Abstract
The term ‘horizontal transfer (HT)’ refers to the transfer of genetic material between two reproductively isolated organisms. HT is thought to occur rarely in eukaryotes compared to vertical inheritance, the transmission of DNA from parent to offspring. In a recent study we have provided evidence that a family of DNA transposons, called SPACE INVADERS or SPIN, independently invaded horizontally the genome of seven distantly related tetrapod species and subsequently amplified to high copy number in each of them. This discovery calls for further investigations to better characterize the extent to which genomes have been shaped through HT events. In this addendum, we briefly discuss some general issues regarding the study of HT and further speculate on the sequence of events that could explain the current taxonomic distribution of SPIN. We propose that the presence of SPIN in the opossum (Monodelphis domestica), a taxon endemic to South America, reflects a transoceanic HT event that occurred from Old to New World, between 46 and 15 million years ago.
Key words: evolution, genome, transposable elements, horizontal transfer, tetrapods, mobile genetic elements
Horizontal transfer (HT) of DNA occurs when a piece of DNA from a donor species gets inserted into the genome of a reproductively isolated recipient species. Such transfers are common in prokaryotes where they are central to the diversification and evolution of taxa,1 but seemingly scarce in eukaryotes where vertical inheritance from parent to offspring is the accepted paradigm. Notwithstanding some cases involving prokaryotic or endosymbiotic donor species,2–5 the most convincing examples of HT described in multicellular eukaryotes are transfers of mobile DNA elements.6–10
In a recent paper,11 we provided evidence that a DNA transposon called SPACE INVADERS, or SPIN, has been horizontally introduced in distantly related tetrapods, including five mammalian lineages (bushbaby, little brown bat, murine rodents, opossum, tenrec), the African clawed frog and the green anole lizard. Analysis of SPIN transposons across species reveals a combination of features that is incompatible with the traditional hypothesis of vertical inheritance. They show a patchy taxonomic distribution, a high level of inter-specific sequence similarity (>90%) despite the 350 million years (my) of independent evolution separating the most divergent taxa (mammals and amphibians), and no evidence of purifying selection acting to preserve the integrity of these sequences since their amplification. Furthermore, we were not able to detect any shared orthologous copies (i.e., present at the same genomic locus) between any two of the seven hosts, a finding that is strongly indicative of lineage-specific insertions. These data unmistakably point to independent germline infiltration of the seven tetrapod lineages by an essentially identical ancestral SPIN transposon that subsequently amplified to reach high copy number in each of these genomes (nearly 100,000 copies per haploid genome in the tenrec). In addition, our time estimates of these various transfers showed that there was a seemingly global wave of SPIN invasion 15–46 mya.
Jumping and Non-Jumping Genes
Our ability to observe HT in multicellular eukaryotes implies that (i) a piece of DNA was excised or copied from the genome of a donor species, (ii) it was inserted into the germline genome of the recipient species, and (iii) it has reached fixation in the new host species. In this regard, the very nature of transposons clearly makes them much more likely to be successful at HT than other pieces of DNA (e.g., genes). First, they are able to autonomously excise from and reintegrate into chromosomes,12 meaning that no other mechanisms are necessary to explain these two essential steps of HT. Second, they can rapidly generate many copies of themselves within a genome, and therefore counteract forces such as drift and/or selection that in many cases would lead to the loss of a single copy gene introduced in a new host.13 HTs of DNA transposons are therefore expected to be more frequent and widespread than HT of cellular genes, a hypothesis that could now be tested on a large evolutionary scale by systematic screening of the large diversity of eukaryotic genome sequences becoming available.
Finding the Bug
Various hypotheses involving intermediate vectors such as viruses and/or bacteria have been put forward to explain how a transposon could be horizontally transferred from one host to another.7 Although several examples of transposons integrated within viral genomes have been reported, there has been no demonstration that these captured transposons can be transferred to a new host species. One of the most intriguing examples is the discovery of a SINE retroposon normally found in snakes that inserted into the genome of a poxvirus known to infect gerbil rodents in West Africa.14 Although there is no evidence that the SINE has infiltrated the gerbil genome, this case provides proof of principle that poxviruses can shuttle transposons between distantly related vertebrates. In fact, poxviruses meet several essential criteria for being considered good HT vector candidates. They are known to infect a wide range of invertebrate and vertebrate taxa, many cases of cross-species infections have been reported, and their tissue tropism does not seem to rely on any cell receptor.15,16 Although they can only replicate in specific cell types, poxviruses can potentially enter many others, including germ cells. In addition, their large genomes are known to be particularly efficient at capturing host genes.17
Were poxviruses the vectors of SPIN HTs? It is conceivable, but given the antiquity of these HTs (see above), it is most likely that the underlying mechanism/vector will never be identified. Indeed, whatever the timing of the HT, pinning down the vector always appears to be a daunting task for two reasons. First we know little about virus biodiversity,18 and secondly a successful HT might not necessarily require insertion of the transposon into the viral genome. Spurious co-packaging of a transposon nucleoprotein complex in viral particles might be sufficient to deliver an active transposon inside the new host cell, and in such cases, no detectable mark of the transposon would remain in the virus after the transfer.
Transoceanic Movement?
Perhaps one of the most puzzling aspects of the SPIN HTs is the probable geographic distribution of the host taxa at the time of the transfer. While the tenrec, bushbaby and African clawed frog are endemic to Africa or Madagascar (tenrec),19,20 the opossum (genus Monodelphis) is endemic to South America19 and the green anole to Central/South America.21 The genus Myotis (represented by the little brown bat M. lucifugus) has a worldwide distribution but did not migrate to North America before 14 million years ago (mya), and reached South America only 7–10 mya.22 Species of the Muroidea superfamily (mice and rats) are also found worldwide but they did not disperse in South America before 6 mya.23 Indeed South America was an island continent throughout most of the Tertiary period, from about 65 to 4 mya.24 These observations imply that SPIN transposons infiltrated the germline of species inhabiting at least two different continents, Africa and America, and somehow were able to travel across the ocean. How could this transoceanic movement happen? One possibility could be that a vector (possibly viral) carrying SPIN reached South America via hitchhiking on the African ancestors of the New World monkeys or those of caviomorph rodents. Both are believed to have colonized South America through transoceanic dispersal between 37 and 16 my ago, and between 45 and 36 my ago respectively, i.e., during periods overlapping widely with the wave of SPIN amplifications (15–46 my ago).25 This hypothesis does not necessarily imply that SPIN transposons invaded the germline of South American rodents and/or monkeys, but it will be interesting to check for the presence of SPIN in these taxa.
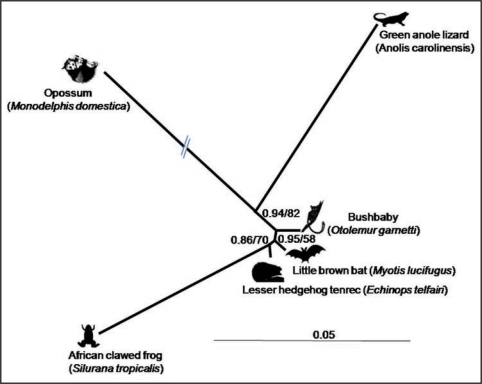
Another puzzling question is whether SPIN was transferred to all its different vertebrate hosts via the same route (e.g., via intercontinental viral pandemics), whether it “jumped” from one host to the other, or whether there was a mix of both routes. Whatever the answers to these questions are, it is interesting to note that the opossum and lizard SPINs are phylogenetically closer to each other than to the other SPIN elements, consistent with the idea that they evolved from the same, possibly American, SPIN ancestor (Fig. 1). A finer description of the taxonomic distribution of SPIN using experimental methods might help decipher this puzzle.
Figure 1.
Unrooted phylogeny of the autonomous SPIN DNA transposons.11 This shows that the SPIN of lizard and opossum—two American taxa—are closely related and therefore that they most probably derive from the same American ancestor. Sequences were aligned using BioEdit.26 The alignment was 1,841 bp after removing ambiguous and gapped positions. Phylogenetic analyses were performed using MrBayes27 and PHYML.28 Posterior probability and bootstrap values are indicated at each node. Only one or few full-length SPIN copies were found in opossum, green anole lizard and African clawed frog.11 Thus for these three species the SPIN sequences used for phylogenetic analysis correspond to individual SPIN copies, while the sequences used for bat, bushbaby and tenrec are consensus sequences derived from multiple full-length SPIN copies. This discrepancy explains in part why the branches of the frog, lizard and opossum are much longer than those of the other three species. Note the branch of the opossum SPIN has been reduced by half for the purpose of this illustration.
Acknowledgements
Research in the Feschotte lab is funded by grant R01-GM077582 from the National Institutes of Health.
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http:// www.landesbioscience.com/journals/cib/article/7720
References
- 1.Ochman H, Lawrence JG, Groisman EA. Lateral gene transfer and the nature of bacterial innovation. Nature. 2000;405:299–304. doi: 10.1038/35012500. [DOI] [PubMed] [Google Scholar]
- 2.Gladyshev EA, Meselson M, Arkhipova IR. Massive horizontal gene transfer in Bdelloid rotifers. Science. 2008;320:1210–1213. doi: 10.1126/science.1156407. [DOI] [PubMed] [Google Scholar]
- 3.Hotopp JC, Clark ME, Oliveira DC, Foster JM, Fischer P, Torres MC, et al. Widespread lateral gene transfer from intracellular bacteria to multicellular eukaryotes. Science. 2007;317:1753–1756. doi: 10.1126/science.1142490. [DOI] [PubMed] [Google Scholar]
- 4.Loftus B, Anderson I, Davies R, Alsmark UC, Samuelson J, Amedeo P, et al. The genome of the protist parasite Entamoeba histolytica. Nature. 2005;433:865–868. doi: 10.1038/nature03291. [DOI] [PubMed] [Google Scholar]
- 5.Morrison HG, McArthur AG, Gillin FD, Aley SB, Adam RD, Olsen GJ, et al. Genomic minimalism in the early diverging intestinal parasite Giardia lamblia. Science. 2007;317:1921–1926. doi: 10.1126/science.1143837. [DOI] [PubMed] [Google Scholar]
- 6.Syvanen M, Kado C, editors. Horizontal Gene Transfer. London: Academic; 2002. [Google Scholar]
- 7.Silva JC, Loreto EL, Clark JB. Factors that affect the horizontal transfer of transposable elements. Curr Issues Mol Biol. 2004;6:57–71. [PubMed] [Google Scholar]
- 8.de Boer JG, Yazawa R, Davidson WS, Koop BF. Bursts and horizontal evolution of DNA transposons in the speciation of pseudotetraploid salmonids. BMC Genomics. 2007;8:422. doi: 10.1186/1471-2164-8-422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diao X, Freeling M, Lisch D. Horizontal transfer of a plant transposon. PLoS Biol. 2006;4:5. doi: 10.1371/journal.pbio.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kordis D, Gubensek F. Horizontal SINE transfer between vertebrate classes. Nat Genet. 1995;10:131–132. doi: 10.1038/ng0695-131. [DOI] [PubMed] [Google Scholar]
- 11.Pace JK, II, Gilbert C, Clark MS, Feschotte C. Repeated horizontal transfer of a DNA transposon in mammals and other tetrapods. Proc Natl Acad Sci USA. 2008;105:17023–17028. doi: 10.1073/pnas.0806548105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Craig NL, Craigie R, Gellert M, Lambowitz AM. Mobile DNA II. Washington, DC: American Society for Microbiology Press; 2002. [Google Scholar]
- 13.Le Rouzic A, Capy P. The first steps of transposable elements invasion: parasitic strategy vs. genetic drift. Genetics. 2005;169:1033–1043. doi: 10.1534/genetics.104.031211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Piskurek O, Okada N. Poxviruses as possible vectors for horizontal transfer of retroposons from reptiles to mammals. Proc Natl Acad Sci USA. 2007;104:12046–12051. doi: 10.1073/pnas.0700531104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.McFadden G. Poxvirus tropism. Nat Rev Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Werden SJ, Rahman MM, McFadden G. Poxvirus host range genes. Adv Virus Res. 2008;71:135–171. doi: 10.1016/S0065-3527(08)00003-1. [DOI] [PubMed] [Google Scholar]
- 17.Bratke KA, McLysaght A. Identification of multiple independent horizontal gene transfers into poxviruses using a comparative genomics approach. BMC Evol Biol. 2008;27:8–67. doi: 10.1186/1471-2148-8-67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breitbart M, Rohwer F. Here a virus, there a virus, everywhere the same virus? Trends Microbiol. 2005;13:278–284. doi: 10.1016/j.tim.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 19.Wilson DE, Reeder DM, editors. Mammal species of the world: a taxonomic and geographic reference 2. Washington and London: Smithsonian Institution Press; 2005. [Google Scholar]
- 20.Evans BJ, Kelley DB, Tinsley RC, Melnick DJ, Cannatella DC. A mitochondrial DNA phylogeny of African clawed frogs: phylogeography and implications for polyploid evolutio. Mol Phylogenet Evol. 2004;33:197–213. doi: 10.1016/j.ympev.2004.04.018. [DOI] [PubMed] [Google Scholar]
- 21.Roughgarden J, editor. Anolis lizards of the Caribbean. Ecology, evolution and plate tectonics. New York and Oxford: Oxford University Press; 1995. [Google Scholar]
- 22.Stadelmann B, Lin L-K, Kunz TH, Ruedi M. Molecular phylogeny of New World Myotis (Chiroptera, Vespertilionidae) inferred from mitochondrial and nuclear DNA genes. Mol Phylogenet Evol. 2007;43:32–48. doi: 10.1016/j.ympev.2006.06.019. [DOI] [PubMed] [Google Scholar]
- 23.Steppan SJS, Adkins RM, Anderson J. Phylogeny and divergence-date estimates of rapid radiations in muroid rodents based on multiple nuclear genes. Syst Biol. 2004;53:533–553. doi: 10.1080/10635150490468701. [DOI] [PubMed] [Google Scholar]
- 24.Marshall LG, Butler RF, Drake RE, Curtis GH, Tedford RH. Calibration of the great American interchange. Science. 1979;204:272–279. doi: 10.1126/science.204.4390.272. [DOI] [PubMed] [Google Scholar]
- 25.Poux C, Chevret P, Huchon D, De Jong WW, Douzery EJP. Arrival and diversification of caviomorph rodents and platyrrhine primates in South America. Syst Biol. 2006;55:228–244. doi: 10.1080/10635150500481390. [DOI] [PubMed] [Google Scholar]
- 26.Hall T. BioEdit version 5.0.6. 2004. Available at, http://www.mbio.ncsu.edu/ BioEdit/bioedit.html.
- 27.Huelsenbeck JP, Ronquist F. MrBayes: Bayesian inference of phylogenetic tree. Bioinformatics. 2001;17:754–755. doi: 10.1093/bioinformatics/17.8.754. [DOI] [PubMed] [Google Scholar]
- 28.Guindon S, Gascuel O. A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]