Cells were treated with 1 mM hydrogen peroxide for various time periods, after which the hydrogen peroxide-containing medium was removed by centrifugation at 4°C. Lactacystin was added to a final concentration of 5 μM, when indicated.
Isolation of nuclei.
The nuclei were isolated by a modification of the method of Emig et al. (20). Any remaining cytosolic contamination was removed by repeated washing procedures until no cytosolic marker enzymes could be detected. In addition, we analyzed the final nuclear preparations for remaining cells and cellular debris by microscopy, and only “clean” nuclear preparations were used. Lysis of nuclei was performed by freeze-thawing cycles in 1 mM DTT. Nuclei were damaged by exposure to 1 mM hydrogen peroxide. Lactacystin and 3-aminobenzamide (3-ABA) were added at a final concentration of 5 μM and 1 mM, respectively, when indicated.
Histone Oxidation.
Histones (2 mg/ml, Boehringer Mannheim) were incubated with various concentrations of hydrogen peroxide in 20 mM phosphate buffer (pH 7.4) for 2 h at 25°C. Samples of 1.0 ml then were dialyzed twice, for 3 h each time, at 4°C against 2 liters of 10 mM phosphate buffer, containing 10 mM KCl. The dialysis solution then was changed again, and the samples were dialyzed overnight for an additional 16 h at 4°C.
20S Proteasome Isolation.
The 20S proteasome was isolated according to Hough et al. (21). For proteasome isolation cells were lysed in 1 mM DTT. After removal of membranes and nonlysed cells by centrifugation, the 20S proteasome was isolated by DEAE-chromatography, sucrose-density gradient ultracentrifugation, and separation on a Mono Q column by using an FPLC system.
Proteolysis Measurements.
Suc-LLVY-MCA degradation. The peptidase activity of nuclear lysates toward the fluorogenic peptide suc-LLVY-MCA was measured by incubation in 50 mM Tris⋅HCl (pH 7.8), 20 mM KCl, 5 mM MgOAc, and 0.5 mM DTT with 200 μM suc-LLVY-MCA for 1 h at 37°C in the presence and absence of 5 μM lactacystin. The reaction was stopped by addition of an equal volume of ice-cold ethanol and 10 vol of 0.125 M borate. The fluorescence was determined at 380 nm excitation/440 nm emission.
Fluorescamine assay.
Fluorescamine reacts quantitatively with primary amines, forming a fluorescent product. The concentration of primary amines in trichloroacetic acid (TCA)-soluble fractions reflects the rate of proteolysis of the substrate protein by the proteasome (i.e., liberation of acid-soluble, “free” primary amines by proteolysis). The degradation of histones was measured by incubation of 40 μg substrate protein with 0.6 μg of proteasome in a proteolysis buffer containing 50 mM Hepes (pH 7.8), 20 mM KCl, 5 mM MgOAc, and 1 mM DTT for 2 h at 37°C. The reaction was stopped by addition of an equal volume of TCA (20%). After centrifugation (15 min at 12,000 × g) supernatants containing primary amines were neutralized with 1 M Hepes (pH 7.8). Fluorescamine (0.3 mg/ml in acetone) was added during rigorous shaking, and the fluorescence was determined at 390 nm excitation/470 nm emission. Leucine was used as a standard to determine amino acid liberation by proteolysis. Proteolysis rates were calculated as the difference between sample values and blank values.
Degradation of radiolabeled histones.
Histones were radiolabeled by reductive methylation with [3H]formaldehyde and sodium cyanoborhydride as described by Jentoft and Deanborn (22), and then extensively dialyzed. Proteolysis of undamaged and H2O2-modified histones by (proteases in) nuclear lysates was measured in 50 mM Tris⋅HCl (pH 7.8), 20 mM KCl, 5 mM MgOAc, and 0.5 mM DTT. Percent histone degradation was determined (23) by liquid scintillation in supernatants of TCA-precipitated proteins, by the formula: (TCA-soluble radioactivity − background radioactivity)/(initial TCA-precipitable radioactivity − background radioactivity) × 100.
Analysis of 20S Proteasome by Nondenaturing PAGE.
Analysis of the 20S proteasome was performed by using nondenaturing PAGE consisting of a 3% stacking and a 6% separating gel according to Hough et al. (21). The separation was performed for 650 Vh at 4°C. Directly after the run the gel was incubated in 50 mM Tris (pH 7.8), 25 mM KCl, 10 mM NaCl, 1 mM MgCl2, 1 mM DTT, 0.1 mM EDTA, and 10% glycerol for 15 min at 37°C, mounted on a light box (emitting light: 366 nm) and overlaid with a 200 μM suc-LLVY-MCA-peptide solution. The fluorescence was photographed after 15–45 min of incubation.
RESULTS AND DISCUSSION
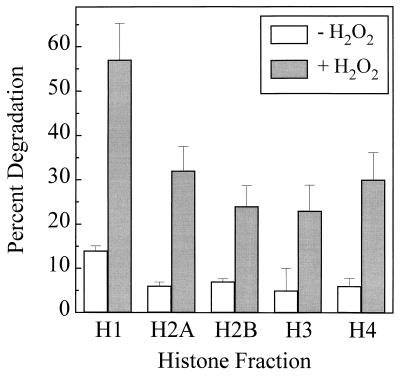
K562 cell proteins were metabolically labeled by incorporation of [3H]leucine over 5 days (see Materials and Methods). Intact K562 cells then were exposed to H2O2 for 30 min, and the degradation of histone proteins was measured. Trypan blue exclusion studies revealed no significant loss in viability of hydrogen peroxide-treated cells in comparison to controls, and no evidence of nuclear disruption was observed in any of our preparations. Histone H1 underwent 57.5% degradation with peroxide treatment, whereas histones H2A, H2B, H3, and H4 all exhibited degradation of between 24% and 34% (Fig. 1). Thus, histone proteins appear to be highly susceptible to oxidant-induced proteolysis in intact cells.
Figure 1.
Degradation of histones in intact human K562 cells stressed by exposure to hydrogen peroxide. K562 cells were grown in RPMI medium 1640 supplemented with 10% FCS, with the addition of [3H]leucine for 5 days. Next, the cells were washed and either exposed or sham-exposed to 1.0 mM H2O2 in PBS. After 30 min of exposure, new culture medium was added, and the cells were harvested, lysed, and subjected to acetic acid-urea-Triton-PAGE as described (25). The gels were Coomassie-stained, and the band patterns were compared with those of isolated histones (Boehringer Mannheim). The band for each histone was excised from the gels and counted by liquid scintillation. Two electrophoresis gels were run for each sample and averaged. Four such independent experiments were conducted, and the values shown for each histone are the grand means ± SE of the four experiments.
We next designed a series of experiments to test whether increased histone turnover after oxidant stress could be the result of protease activation/induction, histone protein modification to form better proteolytic substrates, or both. To investigate the capacity of the nuclear proteolytic system of H2O2-treated cells we isolated nuclei, prepared lysates, and measured the degradation of the fluorogenic peptide suc-LLVY-MCA, a known substrate of the 20S proteasome. To prevent the influence of other peptidases we measured the cleavage of the peptide in the absence and the presence of lactacystin, a highly specific proteasome inhibitor (24), which does not alter the activity of other proteases at the concentrations used.
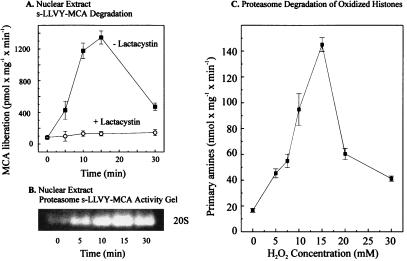
A time-dependent increase in lactacystin-sensitive peptidase activity occurs with H2O2 treatment, reaching a maximum 18-fold increase after 15-min exposure, and declining thereafter (Fig. 2A). To further test the probable role of the proteasome in this process we performed a nondenaturing one-dimensional PAGE for proteasome analysis, as described by Hough et al. (21). By using the fluorogenic peptide suc-LLVY-MCA to run an activity stain of the gel we discovered a clear activation of the proteasome (Fig. 2B). We next studied the effect of oxidative modification on the susceptibility of histones as substrates for proteolysis when added to untreated nuclear lysates. We previously have observed oxidation of individual, purified histones in response to hydrogen peroxide treatment (25). In the present studies actual oxidation of the isolated histones, after H2O2 treatment, was measured by derivatization of protein-bound carbonyl groups by using acetic acid-urea-Triton electrophoresis followed by an immunoblot and development with antidinitrophenylhydrazine antibodies. Significant oxidation of purified, hydrogen peroxide-treated histones was observed by this technique, particularly for histone H1 (data not shown). As shown in Fig. 2C, nuclear lysates preferentially degraded these oxidized histones, as long as the oxidation was not too severe. This pattern of increased proteolytic susceptibility after relatively mild oxidation and decreased susceptibility after more extreme oxidative modification has been observed with a wide variety of other proteins. Substantial data exist to support the concept that mild oxidation exposes hydrophobic residues that render proteins susceptible to degradation, whereas more extreme oxidation promotes hydrophobic aggregation, ionic bonds, and covalent crosslinks, all of which increase promote and diminish proteolytic susceptibility (11–17, 25).
Figure 2.
Activation of nuclear proteolysis and increased proteolytic susceptibility of histones after hydrogen peroxide treatment. K562 cells were grown and exposed to H2O2 as described in the legend to Fig. 1. Cell nuclei then were isolated and lysed, and degradation of the fluorogenic peptide suc-LLVY-MCA was measured as described in Materials and Methods. (A) The lactacystin-sensitive suc-LLVY-MCA degradation as a function of time of K562 cell exposure to 1 mM hydrogen peroxide. The values represent the means of six independent measurements with SD less than 10%. (B) A representative suc-LLVY-MCA degradation activity stain of a one-dimensional polyacrylamide nondenaturing proteasome electrophoresis gel, according to ref. 21, also as a function of time of K562 cell exposure to 1 mM hydrogen peroxide. (C) The proteolytic susceptibility of H2O2-modified histones, incubated with the purified 20S proteasome. [3H]-labeled histones (at 2.0 mg protein/ml, Boehringer Mannheim) were radiolabeled and exposed (or sham-exposed) to hydrogen peroxide in vitro as described in Materials and Methods. The x axis shows the concentrations of hydrogen peroxide used to oxidize histones. After extensive dialysis, proteasome was added for a 2-h incubation, and proteolysis was measured by the fluorescamine assay for release of free amino acids, after incubation of 40 μg histone substrate protein with 0.6 μg proteasome (see Materials and Methods). All values in C represent the means ± SD of three independent experiments.
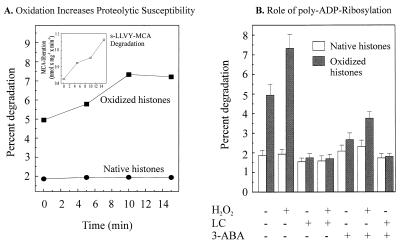
The experiments of Fig. 2 suggest that both substrate activation and protease activation may occur simultaneously in the nuclei of H2O2-treated cells. To further test these two possibilities we studied the proteolysis of oxidized and nonoxidized histones, in lysates harvested from H2O2-treated and nontreated nuclei. Fig. 3A compares two processes: the degradation of oxidized histones vs. native histones, and the effect of time of exposure to hydrogen peroxide of intact isolated nuclei from which lysates then were prepared. With no exposure to peroxide the (proteasome in the) nuclear extracts at time zero are still able to recognize and preferentially degrade oxidatively modified histones. With exposure to peroxide, however, extracts from exposed nuclei acquired an increasing ability to degrade oxidized histones; evidence that proteasome is activated during nuclear exposure to oxidants.
Figure 3.
Proteasome-dependent selective degradation of oxidized histones in nuclear extracts and the role of poly-ADP ribosylation. (A) The degradation of undamaged and H2O2-modified histones and suc-LLVY-MCA by lysates of H2O2-treated isolated nuclei from K562 hematopoietic cells. Nuclei were isolated, exposed to 1 mM H2O2 for times from 0 to 15 min (or sham exposed), lysed, and used to measure the proteolytic susceptibility of both untreated and H2O2-treated [3H]-labeled histones (added to the nuclear lysates) exactly as described in Materials and Methods. Isolated [3H]-labeled histones were oxidatively modified (or used as unmodified “native” histones) as described in Materials and Methods. The time scale represents the length of nuclei exposure (or sham-exposure) to H2O2 and the data points represent the means of three independent experiments (for which SD were always less than 10%). (Inset) The increase in the suc-LLVY-MCA degrading activity of nuclear lysates after treatment of nuclei with 1 mM H2O2 (for details see Fig. 2). (B) The degradation of H2O2-modified histones by K562 cell nuclear lysates is stimulated by pretreatment of nuclei with H2O2 but inhibited by both lactacystin and 3-ABA. Nuclei were isolated, exposed to 1 mM H2O2 for 15 min (or sham exposed), lysed, and used to measure the proteolytic susceptibility of both untreated and H2O2-treated [3H]-labeled histones, as described in Materials and Methods, in the presence or absence of the inhibitors 5 μM lactacystin (LC) or 1 mM 3-ABA. Where used, oxidized histones were treated with 15 mM hydrogen peroxide. All values represent the means ± SD of three independent experiments.
As shown in Fig. 3A, oxidatively damaged histones were degraded 2.8-fold better than nonoxidized histones in lysates from nontreated nuclei. When we substituted lysates from H2O2-treated nuclei, however, the degradation of oxidized histones increased (as a function of the time of H2O2 exposure) to 4.2-fold, whereas the proteolysis of native histones was unchanged (Fig. 3A). Statistical analysis revealed that the differences between 2.8-fold and 4.2-fold increases in proteolysis were indeed statistically significant (P < 0.05) as tested by the Wilcoxon rank sum test. That activation of proteasome occurred under these conditions also was strongly suggested by the increase in suc-LLVY-MCA degrading capacity (Fig. 3A, Inset).
To further examine the probable role of nuclear proteasome in histone turnover during oxidant stress we performed experiments with the proteasome inhibitor lactacystin. Lactacystin essentially completely suppressed the preferential proteolysis of oxidized histones, without significantly altering the degradation of nonoxidized histones, in lysates from both H2O2-treated nuclei and nontreated nuclei (Fig. 3B). From Figs. 1–3 one can conclude that nuclear proteasome is able to recognize and selectively degrade oxidized histones, and that an activation or induction of nuclear proteasome occurs during oxidative stress.
We next turned our attention to the potential mechanism of proteasome activation or induction. It immediately seemed unlikely to us that significant de novo synthesis of the long-lived, multisubunit proteasome complex would occur during the 10–15 min required for maximal activity increases observed in Figs. 2 A and B and 3A. We therefore began to look more seriously at the possibility of enzyme activation. Although proteasome can be activated by various chemical and physical stresses (23), these possibilities did not seem to apply to the condition of H2O2 treatment. Serendipitously we had been working separately on studies of poly-ADP ribosylation, which is considered to be a general response of nuclei to oxidative stress (26). The nuclear enzyme poly-ADP ribose polymerase (PARP) transfers ADP ribose moieties from NAD+ to histones and several other nuclear proteins (27). Because PARP and proteasome both reside in the nucleus, PARP appears to ADP-ribosylate many nuclear proteins during oxidative stress and proteasome modification by other binding agents (e.g., SDS) can activate proteasome, we wondered whether PARP might actually be responsible for the rapid activation of nuclear proteasome during H2O2 exposure.
To examine this question we performed experiments with 3-ABA, a strong and selective inhibitor of PARP (28), to obtain nuclei both with and without poly-ADP-ribosylated proteins. Incubation with 3-ABA blocked the degradation of oxidized histones in nuclear lysates, whereas the degradation of undamaged histones was not impaired by 3-ABA (Fig. 3B). The proteasome itself was not inhibited by 3-ABA at any concentration used in this study (data not shown). 3-ABA is by far the best characterized and most commonly used inhibitor of PARP. Therefore we used this inhibitor in the bulk of the experiments. Furthermore, we did not see other nonspecific effects of 3-ABA at the concentrations used. Other inhibitors, such as 6(5H)phenanthridinone or 4-amino-1,8-naphtalimide, sometimes can have more drastic side effects, like protein kinase inhibition, etc. Nevertheless, we also tested 1.5 μM 6(5H)phenanthridinone and 1.0 μM 4-amino-1,8-naphtalimide in the experiments reported in Fig. 3B and found similar inhibitory results as for 3-ABA (data not shown). These results further support the proposal that PARP may be responsible for the rapid activation of nuclear proteasome during H2O2 exposure.
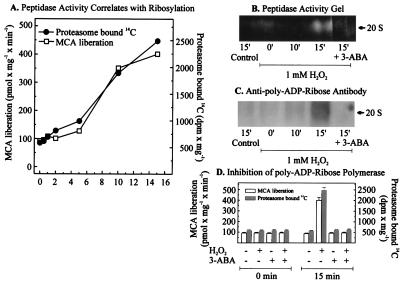
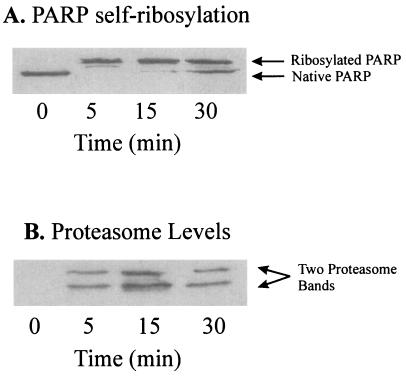
In further experiments we examined whether the 20S proteasome itself is a substrate for poly-ADP ribosylation by PARP. For these studies isolated nuclei were treated with 1 mM hydrogen peroxide in the presence of [14C]-NAD+. Under these conditions incorporation of 14C into proteins indicates poly-ADP ribosylation because of the transfer of ADP ribose moieties from NAD+ (28). Analysis of the proteasome was performed by a nondenaturing one-dimensional polyacrylamide electrophoresis as described by Hough et al. (21). As shown in Fig. 4A, H2O2 caused a time-dependent increase in nuclear proteasome activity, which was paralleled by an increasing incorporation of radioactivity derived from [14C]-NAD+. Further evidence for the concurrent activation and poly-ADP ribosylation of the proteasome is provided by activity staining of nondenaturing electrophoresis gels (Fig. 4B) and by immunoblotting using a mAb directed against poly-ADP ribose (Fig. 4C). Importantly, the PARP inhibitor 3-ABA was able to block 14C incorporation into the proteasome (Fig. 4D), prevented the detection of a signal from the anti-poly-ADP ribose antibody (Fig. 4C), and blocked the increased proteolytic activity toward the fluorogenic peptide suc-LLVY-MCA (Fig. 4 B and D). Similar effects to those reported for 3-ABA also were observed with 1.5 μM 6(5H)phenanthridinone and 1.0 μM 4-amino-1,8-naphtalimide in the experiments reported in Fig. 4 (data not shown).
Figure 4.
Poly-ADP ribosylation of the proteasome in nuclei of K562 human hematopoietic cells after hydrogen peroxide treatment increases its activity. (A) Proteasome peptidase activity correlated with the degree of poly-ADP ribosylation, measured as incorporated 14C from 14C-NAD+ after bolus addition of 1 mM hydrogen peroxide to isolated nuclei from K562 cells. Nuclei of K562 cells were isolated by a modified method of Emig et al. (20) and incubated with 1 mM hydrogen peroxide in 100 mM phosphate buffer (pH 7.8), 200 μM NAD+, 10 mM MgCl2, and 5 mM DTT, containing 10,000 dpm 14C-labeled NAD+. (B and C) The analysis of lysates from K562 cell nuclei after H2O2 treatment. The lysates were analyzed by nondenaturing one-dimensional polyacrylamide electrophoresis according to ref. 21. (B) An activity staining and (C) an immunoblot of the proteasome, using an anti-poly-ADP ribose antibody (Biomol, Plymouth Meeting, PA). (D) The effect of 3-ABA on the proteasome activity and the [14C] incorporation, after hydrogen peroxide treatment. Experimental conditions were as in Fig. 3A except for the use of 3-ABA (1 mM).
From the results presented so far it appears probable that the oxidant stress-induced activation of the proteasome is a specific activation by poly-ADP ribosylation, rather than a nonspecific activation by denaturing or modifying agents, such as has been reported for high temperature and detergents (23). Hydrogen peroxide is able to induce oxidative DNA damage, including single-strand breaks in nuclei of K562 cells, leading to the activation of the PARP. Furthermore tumor cells are known to have a higher PARP activity and a higher basal poly-ADP ribose content than normal cells (29), which could explain the strong activation of poly-ADP ribose synthesis in nuclei of K562 cells after exposure to hydrogen peroxide.
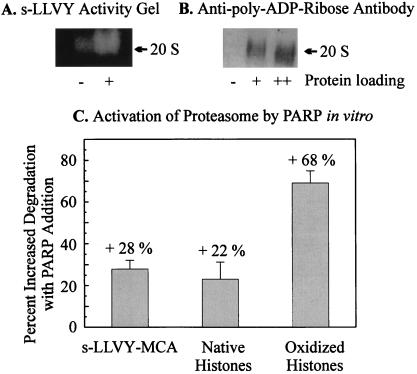
Because we could show that proteasome is poly-ADP ribosylated in isolated nuclei from K562 cells we next tested whether an isolated system of proteasome, PARP, and their substrates is necessary and sufficient for the poly-ADP ribose-mediated up-regulation of proteasomal activity, in the absence of other cellular components. By using the conditions of Banasik et al. (27) for in vitro poly-ADP ribosylation of proteins we were indeed able to demonstrate the activation of isolated proteasome by activity staining of nondenaturing electrophoresis gels (Fig. 5A) and nondenaturing-PAGE followed by immunoblots with the monoclonal anti-poly-ADP ribose antibody (Fig. 5B). In vitro poly-ADP ribosylated proteasome shows an increased proteolytic activity toward oxidized histones and to the suc-LLVY-MCA-peptide (Fig. 5C).
Figure 5.
Activation of the isolated 20S proteasome by in vitro poly-ADP ribosylation. (A) Activation of the isolated 20S proteasome after in vitro poly-ADP ribosylation. In vitro poly-ADP ribosylation of the 20S proteasome was performed according to Banasik et al. (27). Purified proteasome (0.15 mg/ml) was incubated with 1 μg/ml of PARP and 10 μg/ml of DNA (sonicated 10 times for 20 s) in a medium consisting of 100 mM Tris, 200 μM NAD+, 10 mM MgCl2, and 5 mM DTT. Controls were incubated without NAD+. After 10 min the reaction was stopped by addition of 1 mM 3-ABA, and the mixture was rapidly cooled on ice. Peptidase activity was determined after nondenaturing one-dimensional polyacrylamide electrophoresis as described in Fig. 4. (A) The activation of the peptidase activity of the proteasome after overlaying the gel with 200 μM suc-LLVY-MCA. (B) The immunoblot analysis of the gel after blotting on a nitrocellulose membrane by using the anti-ADP ribose antibody (Biomol) used in Fig. 4. The lanes indicated by + and ++ are the same probes loaded with different amounts of protein. Proteolytic activities in C were analyzed before and after in vitro poly-ADP ribosylation, using undamaged and H2O2-modified histones and the peptide suc-LLVY-MCA as substrates (see Figs. 1–4).
To test for a possible interaction of poly-ADP ribosylated PARP with proteasome in intact cells, we used the anti-DNA binding domain (DBD)-PARP antibody so that only unbound (DNA free) and poly-ADP ribosylated PARP should react with the antibody. Immunoprecipitation experiments with nuclear extracts from H2O2-treated cells revealed a clear band-shift for PARP, indicative of an (auto)poly-ADP ribosylation of the enzyme (Fig. 6A). As a final check on our hypothesis of PARP stimulation of nuclear proteasome, we tested whether a mAb directed against the DBD (anti-DBD) of PARP could coprecipitate “core” proteasome subunits from the H2O2-treated intact K562 cells. As shown in Fig. 6B, the anti-DBD-PARP antibody coprecipitated proteins, which crossreacted with a polyclonal antibody to the core proteasome. Further tests of this material revealed an extremely high proteolytic activity against the proteasome substrate suc-LLVY-MCA (data not shown). Importantly, the PARP activation shown in Fig. 6A occurred on exactly the same time scale as the increased binding of PARP to proteasome shown in Fig. 6B.
Figure 6.
Immunoprecipitation of the proteasome with a monoclonal anti-DBD-PARP antibody in nuclear extracts from H2O2-treated K562 cells. Intact K562 human chronic myelogenous leukemia cells were grown, harvested, treated with 1 mM H2O2, and then subjected to isolation and lysis of nuclei, as described in Materials and Methods. Immunoprecipitation was conducted with 100 μg of lysate protein, 100 μl of Tris/NaCl buffer (containing 50 mM Tris, plus 100 μM NaCl, at pH 8.0), and 1 μl of a mAb directed against the DBD (anti-DBD) of PARP (at 5 mg/ml), by a modification of the procedure of Zwilling et al. (30). The primary antibody reaction was performed in 50 mM Tris (pH 7.8) containing 150 mM NaCl (Tris buffered saline), 0.1% BSA, and 1.0 μl of rabbit-anti-DBD-IgG (5 μg/μl stock) for 1 h at 4°C in a total volume of 200 μl. For secondary antibody reactions 1.0 μl of donkey-anti-rabbit-IgG (5 μg/μl stock) was added and incubated for 1 h at 4°C. Binding of the secondary antibody was detected by using an ECL kit. Immunoprecipitation was initiated by addition of 20 μl of protein A-Sepharose (equilibrated 1:1 with a pH 8.0 solution of 50 mM Tris, 150 mM NaCl, and 0.1% BSA) for a 1-h incubation at 4°C. Controls were incubated with the secondary antibody and the protein A-Sepharose only. The Sepharose was centrifuged (12,000 × g for 30 min) and washed five times with Tris buffered saline. Sepharose-bound proteins were separated by 15% SDS/PAGE, transblotted onto nitrocellulose membranes, and incubated either with a polyclonal goat-anti-PARP-IgG (A) or a polyclonal rabbit-anti-core-proteasome-IgG (B). Control samples containing secondary antibody plus Sepharose A, or Sepharose A alone, produced no visible bands in the gels (data not shown). The results shown are a representative immunoblot from a series of several independent experimental repeats.
The results presented indicate that binding occurs between nuclear proteasome, PARP, and poly-ADP ribose, in response to oxidative stress. Furthermore, this binding appears to be necessary and sufficient to account for proteasome activation. Naturally, it will be important to identify the nature of the interactions between nuclear proteasome, PARP, and poly-ADP ribose. We have performed some preliminary studies using denaturing SDS/PAGE in an attempt to determine which proteasome subunits are involved in binding (data not shown). Unfortunately, our results were not instructive, perhaps because the 14 different proteasome subunits (held together by noncovalent interactions) range in size from 21 kDa to 32 kDa (18) and would not be expected to greatly alter the gel mobility of the 110-kDa PARP. One intriguing possibility we are pursuing is that autoribosylation of PARP during oxidative stress may provide the enzyme with a “bristling array” of molecular “fishing hooks” with which to snare the nuclear proteasome and effect its activation.
Poly-ADP ribosylation is strongly related to DNA repair (31). PARP molecules bind tightly to DNA strand breaks and auto-poly-ADP ribosylation of the protein then effects its release and allows access to lesions for DNA repair enzymes (31). The main function of poly-ADP ribosylation in the nucleus is still not clear. Binding of PARP to strand breaks may represent a signal that switches off DNA replication and transcription to ensure that lesions are not replicated before repair, and a “nick protection” during replication or cellular differentiation also has been proposed (31). Another function of poly-ADP ribosylation is the “shuttling” of histones by detachment and reattachment of histones to chromatin through reversible poly-ADP ribose synthesis and degradation, as experimentally shown by Realini and Althaus (8). PARP binds closely to DNA strand breaks and therefore should be able to initiate the poly-ADP ribosylation and activation of the proteasome in direct proximity to chromatin lesions, contributing to the specific degradation of damaged histones. The binding of PARP to DNA lesions and a strong activation after administration of various antitumor drugs has been reported in vitro (29). Increased poly-ADP ribosylation also has been suggested as an adaptive response of tumor cells to long-term antitumor drugs, which may be responsible for the development of chemotherapeutic resistance (7, 28). Thus the interaction between PARP and the nuclear proteasome may well play an important role in the secondary antioxidant defenses of K562 human myelogenous leukemia cell nuclei against histone damage during oxidative stress.
Acknowledgments
We thank Prof. S. Omura (Kitasato Institute, Tokyo, Japan) for kindly providing lactacystin, Prof. Keiji Tanaka (Tokyo Metropolitan Institute of Medical Science, Japan) for the kind gift of polyclonal antibodies to proteasome, and Prof. Manfred Schweiger (Free University of Berlin, Germany) for his kind gift of the monoclonal anti-DBD-PARP antibody. This work was supported by a fellowship from the Ernst-Schering-Research Foundation to O.U., by the Deutsche Forschungsgemeinschaft (Grant SFB 507/A7) and the Charité Research Fond grants to T.G., and by National Institutes of Health/National Institute on Environmental Health Sciences Grant ES 03598 to K.J.A.D.
ABBREVIATIONS
- PARP
poly-ADP ribose polymerase
- DBD
DNA binding domain
- TCA
trichloroacetic acid
- 3-ABA
3-aminobenzamide
References
- 1.Sinha B K, Mimnaugh E G. Free Radical Biol Med. 1990;8:567–581. doi: 10.1016/0891-5849(90)90155-c. [DOI] [PubMed] [Google Scholar]
- 2.Davies K J A, Doroshow J H. J Biol Chem. 1986;261:3060–3067. [PubMed] [Google Scholar]
- 3.Doroshow J H, Davies K J A. J Biol Chem. 1986;261:3068–3074. [PubMed] [Google Scholar]
- 4.Marcillat O, Zhang Y, Davies K J A. Biochem J. 1989;259:181–189. doi: 10.1042/bj2590181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Feinstein E, Canaani E, Weiner L M. Biochemistry. 1993;32:13156–13161. doi: 10.1021/bi00211a026. [DOI] [PubMed] [Google Scholar]
- 6.Goria-Gatti L, Iannone A, Tomasi A, Poli G, Albano E. Carcinogenesis. 1992;13:799–805. doi: 10.1093/carcin/13.5.799. [DOI] [PubMed] [Google Scholar]
- 7.Sinha B K, Mimnaugh E G. Free Radical Biol Med. 1990;8:567–581. doi: 10.1016/0891-5849(90)90155-c. [DOI] [PubMed] [Google Scholar]
- 8.Realini C A, Althaus F R. J Biol Chem. 1992;267:18858–18865. [PubMed] [Google Scholar]
- 9.Rivett A J. J Biol Chem. 1985;26:300–305. [PubMed] [Google Scholar]
- 10.Rivett A J. J Biol Chem. 1985;26:12600–12606. [PubMed] [Google Scholar]
- 11.Davies K J A. J Biol Chem. 1987;262:9895–9920. [PubMed] [Google Scholar]
- 12.Grune T, Reinheckel T, Davies K J A. J Biol Chem. 1995;270:2344–2351. doi: 10.1074/jbc.270.5.2344. [DOI] [PubMed] [Google Scholar]
- 13.Grune T, Reinheckel T, Davies K J A. J Biol Chem. 1996;271:15504–15509. doi: 10.1074/jbc.271.26.15504. [DOI] [PubMed] [Google Scholar]
- 14.Grune T, Reinheckel T, Davies K J A. FASEB J. 1997;11:526–534. [PubMed] [Google Scholar]
- 15.Conconi M, Szweda L I, Levine R L, Stadtman E R, Friguet B. Arch Biochem Biophys. 1996;331:232–240. doi: 10.1006/abbi.1996.0303. [DOI] [PubMed] [Google Scholar]
- 16.Berlett B S, Stadtman E R. J Biol Chem. 1997;272:20313–20316. doi: 10.1074/jbc.272.33.20313. [DOI] [PubMed] [Google Scholar]
- 17.Chao C-C, Ma Y-S, Stadtman E R. Proc Natl Acad Sci USA. 1997;94:2969–2974. doi: 10.1073/pnas.94.7.2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Coux O, Tanaka K, Goldberg A L. Annu Rev Biochem. 1996;65:801–847. doi: 10.1146/annurev.bi.65.070196.004101. [DOI] [PubMed] [Google Scholar]
- 19.Kumatori A, Tanaka K, Inamura N, Sone S, Ogura T, Matsumoto T, Tachikawa T, Shin S, Ichihara A. Proc Natl Acad Sci USA. 1990;87:7071–7075. doi: 10.1073/pnas.87.18.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Emig S, Schmalz D, Shakibaei M, Buchner K. J Biol Chem. 1995;270:13787–13793. doi: 10.1074/jbc.270.23.13787. [DOI] [PubMed] [Google Scholar]
- 21.Hough R, Pratt G, Rechsteiner M J. J Biol Chem. 1987;262:8303–8313. [PubMed] [Google Scholar]
- 22.Jentoft N, Dearborn D G. J Biol Chem. 1979;254:4359–4365. [PubMed] [Google Scholar]
- 23.Arribas J, Castano J G. J Biol Chem. 1990;265:13969–13973. [PubMed] [Google Scholar]
- 24.Omura S, Fujimoto T, Otoguro K, Matsuzaki K, Moriguchi R, Tanaka H, Sasaki Y. J Antibiot. 1991;44:113–116. doi: 10.7164/antibiotics.44.113. [DOI] [PubMed] [Google Scholar]
- 25.Ullrich O, Sitte N, Sommerburg O, Sandig V, Davies K J A, Grune T. Arch Biochem Biophys. 1999;362:211–216. doi: 10.1006/abbi.1998.1031. [DOI] [PubMed] [Google Scholar]
- 26.Cerutti P A. Science. 1985;227:375–381. doi: 10.1126/science.2981433. [DOI] [PubMed] [Google Scholar]
- 27.Banasik M, Komura H, Shimoyama M, Ueda K. J Biol Chem. 1992;267:1569–1575. [PubMed] [Google Scholar]
- 28.Urade M, Sugi M, Miyazaki T. Cancer. 1988;61:1501–1507. doi: 10.1002/1097-0142(19880415)61:8<1501::aid-cncr2820610805>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 29.Malapetsa A, Noe A J, Poirier G G, Desnoyers S, Berger N A, Panasci L C. Mutat Res. 1996;362:41–50. doi: 10.1016/0921-8777(95)00030-5. [DOI] [PubMed] [Google Scholar]
- 30.Zwilling S, König H, Wirth T. EMBO J. 1995;14:1198–1298. doi: 10.1002/j.1460-2075.1995.tb07103.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satoh M S, Lindahl T. Nature (London) 1992;356:356–358. doi: 10.1038/356356a0. [DOI] [PubMed] [Google Scholar]