Abstract
A chromodomain is a domain contained in various proteins involved in chromatin remodeling and the regulation of gene expression in eukaryotes during development. Chromodomains perform a wide range of diverse functions including chromatin targeting and interactions between different proteins, RNA and DNA. The chromodomains also have been found as an additional domain at the C-terminal region of Polyproteins (Pol) encoded by transposable elements, which belong to the Gypsy LTR retrotransposons superfamily. Chromoviruses or chromodomain-containing Gypsy LTR retrotransposons form the most widespread clade of Gypsy LTR retrotransposons and can be found in diverse eukaryotes including plants, fungi and vertebrates. The recent finding suggested that chromodomains can be responsible for the targeted integration of LTR retrotransposons and, thus, should be favorable for mobile elements by allowing them to avoid negative selection arising from insertion into coding regions.
Key words: chromodomain, LTR retrotransposons, targeted integration, plants, evolution
Chromatin Organization Modifiers and Chromatin Regulation
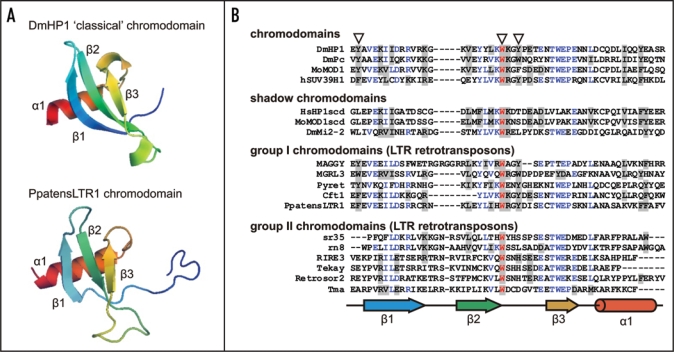
The chromodomains (chromatin organization modifiers) are protein structural domains of about 40–50 amino acid residues commonly found in proteins associated with the remodeling of chromatin.1 Chromodomains have been identified in a variety of proteins including factors that activate and repress transcription. More than 40 examples of chromodomains are known so far.2 Chromodomains were originally identified as a conserved sequence motif between Polycomb, a protein required for homeotic gene repression, and heterochromatin protein-1 (HP1), a component of centromeric heterochromatin, from Drosophila melanogaster;3 both are regulators of chromatin structure and involved in epigenetic repression. The chromodomains are highly conserved among eukaryotes, and are represented in a large number of various proteins in many genomes. All chromodomains are believed to have similar three-dimensional structure. They consist of an N-terminal three-stranded anti-parallel β-sheet (β-sandwich), which packs against the C-terminal α-helix (Fig. 1). A hydrophobic groove is formed on one side of the β-sheet, which is composed of conserved nonpolar residues.5,6
Figure 1.
Structures of chromodomains from the HP1 protein (Drosophila melanogaster; pdb:1q3l)4 and PpatensLTR1 (Gypsy LTR retrotransposon from moss Physcomitrella patens) (A); and multiple alignment of ‘classical’ chromodomains and ‘shadow’ chromo-like motifs from functional proteins as well as group I and group II Gypsy LTR retrotransposon chromodomains (B). The PpatensLTR1 chromodomain structure was predicted by (PS)2.5 The residues that constitute the aromatic pocket are indicated by arrows at the top. The secondary structural elements are shown at the bottom: arrows indicate β-strands; cylinder indicates α-helix. The GenBank accession numbers for the sequences are as follows: DmHP1—M57574; DmPc—P26017; MoMOD1 and MoMOD1scdv—P23197; hSUV39H1—S47004; HsHP1scd—AAB26994; DmMi2-2—AF119716; MAGGY—L35053; MGRL3—AF314096; Pyret—AB062507; Cft1—AF051915; PpatensLTR1—XM_001752430; sr35—AC068924; rn8—AK068625; RIRE3—AC119148; Tekay—AF050455; RetroSor2—AF061282; Tma—AF147263.
In general, chromodomain-containing proteins are responsible for aspects of gene regulation related to chromatin remodeling and formation of heterochromatin regions.7 Several studies have demonstrated that chromodomains are responsible for chromatin targeting and for interactions between various proteins as a stand-alone protein interaction module,8 as a RNA-binding module,9 and, most recently, as a DNA-binding module.10 Chromodomain-containing proteins are found in association with the pericentromeric heterochromatin in many eukaryotes. Some chromodomain-containing genes have multiple alternative-splicing isoforms that omit the chromodomain entirely.11
There are two major groups of chromodomains found in eukaryotic chromodomain-containing proteins. So-called ‘classical’ chromodomains carry the characteristic chromo-box motif [Yf] [LFY][LIV]K[Wy][kr] (single letter code, capital letters standing for the most prominent amino acid).2 The second group of chromodomains is more variable and, includes chromo-related domains, which are well conserved in their central part (strand β2), but deviate significantly in other regions (strand β1 and α-helix) (Fig. 1). The best-known protein with both types of chromodomains is HP1.
HP1 is a hallmark of constitutive heterochromatin, a condensed and highly repressive type of chromatin that organizes the repetitive pericentromeric DNA.12,13 This protein contains an N-terminal ‘classical’ chromodomain and a C-terminal ‘shadow’ chromo-related domain.2 Mutational analysis showed that the N-terminal chromodomain of HP1 is essential for direct heterochromatin binding, whereas the ‘shadow’ domain is necessary for nuclear localization.14 So far the function of Drosophila HP1 N-terminal chromodomain is the most investigated of all known chromodomains. This domain of HP1 is sufficient for specific interactions with the histone H3 tail, in a manner that depends on the methylation of Lys9, dimethyllysine (Me2K9) or trimethyllysine (Me3K9). The MeK recognition involves a conserved aromatic pocket, whereas interactions between H3 tail and the chromodomain consist of a series of backbone hydrogen bonds and complementary surfaces formed between their side chains.4,15 The aromatic pocket is formed by three conserved aromatic residues Y24, W45 and Y48. The ‘shadow’ chromodomain of HP1 binds to the pentapeptide motif [PL][WRY]V[MIL][MLV] that been seen in non-histone proteins which interact with HP1.16,17 Interestingly, HP1 may form homodimers and subsequently compact chromatin into higher order heterochromatin structures when bound to histone H3 MeK.18 Collectively this structure would contribute to an overall transcriptionally repressive chromatin environment.
The fact that the aromatic cage residues as well as the residues that responsible for H3 tail sequence specificity can be found in the number of chromodomains from other chromodomain-containing proteins suggests a common function. The studies did not reveal a universal principle by which all chromodomains act, but uncovered a potential for diverse molecular interactions.19 For example, the Polycomb chromodomains interacts with H3 MeK27,20,21 whereas the tandem chromodomains of human chromo-helicase/ATPase DNA-binding protein 1 (CHD1) act cooperatively to specifically bind H3 MeK4,22 a mark characteristic of euchromatin. MOF protein is histone acetylase which targeted to the male X chromosome in Drosophila where it acetylates histone H4 at Lys16.23 This acetylation is critical to the activation of X-linked genes as a mechanism to compensate for the reduced gene dosage in male versus female somatic cells.24 MOF uses its chromodomains to bind roX RNA.9
Chromoviruses as LTR Retrotransposons with a Chromodomain
Surprisingly, the chromodomains also have been found as an additional domain at the C-terminal region of Polyproteins (Pol) encoded by transposable elements from Gypsy LTR retrotransposon superfamily.25–27 Transposable elements (TEs) have the ability to replicate and spread in genome. They were found virtually in all investigated eukaryotes and represent the ubiquitous components of eukaryotic genomes. For example, at least 46% of the human genome is represented by TE-derived sequences28 and more than 70% of the plants genomes can be composed by TEs.29 The relative abundance and diversity of TEs have contributed to the differences in the structure and size of eukaryotic genomes, especially in the plant kingdom.30 Transposable elements have had a profound influence on the evolution of eukaryote genomes.31 Recent evidence suggests that TEs may provide the genome with potent agents to generate genetic and genomic plasticity. Insertions of TEs near genes can lead to alterations in gene expression patterns—since the elements usually contain transcriptional regulatory sequences—while insertions within genes can directly alter gene structure. Recombination between elements at different sites can lead to large-scale chromosomal rearrangements. TEs may have reshaped the human genome by ectopic rearrangements, by creating new genes, and by modifying and shuffling existing genes.28 In some cases, TEs perform critical biological functions in their host. For example, the preferential insertion of some retrotransposons in Drosophila at telomeric locations has removed the need for telomerase function.32
Two classes of TEs are known currently. Class II TEs, or DNA transposons, utilize DNA-based modes of transposition including ‘cut-and-paste’ mechanism, rolling-circle replication, and a mechanism which involves DNA polymerase and is not yet well understood.33–35 Class I, or retrotransposons, are mobile genetic elements that use reverse transcription in their transposition. Five orders of retrotransposons are recognized: those having long terminal repeats (LTRs) (LTR retrotransposons); those lacking LTRs (non-LTR retrotransposons); DIRS retrotransposons; Penelope-like retrotransposable elements; and short interspersed nuclear elements (SINEs).36
LTR retrotransposons contain long terminal repeats (LTRs) in direct orientation. The mechanism of transposition is similar to that of retroviruses. Full-sized autonomous LTR retrotransposons contain two genes, gag and pol, or a fused gag-pol gene. The gag gene encodes an RNA-binding protein similar to the retrovirus capsid protein; the pol gene encodes a protein that has protease (PR), reverse transcriptase (RT), ribonuclease H (RNaseH) and integrase (Int) activities. According to the modern classification, LTR retrotransposons are divided into two superfamilies: Copia (Pseudoviridae) and Gypsy (Metaviridae).36–39 Gypsy LTR retrotransposons are similar in structure to retroviruses. Particularly, in Gypsy LTR retrotransposons and retroviruses the pol gene domains are located in the order PR-RT-RNaseH-Int. Moreover, a few Gypsy LTR retrotransposons carry additional third gene env, encoding an envelope-like protein (Fig. 2).36
Figure 2.
Chromoviruses in plants: structural organization of PpatensLTR1 from Physcomitrella patens (A); a schematic evolutionary tree of plants chromoviruses reconstructed from a multiple alignment of reverse transcriptase DNA fragment (B); and distribution of various chromovirus clades (C). Abbreviations: 5′ and 3′ LTRs—5′ and 3′ long terminal repeats; PR—aspartyl protease, RT—reverse transcriptase, RNaseH—ribonuclease H, Int—integrase, chromo—chromodomains. The plant phylogenetic tree is after Bowman et al.40 with minor modifications. The divergence time estimations are shown after Hedges.41 For a more detailed evolutionary tree of plant chromoviruses, see Novikova et al.42
A number of Gypsy LTR retrotransposons include a chromodomain as an additional domain at the C-terminus of integrase, suggesting that chromodomains might control integration. Such Gypsy elements were classified as chromoviruses.25–27 Their integrase with a chromodomain was named chromo-integrase. Among Gypsy LTR retrotransposons, the chromoviruses are the most widespread group, present in genomes of plants, fungi and vertebrates.27 Although the genomic data for basal eukaryotes is very limited, it is believed that chromoviruses are absent in the genomes of the most basal eukaryotic lineages (Diplomonadida, Euglenozoa, Alveolata and Bacillariophyta). It was proposed that chromoviruses appeared no earlier than in Cercozoa.27
There are two distinct groups of retrotransposon chromodomains. Group I is characterized by the presence of the three conserved aromatic residues, which are the same as the conserved residues form aromatic cage of ‘classical’ N-terminal chromodomain from HP1.4,15,43 This group of chromodomains was found in diverse eukaryotic LTR retrotransposons including fungal and vertebrate Gypsy elements. Representatives of the group II of chromodomains lack the first conserved aromatic residue and usually the third. Group II was identified only in plant retrotransposons. The structure-based alignment allows attributing the group II retrotransposons chromodomain to ‘shadow’ chromodomain-like motifs (data not shown; see Fig. 1).
Chromoviruses in Plants
The chromosomes of plants are littered with retrotransposons that, in many cases, constitute as much as 80% of plant genomes. LTR retrotransposons have been especially successful colonizers of plant chromosomes. Examination of their function, evolution and dispersal is essential to the understanding the evolution of eukaryotic genomes. A number of studies were performed in attempt to describe the diversity of Gypsy and Copia retrotransposons in the plant kingdom. Comprehensive studies of LTR retrotransposons in plant genomes are regularly published.44–47 However, these works concerned mainly agricultural species or model organisms and the majority of the known families of LTR retrotransposons have been found and fully characterized in angiosperms while other large groups are less covered.30 The recently published survey of chromodomain-containing Gypsy LTR retrotransposons from mosses performed in our laboratory suggested that the diversity of plants LTR retrotransposons is highly underestimated and the evolutionary history of Gypsy LTR retrotransposons from plants is still not fully resolved.42
The phylogenetic analysis of number of chromodomain-containing Gypsy LTR retrotransposons showed that they represented by four separate clades in the genomes of higher (seed) plants: Tekay, Galadriel, CRM and Reina (Fig. 2).27 At least two additional clades, indicated as A and B on the Figure 2, can be found in genomes of non-seed plants such as mosses,42 ferns and lycophytes (unpublished data). The analyses of whole genomic sequence of moss Physcomitrella patens (Bryophyta, Funariales) along with experimental investigation of diverse moss species allowed us to describe several new chromodomain-containing Gypsy LTR retrotransposons belonging to clades A and B. The most interesting finding in our investigation of Gypsy LTR retrotransposons from non-seed plants is that two types of retrotransposon chromodomains (group I in clade A and group II in clade B) indeed are represented in non-seed plant genomes.42 At the same time, only group II chromodomains are known in angiosperm and gymnosperm Gypsy LTR retrotransposons.27,43 It has to be noted that representatives of CRM clade have no detectable chromodomains. Instead, they showed the presence of the conservative protein domain, which was named ‘CR motif’. The role of this motif is unkown.43
The information on the role of chromodomains in retrotransposition of LTR retrotransposons is limited. The chromo-integrase of Tf1 LTR retrotransposon from Schizosaccharomyces pombe lacking the chromodomain demonstrated a significantly higher activity and substantially reduced substrate specificity. This suggests that both the activity and specificity of enzymes can be modulated by their chromodomains.48 Conversely, the transposition activity of MAGGY retrotransposon of the rice blast fungus Magnaporthe oryzae dramatically decreased with the loss or alteration of chromodomain.49 As was demonstrated recently, MAGGY chromodomain, which belonged to the group I chromodomains, interacts with H3 Me2K9 and H3 Me3K9 similarly to HP1 ‘classical’ chromodomain. Moreover, it was proposed that chromodomains can target integration of chromoviruses into heterochromatic regions.43 Almost nothing is known about activity and role of group II chromodomains from plant retrotransposons. The mostly heterochromatic distribution of plant chromoviruses along with data suggesting the localization of chromodomain-YFP fused protein in heterochromatin can be used as an indirect evidence for recognizing heterochromatin and directing the integration role of group II chromodomains.43 Nevertheless, the mechanisms by which chromodomains group II act are still unknown.
As a result of potentially deleterious effects of transposable element proliferation, host organisms have often evolved mechanisms that limit TEs activity, such as specific methylation. Similarly, since TEs are, in general, only rarely transmitted across species boundaries, their continued existence is usually dependent upon the continued survival of their hosts. As a result, transposable elements themselves often appear to have evolved mechanisms, such as directing their integration to specific parts of the genome, which keep the damage they cause to a minimum.50 Selective targeting to heterochromatin provided by chromodomains can be favorable for mobile elements by allowing them to avoid negative selection arising from their insertion into coding regions.43,51 The heterochromatin-targeted integration of mobile elements should be essentially beneficial for those organisms which are predominantly haploid in their life cycle such as fungi and non-seed plants.
Future Trends
The further studies of retrotransposon chromodomains role should provide insights not only into the mechanism of targeted integration of LTR retrotransposons but also to general evolutionary issues. Several evolutionary questions can be addressed by such investigations. For example, how the shift from LTR retrotransposons with group I chromodomains to those with group II containing has effected the organization of plant genomes? Is the difference in chromodomains activity played key role in this shift? Why do mosses contain predominantly group I retrotransposon chromodomains while seed plants contain only group II retrotransposon chromodomains? Is there a difference in beneficial features provided for LTR retrotransposons by group I and group II chromodomains?
We have a unique opportunity for in vivo investigation of the activity of the retrotransposon group I and group II chromodomains in a homogenous system. Moss Physcomitrella patens can be used as a model organism for the analysis of retrotransposon chromodomains activity and targeted integration. The haploidy of the dominant gametophyte stage in moss development makes mosses attractive material for genetic studies because isolation of mutants and genetic analysis are more straightforward than in species with a dominant diploid phase. The potential of mosses as model systems to study plant biological processes is reinforced by their suitability for cell lineage analysis and similar responses to plant growth factors and environmental stimuli to those observed in other plants.52 Moreover, the P. patens genome sequence has been published recently, which provides the basis for a comprehensive survey of transposable elements, and investigation of retrotransposon distribution in genomic sequences and their target site preferences.53
Acknowledgements
Author thanks Dr. Marie-Angèle Grandbastien (Institut Jean-Pierre Bourgin, INRA, France) for the helpful comments and Dr. Victor Fet (Marshall University, West Virginia, USA) for his stylistic suggestions.
Abbreviations
- HP1
heterochromatin protein-1
- MeK
methyllysine
- H3 Me2K9
dimethyllysine at position 9 of histone H3
- H3 Me3K9
trimethyllysine at position 9 of histone H3
- CHD1
human chromo-helicase/ATPase DNA-binding protein 1
- Pol
polyprotein
- TEs
transposable elements
- LTRs
long terminal repeats
- PR
protease
- RT
reverse transcriptase
- RNaseH
ribonuclease H
- Int
integrase
- YFP
yellow fluorescent protein
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/7702
References
- 1.Koonin EV, Zhou S, Lucchesi JC. The chromo superfamily: new members, duplication of the chromo domain and possible role in delivering transcription regulators to chromatin. Nucleic Acids Res. 1995;23:4229–4233. doi: 10.1093/nar/23.21.4229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Aasland R, Stewart AF. The chromo shadow domain, a second chromo domain in heterochromatin-binding protein 1, HP1. Nucleic Acids Res. 1995;23:3168–3174. doi: 10.1093/nar/23.16.3168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Paro R, Hogness DS. The Polycomb protein shares a homologous domain with a heterochromatin-associated protein of Drosophila. Proc Natl Acad Sci USA. 1991;88:263–267. doi: 10.1073/pnas.88.1.263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jacobs SA, Khorasanizadeh S. Structure of HP1 chromodomain bound to a lysine 9-methylated histone H3 tail. Science. 2002;295:2080–2083. doi: 10.1126/science.1069473. [DOI] [PubMed] [Google Scholar]
- 5.Chen CC, Hwang JK, Yang JM. (PS)2: protein structure prediction server. Nucleic Acids Res. 2006;34:152–157. doi: 10.1093/nar/gkl187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ball LJ, Murzina NV, Broadhurst RW, Raine AR, Archer SJ, Stott FJ, et al. Structure of the chromatin binding (chromo) domain from mouse modifier protein 1. EMBO J. 1997;16:2473–2481. doi: 10.1093/emboj/16.9.2473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Brehm A, Tufteland KR, Aasland R, Becker PB. The many colours of chromodomains. Bioessays. 2004;26:133–140. doi: 10.1002/bies.10392. [DOI] [PubMed] [Google Scholar]
- 8.Lachner M, O'Carroll D, Rea S, Mechtler K, Jenuwein T. Methylation of histone H3 lysine 9 creates a binding site for HP1 proteins. Nature. 2001;410:116–120. doi: 10.1038/35065132. [DOI] [PubMed] [Google Scholar]
- 9.Akhtar A, Zink D, Becker PB. Chromodomains are protein-RNA interaction modules. Nature. 2000;407:405–409. doi: 10.1038/35030169. [DOI] [PubMed] [Google Scholar]
- 10.Bouazoune K, Mitterweger A, Längst G, Imhof A, Akhtar A, Becker PB, et al. The dMi-2 chromodomains are DNA binding modules important for ATP-dependent nucleosome mobilization. EMBO J. 2002;21:2430–2440. doi: 10.1093/emboj/21.10.2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tajul-Arifin K, Teasdale R, Ravasi T, Humel DA, Mattick JS. Identification and analysis of chromodomain-containing proteins encoded in the mouse transcriptome. Genome Res. 2003;13:1416–1429. doi: 10.1101/gr.1015703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Elgin SC. Heterochromatin and gene regulation in Drosophila. Curr Opin Genet Dev. 1996;6:193–202. doi: 10.1016/s0959-437x(96)80050-5. [DOI] [PubMed] [Google Scholar]
- 13.Grewal SI, Elgin SC. Heterochromatin: new possibilities for the inheritance of structure. Curr Opin Genet Dev. 2002;12:178–187. doi: 10.1016/s0959-437x(02)00284-8. [DOI] [PubMed] [Google Scholar]
- 14.Platero JS, Hartnett T, Eissenberg JC. Functional analysis of the chromo domain of HP1. EMBO J. 1995;14:3977–3986. doi: 10.1002/j.1460-2075.1995.tb00069.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jacobs SA, Taverna SD, Zhang Y, Briggs SD, Li J, Eissenberg JC, Allis CD, Khorasanizadeh S. Specificity of the HP1 chromo domain for the methylated N-terminus of histone H3. EMBO J. 2001;20:5232–5241. doi: 10.1093/emboj/20.18.5232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Smothers JF, Henikoff S. The HP1 chromo shadow domain binds a consensus peptide pentamer. Curr Biol. 2000;10:27–30. doi: 10.1016/s0960-9822(99)00260-2. [DOI] [PubMed] [Google Scholar]
- 17.Brasher SV, Smith BO, Fogh RH, Nietlispach D, Thiru A, Nielsen PR, et al. The structure of mouse HP1 suggests a unique mode of single peptide recognition by the shadow chromo domain dimer. EMBO J. 2000;19:1587–1597. doi: 10.1093/emboj/19.7.1587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, et al. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Eissenberg JC. Molecular biology of the chromo domain: an ancient chromatin module comes of age. Gene. 2001;275:19–29. doi: 10.1016/s0378-1119(01)00628-x. [DOI] [PubMed] [Google Scholar]
- 20.Fischle W, Wang Y, Jacobs SA, Kim Y, Allis CD, Khorasanizadeh S. Molecular basis for the discrimination of repressive methyl-lysine marks in histone H3 by Polycomb and HP1 chromodomains. Genes Dev. 2003;17:1870–1881. doi: 10.1101/gad.1110503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Min J, Zhang Y, Xu RM. Structural basis for specific binding of Polycomb chromodomain to histone H3 methylated at Lys 27. Genes Dev. 2003;17:1823–1828. doi: 10.1101/gad.269603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Flanagan JF, Mi LZ, Chruszcz M, Cymborowski M, Clines KL, Kim Y, et al. Double chromodomains cooperate to recognize the methylated histone H3 tail. Nature. 2005;438:1181–1185. doi: 10.1038/nature04290. [DOI] [PubMed] [Google Scholar]
- 23.Akhtar A, Becker PB. Activation of transcription through histone H4 acetylation by MOF, an acetyltransferase essential for dosage compensation in Drosophila. Mol Cell. 2000;5:367–375. doi: 10.1016/s1097-2765(00)80431-1. [DOI] [PubMed] [Google Scholar]
- 24.Lucchesi JC. Dosage compensation in Drosophila and the ‘complex’ world of transcriptional regulation. Bioessays. 1996;18:541–547. doi: 10.1002/bies.950180705. [DOI] [PubMed] [Google Scholar]
- 25.Malik HS, Eickbush TH. Modular evolution of the integrase domain in the Ty3/gypsy class of LTR retrotransposons. J of Virol. 1999;73:5186–5190. doi: 10.1128/jvi.73.6.5186-5190.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Marín I, Lloréns C. Ty3/Gypsy retrotransposons: description of new Arabidopsis thaliana elements and evolutionary perspectives derived from comparative genomic data. Mol Biol Evol. 2000;17:1040–1049. doi: 10.1093/oxfordjournals.molbev.a026385. [DOI] [PubMed] [Google Scholar]
- 27.Gorinsek B, Gubensek F, Kordis D. Evolutionary genomics of chromoviruses in eukaryotes. Mol Biol Evol. 2004;21:781–798. doi: 10.1093/molbev/msh057. [DOI] [PubMed] [Google Scholar]
- 28.Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 29.Bennetzen JL, SanMiguel P, Chen M, Tikhonov A, Francki M, Avramova Z. Grass genomes. Proc Natl Acad Sci USA. 1998;95:1975–1978. doi: 10.1073/pnas.95.5.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vitte C, Panaud O. LTR retrotransposons and flowering plant genome size: emergence of the increase/decrease model. Cytogenet Genome Res. 2005;110:91–107. doi: 10.1159/000084941. [DOI] [PubMed] [Google Scholar]
- 31.Kidwell MG, Lisch DR. Perspective: transposable elements, parasitic DNA and genome evolution. Evolution. 2001;55:1–24. doi: 10.1111/j.0014-3820.2001.tb01268.x. [DOI] [PubMed] [Google Scholar]
- 32.Pardue ML, Rashkova S, Casacuberta E, DeBaryshe PG, George JA, Traverse KL. Two retrotransposons maintain telomeres in Drosophila. Chromosome Res. 2005;13:443–453. doi: 10.1007/s10577-005-0993-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kapitonov VV, Jurka J. Helitrons on a roll: eukaryotic rolling-circle transposons. Trends Genet. 2007;23:521–529. doi: 10.1016/j.tig.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 34.Pritham EJ, Putliwala T, Feschotte C. Mavericks, a novel class of giant transposable elements widespread in eukaryotes and related to DNA viruses. Gene. 2007;390:3–17. doi: 10.1016/j.gene.2006.08.008. [DOI] [PubMed] [Google Scholar]
- 35.Feschotte C, Pritham EJ. DNA transposons and the evolution of eukaryotic genomes. Annu Rev Genet. 2007;41:331–368. doi: 10.1146/annurev.genet.40.110405.090448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8:973–982. doi: 10.1038/nrg2165. [DOI] [PubMed] [Google Scholar]
- 37.Goodwin TJ, Poulter RT. The DIRS1 group of retrotransposons. Mol Biol Evol. 2001;18:2067–2082. doi: 10.1093/oxfordjournals.molbev.a003748. [DOI] [PubMed] [Google Scholar]
- 38.Peterson-Burch BD, Voytas DF. Genes of the Pseudoviridae (Ty1/copia retrotransposons) Mol Biol Evol. 2002;19:1832–1845. doi: 10.1093/oxfordjournals.molbev.a004008. [DOI] [PubMed] [Google Scholar]
- 39.Havecker ER, Gao X, Voytas DF. The diversity of LTR retrotransposons. Genome Biol. 2004;5:225. doi: 10.1186/gb-2004-5-6-225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bowman JL, Floyd SK, Sakakibara K. Green genes-comparative genomics of the green branch of life. Cell. 2007;129:229–234. doi: 10.1016/j.cell.2007.04.004. [DOI] [PubMed] [Google Scholar]
- 41.Hedges SB. The origin and evolution of model organisms. Nat Rev Genet. 2002;3:838–849. doi: 10.1038/nrg929. [DOI] [PubMed] [Google Scholar]
- 42.Novikova O, Mayorov V, Smyshlyaev G, Fursov M, Adkison L, Pisarenko O, et al. Novel clades of chromodomain-containing Gypsy LTR retrotransposons from mosses (Bryophyta) Plant J. 2008;56:562–574. doi: 10.1111/j.1365-313X.2008.03621.x. [DOI] [PubMed] [Google Scholar]
- 43.Gao X, Hou Y, Ebina H, Levin HL, Voytas DF. Chromodomains direct integration of retrotransposons to heterochromatin. Genome Res. 2008;18:359–369. doi: 10.1101/gr.7146408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wright DA, Voytas DF. Potential retroviruses in plants: Tat1 is related to a group of Arabidopsis thaliana Ty3/gypsy retrotransposons that encode envelope-like proteins. Genetics. 1998;149:703–715. doi: 10.1093/genetics/149.2.703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wicker T, Stein N, Albar L, Feuillet C, Schlagenhauf E, Keller B. Analysis of a contiguous 211 kb sequence in diploid wheat (Triticum monococcum L.) reveals multiple mechanisms of genome evolution. Plant J. 2001;26:307–316. doi: 10.1046/j.1365-313x.2001.01028.x. [DOI] [PubMed] [Google Scholar]
- 46.Hirochika H, Fukuchi A, Kikuchi F. Retrotransposon families in rice. Mol Gen Genet. 1992;233:209–216. doi: 10.1007/BF00587581. [DOI] [PubMed] [Google Scholar]
- 47.Kumekawa N, Ohtsubo E, Ohtsubo H. Identification and phylogenetic analysis of gypsy-type retrotransposons in the plant kingdom. Genes Genet Syst. 1999;74:299–307. doi: 10.1266/ggs.74.299. [DOI] [PubMed] [Google Scholar]
- 48.Hizi A, Levin HL. The integrase of the long terminal repeat-retrotransposon tf1 has a chromodomain that modulates integrase activities. J Biol Chem. 2005;280:39086–39094. doi: 10.1074/jbc.M506363200. [DOI] [PubMed] [Google Scholar]
- 49.Nakayashiki H, Awa T, Tosa Y, Mayama S. The C-terminal chromodomain-like module in the integrase domain is crucial for high transposition efficiency of the retrotransposon MAGGY. FEBS Lett. 2005;579:488–492. doi: 10.1016/j.febslet.2004.12.017. [DOI] [PubMed] [Google Scholar]
- 50.Sandmeyer S. Targeting transposition: at home in the genome. Genome Res. 1998;8:416–418. doi: 10.1101/gr.8.5.416. [DOI] [PubMed] [Google Scholar]
- 51.Boeke JD, Devine SE. Yeast retrotransposons: finding a nice quiet neighborhood. Cell. 1998;93:1087–1089. doi: 10.1016/s0092-8674(00)81450-6. [DOI] [PubMed] [Google Scholar]
- 52.Cove DJ, Kammerer W, Knight CD, Leech MJ, Martin CR, Wang TL. Developmental genetic studies of the moss, Physcomitrella patens. Symp Soc Exp Biol. 1997;45:31–43. [PubMed] [Google Scholar]
- 53.Rensing SA, Lang D, Zimmer AD, Terry A, Salamov A, Shapiro H, et al. The Physcomitrella genome reveals evolutionary insights into the conquest of land by plants. Science. 2008;319:64–69. doi: 10.1126/science.1150646. [DOI] [PubMed] [Google Scholar]