Abstract
In Saccharomyces cerevisiae, glucose starvation induces key gluconeogenic enzymes such as fructose-1,6-bisphosphatase (FBPase), malate dehydrogenase (MDH2) and phosphoenolpyruvate carboxykinase, while glucose addition inactivates these enzymes. Significant progress has been made identifying mechanisms that mediate the “catabolite inactivation” of FBPase and MDH2. For example, the site of their degradation has been shown to change, depending on the duration of starvation. When glucose is added to short-termed starved cells, these proteins are degraded in the proteasome. However, when glucose is added to long-termed starved cells, they are degraded in the vacuole by a selective autophagy pathway. For the vacuole pathway, these proteins are first imported into novel vesicles called Vid (vacuole import and degradation) vesicles. Following import, Vid vesicles merge with the endocytic pathway. Future experiments will be directed at understanding the molecular mechanisms that regulate the switch from proteasomal to vacuolar degradation and determining the site of Vid vesicle biogenesis.
Key words: catabolite inactivation; fructose-1,6-bisphosphatase; proteasome; vacuole; vacuole import and degradation; autophagy; coatomer proteins
Introduction
Protein levels in a cell reflect a balance between the processes of protein synthesis and protein degradation. For protein degradation, two major pathways have been identified. These are the proteasome dependent and the lysosomal dependent degradation pathways.1–9 The proteasomal system requires the ubiquitination of protein substrates using E1, E2 and E3 enzymes.1–4 These ubiquitinated proteins are then targeted to the proteasome and destroyed.1–4 The other major degradation pathway in cells occurs in the lysosome.5–9 Lysosomes are acidic organelles that contain a variety of hydrolases. In general, lysosomes are responsible for degrading most of the cells long-lived proteins and organelles.5–9 This is in contrast to the proteasome, which primarily degrades short-lived cytosolic proteins.1–4
Lysosomal protein degradation can be regulated by nutrients and hormones. For example, both amino acid starvation and glucagon addition increase lysosomal degradation, while the presence of insulin inhibits this process.8,9 Proteins are targeted for degradation via multiple pathways. One example is macroautophagy, a non-selective degradation process. This pathway, which has been studied extensively, plays an important role in the survival of cells during periods of nutrient deprivation. In addition to its role in starvation, autophagy is required for the normal development of worms, flies, mice and humans.10–14 As such, defects in this process can lead to diseases.10–14
In contrast to the non-discriminate nature of macroautophagy, there are also selective autophagy pathways. One type of lysosomal degradation selects proteins with a specific pentapeptide sequence. Proteins with KFERQ in their sequence are recognized by the heat shock protein Hsc73.15–17 These proteins are then bound by the lysosomal receptor protein LAMP2,18 and transported into the lumen of lysosomes. Chaperone mediated autophagy also requires the assistance of several luminal chaperones,19 in conjunction with the function of cytosolic chaperones. Since, a relatively high percentage of proteins contain the KFERQ sequence, this form of autophagy plays an important role in the degradation of bulk cytosolic proteins during starvation. However, other proteins such as the mutant forms of synuclein can be degraded via chaperone mediated autophagy, along with oxidized proteins and other damaged proteins.20–24 Interestingly, this pathway is impaired during aging24 and it is stimulated by ketone bodies.19
Multiple Protein Targeting Pathways to the Yeast Vacuole
In yeast, the vacuole is the homolog of mammalian cell lysosomes.26–28 Multiple trafficking pathways carry proteins to the vacuole, where they are either degraded or processed. For example, proteins can be endocytosed from the plasma membrane and targeted for degradation in the vacuole.26–28 In contrast, most vacuole resident proteins are synthesized and travel to the vacuole via the Vps pathway.29–31 However, one vacuolar resident, aminopeptidase I, is delivered directly from the cytosol to the vacuole for processing via the Cvt pathway.10–12,32–35 The Cvt and macroautophagy pathway shares many genes. Furthermore, both pathways can be divided into distinct steps, i.e., the nucleation, expansion and sequestration steps. These steps are, in turn, regulated by a number of ATG genes.10–12,32–35 Finally, unique ubiquitin-like conjugation36 and lipidation processes37 play a role in regulating autophagy.
The vacuole is also the site for organelle degradation. For example, peroxisomes can be degraded in the vacuole via two pexophagy pathways that can be distinguished morphologically.38–40 When methyltrophic yeast are grown in methanol and shifted to ethanol, peroxisomes are degraded by macropexophagy.38–40 By contrast, when these cells are shifted from methanol to glucose, peroxisomes are degraded by micropexophagy.38–40
Catabolite Inactivation in Yeast
In Saccharomyces cerevisiae, a number of gluconeogenic enzymes are induced by glucose starvation. Typically, these enzymes are inactivated when cells are replenished with fresh glucose. The most studied example of this “Catabolite Inactivation” is the key gluconeogenic enzyme fructose-1,6-bisphosphatase (FBPase).41–46 In addition to FBPase, other gluconeogenic enzymes including malate dehydrogenase (MDH2) and phosphoenolpyruvate carboxykinase (PEPCK) are inactivated by glucose.45 Likewise, glucose inactivates sugar transporters such as the high affinity glucose transporter,45 the galactose transporter47 and the maltose transporter.48 Peroxisomes are involved in fatty acid metabolism and they are also inactivated by glucose. Thus, catabolite inactivation applies, not only to enzymes involved in sugar transport and gluconeogenesis, but also to peroxisomes involved in fatty acid metabolism. Catabolite inactivation of the maltose transporter and the galactose transporter is mediated by endocytosis of these proteins from the plasma membrane followed by degradation in the vacuole.48–50
Catabolite inactivation of FBPase has been examined extensively. During catabolite inactivation, FBPase is phosphorylated and inactivated via a cAMP mediated process.51–53 In wild-type cells, cAMP levels increase when glucose is added following a period of starvation. However, in the ras2318S mutant, cAMP levels do not increase following glucose stimulation, and FBPase is not phosphorylated.54 Phosphorylation of FBPase most likely occurs at multiple sites. Phosphorylation at serine 11 has been demonstrated following a glucose shift.52 Interestingly, mutation of serine 11 has little or no effect on FBPase degradation.63 FBPase degradation is blocked in the ras2318S mutant, suggesting that phosphorylation events are important for FBPase degradation.54,55 At present, it is unknown whether the phosphorylation of FBPase plays a required role in the degradation of FBPase in the vacuole. Alternatively, the phosphorylation of other molecules may be required for this degradation event.
The Site of FBPase Degradation
The site of FBPase degradation was a matter of debate for several years,56–62 although this has been resolved recently.63 Previously, the Wolf lab showed that FBPase is degraded in the proteasome following glucose addition.56–59 In contrast, our lab demonstrated that FBPase is targeted to the vacuole for degradation.60–62 In our earlier work, we utilized a vacuolar mutant strain deficient in proteinase A (Δpep4) in order to examine FBPase degradation.60–62 We reasoned that FBPase degradation would be blocked in this strain, if the vacuole were the site of degradation. Unfortunately, the Δpep4 mutant did not block FBPase degradation completely, since other proteinases such as proteinase B and proteinase C were still present. Thus, it was difficult to rule out the possibility that other protein degradation pathways might operate under our conditions. To address this issue, the degradation process was re-examined using a Δpep4Δprb1Δprc1 vacuole mutant with deletions of proteinases A, B and C.63 In this mutant, FBPase was degraded normally when cells are starved for 1d. By contrast, FBPase degradation was retarded when this strain was starved for 3d. These results suggest that the vacuole proteinases are not required for FBPase degradation in 1d starved cells, but they are required in 3d starved cells. As expected, in control wild-type cells, FBPase was degraded under all conditions.
To further examine the FBPase degradation process, a vacuole mutant strain was transformed to express FBPase-GFP. Unfortunately, the Δpep4Δprb1Δprc1 mutant exhibited strong autofluorescence. Therefore, FBPase-GFP was expressed in the Δpep4 strain.63 FBPase was found in the cytosol in 3d starved Δpep4 cells prior to glucose shift. However, this protein was detected in the vacuole following a glucose shift, providing the most direct evidence that FBPase is targeted to the vacuole for degradation when prolonged starved cells are shifted to glucose. Thus, our results indicated that the site of FBPase degradation varies, at least in part, due to the duration of glucose starvation. When glucose is added to cells that are starved for a short time (1d), FBPase is degraded in the proteasome. By contrast, when glucose is added to cells that are starved for longer periods of time (3d), FBPase is degraded in the vacuole. These results have been confirmed via a variety of methods. Re-distribution of FBPase from the cytosol to the vacuole has been observed using indirect fluorescence.60 Moreover, FBPase targeting to the vacuole has been reconstituted in vitro using semi-intact permeabilized cells lacking the endogenous FBPase.64 Under the in vitro condition, a fraction of exogenously added FBPase was protected from proteinase K digestion, suggesting that FBPase was sequestered inside organelles. Furthermore, sequestered FBPase was detected in the vacuole in semi-intact cells using indirect immunofluroesence.64
MDH2 Shares the Same Degradation Characteristics as FBPase
The cytosolic protein malate dehydrogenase (MDH2) is inactivated and degraded in response to glucose.65–67 In order to test whether MDH2 was also degraded in the vacuole in prolonged starved cells, we followed the degradation of MDH2 in short term starved and long termed starved Δpep4Δprb1Δprc1 cells using the same strategy mentioned above.63 In the Δpep4Δprb1Δprc1 mutant, MDH2 was degraded normally following 1d of starvation. However, when this mutant was starved for 3d and shifted to glucose, MDH2 degradation was inhibited. Thus, vacuole proteinases are required for MDH2 degradation in 3d starved cells, but not for 1d starved cells. To determine whether MDH2 was indeed targeted to the vacuole for degradation, the Δpep4 vacuole mutant was transformed to express MDH2-GFP. In 3d starved Δpep4 cells, MDH2-GFP was detected in the cytosol before a glucose shift. However, this protein was detected in the vacuole following a glucose shift, providing direct evidence that this protein is also targeted to the vacuole when long termed starved cells are replenished with fresh glucose.
The degradation of MDH2 shares many characteristics with FBPase. For example, both are degraded in the proteasome when glucose is added to short termed starved cells. Furthermore, both are degraded in the vacuole when glucose is added to long termed starved cells. The degradation of both proteins also requires the N-terminal sequence.63 Proline is the first amino acid of MDH2 and FBPase and it is required for degradation under either the proteasome or the vacuolar specific conditions. When the proline residue was changed to a serine residue, degradation of FBPase and MDH2 was blocked in both 1d and 3d starved cells.63 This suggests that the N-terminal proline is required for both proteasomal and vacuole dependent degradation. This observation raises an interesting question regarding how the same residue is utilized for two distinct pathways. One possible explanation is that this residue interacts with the same protein molecules, and these, in turn can interact with distinct binding partners, one for the vacuole and another for the proteasomal pathway. Alternatively, this residue may interact with the same protein molecules or complexes, but these molecules are modified differently depending on the length of starvation.
The Vacuole Import and Degradation Pathway
FBPase is targeted to the vacuole via a selective autophagy pathway. This process requires several genes, collectively called VID (Vacuole Import and Degradation) genes. Mutants defective in FBPase degradation have been isolated from a transposon library, a deletion library, and by screening cells generated by random mutagenesis.68–70 Characterization of these mutants based on FBPase distribution within cells indicated that they can be classified into at least two categories.68 Following a glucose shift, most of the mutants displayed cytosolic staining of FBPase, while a subset of mutants showed FBPase distribution in distinct punctate structures.68 When lysates were fractionated using a size column, at least two peaks were found to contain FBPase. The first peak also contained the plasma membrane protein Pma1p, while the second peak contained Vid vesicles. Kinetic studies indicate that Vid vesicles are intermediate carriers in the FBPase degradation pathway.71 As such, FBPase appears to be associated with these vesicles prior to targeting to the vacuole.
Purified Vid vesicles retain the ability to transport exogenously added FBPase in vitro.72 For the in vitro system, FBPase was purified from wild-type cells that were starved for prolonged periods of time and shifted to glucose for 20 min. Vid vesicles were purified from FBPase deletion strains that were also shifted to glucose for 20 min. In this in vitro system, FBPase sequestration into purified Vid vesicles was dependent on an ATP regenerating system and cytosol.72 Furthermore, the cytosolic heat shock proteins Ssa1p was found to play a role in the import process.72 Cyclophilin A is also required for the import of FBPase into Vid vesicles.73
The biogenesis of Vid vesicles is regulated by the UBC1 (ubiquitin conjugating enzyme 1) gene.74 Thus, in the absence of this gene, the formation of Vid vesicles is substantially reduced. At least one of the VID gene products, Vid24p, localizes to Vid vesicles.75 Vid24p is present at low levels in cells prior to a glucose shift. However, following a shift to glucose containing media, levels of this protein increase. Vid24p is associated with Vid vesicles as a peripheral protein. Although the exact role of Vid24p is unknown, in cells lacking this gene, significant amounts of FBPase were still found in Vid vesicle enriched fractions75,76 This suggests that Vid24p plays a role in a trafficking step subsequent to the import of FBPase into Vid vesicles. Because of its unique distribution, Vid24p is now routinely used as a specific marker for Vid vesicles.
COP I Coatomer Subunits are Present on Vid Vesicles
Although Vid vesicles can be purified from glucose shifted cells, it is not known whether these vesicles are present prior to a glucose shift, or whether they are subsequently derived from precursor membranes. As stated above, Vid24p is a specific marker for Vid vesicles. Unfortunately, this protein has not been useful in identifying the site of Vid vesicle biogenesis. This is because Vid24p levels are low in glucose-starved cells, and as such, it is difficult to detect structures that may associate with this protein prior to their stimulation with glucose. In order to address this issue, we attempted to identify other Vid vesicle proteins, some of which might be constitutively expressed. For these experiments, Vid vesicles were purified and subjected to MALDI analysis. A number of proteins were identified via this approach. Most notably, components of COPI vesicles such as Sec28p, Ret1p, Ret2p and Sec21p were all found on purified Vid vesicles.70 COPI vesicles have a well-known role in the trafficking of vesicles from the Golgi to the ER.77–80 Likewise, they are important in intra Golgi trafficking.77–80 Interestingly, a subset of COPI proteins have been found in endocytic compartments in mammalian cells81–85 and in yeast.86 These proteins appear to actively participate in endosomal trafficking, in mammalian cells81–85 and in multivesicular body sorting in yeast.86
The association of COPI coatomer proteins with Vid vesicles has been demonstrated experimentally.70 Coatomer proteins were observed to co-localize with Vid24p and FBPase. Coatomer proteins were present at low levels during glucose starvation. Following a glucose shift, however, coatomer proteins in Vid vesicle fractions first increased, and then decreased. Thus, there is a transient increase of Vid vesicle coatomer proteins in response to glucose. Interestingly, coatomer proteins were found to form a large protein complex with Vid24p. Interaction of coatomer proteins with Vid24p increased following a glucose shift. Furthermore, they play an important role in recruiting Vid24p to Vid vesicles. For example, in mutants defective in coatomer subunits, Vid24p association with Vid vesicles was reduced.
Coatomer Protein Sec28p Traffics to Endocytic Compartments and to the Vacuole
Since Sec28p is a component of Vid vesicles we have followed the distribution of this protein as a means to track Vid vesicle trafficking.70 When glucose was added to prolonged starved wild-type cells for 15–30 min, Sec28p-GFP was detected in punctuate structures. These structures also contained FM4–64, a lipophilic dye that is internalized from the plasma membrane to early endosomes, late endosomes, and the vacuole membrane.87 When we examined FM distribution in mutants that block our degradation pathway, its distribution varied. Some mutants such as Δypt7 and Δvam3 showed small circles, while others such as Δvph1 showed large FM-labeled circles. In wild-type cells expressing Sec28p-GFP, FM co-localized with Sec28p after a brief glucose shift, suggesting that Sec28p resides on or traffics to endosomes. To identify genes that control the trafficking of Sec28p to endosomes, we screened mutants that failed to show co-localization of Sec28p with FM following a glucose shift. We found that, Sec28p did not co-localize with FM containing endosomes in the Δvam3 mutants, suggesting that Sec28p distribution on endosomes is controlled by VAM3.
The trafficking of Sec28p was also studied using a Δubc1 mutant. Previously, we demonstrated that Vid vesicle formation is regulated by the UBC1 gene.74 As such, in the absence of this gene, Vid vesicle formation is significantly reduced. Because of this, we reasoned that the Δubc1 mutant might accumulate Sec28p at membrane sites that directly or indirectly form Vid vesicles. Sec28p and FM co-localized to vesicles at earlier time points in this mutant. However, following a longer glucose shift, both Sec28p and FM were on the vacuole membrane. The localization of Sec28p to this location indicates that the absence of the UBC1 gene does not adversely affect the anterograde movement of Sec28p to the vacuolar membrane. This suggests that UBC1 controls a step after the delivery of Sec28p to the vacuole membrane.
Coatomer proteins play an important role in the budding of COPI vesicles. During the formation of COPI vesicle, coat proteins are assembled at the budding site. Therefore, if Sec28p plays a similar role in Vid vesicles formation, then this protein should localize to areas that form these buds. Localization to the budding site may be more stable in mutants with defective coatomer subunits. Therefore, we used a ret2-1 mutant that encodes a temperature sensitive product of the 60 kD subunit of the coatomer complex. In the ret2-1 mutant, Sec28p initially co-localized with FM, indicating that Sec28p traffics to endosomes. However, at later time points, FM was on the vacuole membrane and Sec28p was in dots adjacent to the vacuole membrane. This can be explained in two ways. Sec28p is either in vesicles that are ready to fuse with the vacuole membrane, or Sec28p is in vesicles that are budding from the vacuole membrane. To address this issue, we pre-labeled the vacuole membrane with FM in the ret2-1 strain and examined the distribution of Sec28p-GFP. Following a shift to glucose, Sec28p localized to small buds on the vacuole membrane for extended periods of time. Thus, our results suggest that Sec28p vesicles can form from the vacuole membrane.
A Model for the Vid Vesicle Trafficking Pathway
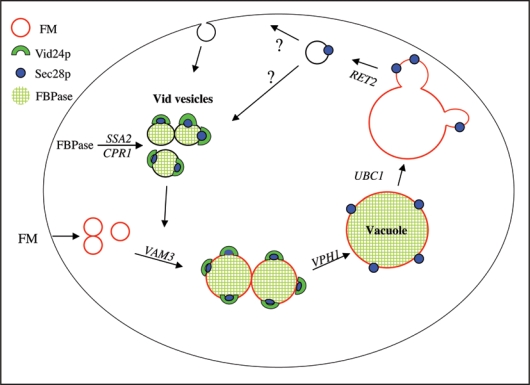
Based on our Sec28p localization studies, we proposed the following model for the Vid trafficking pathway (Fig. 1). Sec28p resides on Vid vesicles as a peripheral protein, where it interacts with other coatomer proteins. The Sec28p complex acts to recruit Vid24p to Vid vesicles. Accordingly, when coatomer genes are mutated, Vid24p association with Vid vesicles is reduced. Following the recruitment of Vid24p, the Vid vesicles converge with the endocytic pathway. This process is regulated by VAM3, since Vid vesicles failed to merge with endosomes in the absence of this gene. Sec28p then moves from endosomes to the vacuole. After fusion, Sec28p is distributed on the vacuole membrane. Sec28p then concentrates in areas of the membrane that later bud off from the vacuole membrane. Coatomer genes such as RET2 also play a required role in Vid vesicle budding. Sec28p concentrates in discrete areas in the ret2-1 mutant, and these vesicles do not bud off from the vacuole.
Figure 1.
A model for the FBPase trafficking pathway. Sec28p and other coatomer proteins associate with vesicles at an early step, and this complex recruits Vid24p to the vesicles. FBPase is imported from the cytosol into Vid vesicles via a process that requires Ssa2p and Cpr1p. Following the import of their cargo, the Vid vesicles converge with the endocytic pathway, in a VAM3 dependent manner. These endosomes then carry their cargo to the vacuole in a step that requires VPH1. Following endosomes fusion, Sec28p remains associated with the vacuole membrane, while the fate of Vid24p remains unknown. In the absence of UBC1, Sec28p is diffusely associated with the membrane. Finally, buds are formed and released from the vacuole in a RET2-dependent manner. The budded vesicles may recycle their membranes back to the plasma membrane, or alternatively, they may become Vid vesicles.
In support of our model, FBPase was observed to follow the same distribution pattern as Sec28p, most notably in mutants that block various steps of the pathway. As mentioned above, the VAM3 gene controls the merger of Vid vesicles with the endocytic pathway. In cells lacking this gene, FBPase was in Vid vesicles and failed to co-localize with FM vesicles. In the Δvph1 mutant, FBPase was found inside large FM-labeled circles. Thus, this suggests that FBPase enters these endocytic compartments as a luminal protein. Along these same lines, we have shown that FBPase is sequestered in the lumen of Vid vesicles. Therefore, this protein should also be found in the lumen of endosomes after Vid vesicles fuse with these structures. Sec28p, on the other hand, remains on the outside of the membrane as a peripheral protein. The same topology can be seen after the fusion of endosomes with vacuoles. FBPase is in the lumen, while Sec28p is on the outside of the membrane. Thus, our results suggest that Vid vesicles undergo a series of fusion events leading to the degradation of FBPase in the lumen of the vacuole.
Prospectives
Based on our studies with Sec28p, we have shown that the Vid vesicle pathway merges with the endocytic pathway. However, a number of questions remain to be answered. For example, are Vid vesicles derived from retrograde vesicles that bud from the vacuole membrane? Alternatively, do retrograde vesicles cycle to the plasma membrane and then form Vid vesicles at this location? If this is the case, are they a distinct type of early endosome? To address the latter possibility, it will be useful to utilize endocytic mutants that block the internalization process. If Sec28p or Vid24p are localized to the plasma membrane when internalization is blocked, it may suggest that Vid vesicles are derived from the plasma membrane. A number of endocytic mutants that block the plasma membrane internalization process are also defective in FBPase degradation. However, the exact mechanisms regarding why internalization is required for FBPase degradation is not clear.
The actin cytoskeleton may play an important role in Vid vesicle formation. Many endocytic mutants affect actin organization at the plasma membrane.88–93 Furthermore, actin patches are the sites of plasma membrane internalization, since these patches co-localize with receptor proteins and the FM dye. In addition, various actin-binding proteins associate with actin patches, albeit with different mobility. Some are relatively immobile, while others move rapidly toward the center of cells.93 If Vid vesicles are derived from the plasma membrane, we may see Sec28p, Vid24p or FBPase targeted to this location. In some mutants, they may be distributed evenly on the plasma membrane, while in others, they may show localized distribution. If this is the case, further experiments will be conducted to determine whether they co-localize with actin patches. If Vid vesicles are indeed derived from the plasma membrane, then the next question is how does FBPase get to the lumen of Vid vesicles. Is it possible that FBPase is translocated or exocytosed to the media and then internalized into Vid vesicles? Is it possible that Vid vesicles are formed first and FBPase is then translocated into Vid vesicles from the cytosol? If this is the case, Vid vesicles may be incompetent for import initially, but become competent at some time point.
It is interesting that the same set of gluconeogenic enzymes including FBPase, and MDH2 are degraded in the proteasome under one condition, but are degraded in the vacuole under different conditions. How do cells decide whether they should be degraded in the proteasome or the vacuole? Differential modification may account for the site of their degradation. As stated above, FBPase is ubiquitinated prior to degradation in the proteasome, while it is phosphorylated before targeting to the vacuole.56,58,59 This may be caused by the activation of particular signaling pathways under certain conditions. For the vacuole dependent pathway, FBPase phosphorylation requires the RAS2 gene and protein kinase A.54,55 When the first proline residue was mutated to serine, FBPase degradation was inhibited in both 1d and 3d cells, suggesting that this residue is important for both proteasome and vacuole degradation pathways. However, these may be other domains or sequences utilized for either the proteasome or the vacuole pathway. In future studies, it would be desirable to screen for FBPase mutants that are defective in 1d versus 3d cells. Mutations that are defective in 1d, but not 3d may be useful to study the proteasomal pathway. Likewise, mutations that show normal FBPase degradation in 1d cells, but defective degradation in 3d cells may uncover sequences required for the vacuole pathway.
It is also interesting that a subset of VID or GID genes are involved in both the proteasomal and vacuole pathways. In our hands, Vid24p is a structural protein on Vid vesicles.75 This protein is required for FBPase trafficking after FBPase is sequestered inside Vid vesicles. However, recent evidence from the Wolf group suggests that Vid24p is also part of the ubiquitin ligase complex. As such, it is involved in the ubiquitination of FBPase that occurs prior to its proteasomal dependent degradation.94 Thus, Vid24p appears to be involved in two very different cellular functions depending on whether FBPase is targeted to the proteasome or the vacuole. Given that Vid24p plays dual roles in the proteasomal versus vacuole degradation pathways, it is reasonable to assume that other VID or GID genes may code for multifunctional proteins. Thus, a major focus of our future research will be to sort out the roles that these proteins play in the proteasomal versus vacuole dependent degradation of FBPase.
Acknowledgements
This work was supported by the NIH GM 59480 and the PA Tobacco Settlement Fund to Hui-Ling Chiang.
Footnotes
Previously published online as a Communicative & Integrative Biology E-publication: http://www.landesbioscience.com/journals/cib/article/7711
References
- 1.Goldberg AL. Functions of the proteasome: from protein degradation and immune surveillance to cancer therapy. Biochem Soc Trans. 2007;35:12–17. doi: 10.1042/BST0350012. [DOI] [PubMed] [Google Scholar]
- 2.Hershko A. The ubiquitin system for protein degradation and some of its roles in the control of the cell division cycle. Cell Death Differ. 2005;12:1191–1197. doi: 10.1038/sj.cdd.4401702. [DOI] [PubMed] [Google Scholar]
- 3.Varshavsky A. Regulated protein degradation. Trends Biochem Sci. 2005;30:283–286. doi: 10.1016/j.tibs.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 4.Hanna J, Finley D. A proteasome for all occasions. FEBS Lett. 2007;581:2854–2861. doi: 10.1016/j.febslet.2007.03.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Seglen PO, Gordon PB, Holen I. Non-selective autophagy. Semin Cell Biol. 1990;1:441–448. [PubMed] [Google Scholar]
- 6.Henell F, Glaumann H. Participation of lysosomes in basal proteolysis in perfused rat liver. Discrepancybetween leupeptin-induced lysosomal enlargement and inhibition of proteolysis. Exp Cell Res. 1985;158:257–262. doi: 10.1016/0014-4827(85)90448-3. [DOI] [PubMed] [Google Scholar]
- 7.Neely AN, Cox JR, Fortney JA, Schworer CM, Mortimore GE. Alterations of lysosomal size and density during rat liver perfusion. Suppression by insulin and amino acids. J Biol Chem. 1977;252:6948–6954. [PubMed] [Google Scholar]
- 8.Mortimore GE, Neely AN, Cox JR, Guinivan RA. Proteolysis in homogenates of perfused rat liver: responses to insulin, glucagon and amino acids. Biochem Biophys Res Commun. 1973;54:89–95. doi: 10.1016/0006-291x(73)90892-9. [DOI] [PubMed] [Google Scholar]
- 9.Mortimore GE, Hutson NJ, Surmacz CA. Quantitative correlation between proteolysis and macro- and microautophagy in mouse hepatocytes during starvation and refeeding. Proc Natl Acad Sci USA. 1983;80:2179–2183. doi: 10.1073/pnas.80.8.2179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie Z, Klionsky DJ. Autophagosome formation: core machinery and adaptations. Nat Cell Biol. 2007;9:1102–1109. doi: 10.1038/ncb1007-1102. [DOI] [PubMed] [Google Scholar]
- 12.Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu Rev Nutr. 2007;7:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- 13.Cecconi F, Levine B. The role of autophagy in mammalian development: cell makeover rather than cell death. Dev Cell. 2008;15:344–357. doi: 10.1016/j.devcel.2008.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaushik S, Cuervo AM. Chaperone-mediated autophagy. Methods Mol Biol. 2008;445:227–244. doi: 10.1007/978-1-59745-157-4_15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chiang HL, Dice JF. Peptide sequences that target proteins for enhanced degradation during serum withdrawal. J Biol Chem. 1988;263:6797–6805. [PubMed] [Google Scholar]
- 16.Chiang H-L, Terlecky SR, Plant CP, Dice JF. A role for a 70 kD heat shock protein in lysosomal degradation of intracellular proteins. Science. 1989;246:382–385. doi: 10.1126/science.2799391. [DOI] [PubMed] [Google Scholar]
- 17.Terlecky SR, Chiang HL, Oslon TS, Dice JF. Protein and peptide binding and stimulation of in vitro lysosomal proteolysis by 73 kD heat shock cognate protein. J Biol Chem. 1992;267:9202–9209. [PubMed] [Google Scholar]
- 18.Cuervo AM, Dice JF. A receptor for the selective uptake and degradation of proteins by lysosomes. Science. 1996;273:501–503. doi: 10.1126/science.273.5274.501. [DOI] [PubMed] [Google Scholar]
- 19.Agarraberes FA, Terlecky SR, Dice JF. An intralysosomal hsp70 is required for a selective pathway of lysosomal protein degradation. J Cell Biol. 1997;137:825–834. doi: 10.1083/jcb.137.4.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Massey A, Kiffin R, Cuervo AM. Pathophysiology of chaperone-mediated autophagy. Int J Biochem Cell Biol. 2004;36:2420–2434. doi: 10.1016/j.biocel.2004.04.010. [DOI] [PubMed] [Google Scholar]
- 21.Cuervo AM. Autophagy: in sickness and in health. Trends Cell Biol. 2004;14:70–77. doi: 10.1016/j.tcb.2003.12.002. [DOI] [PubMed] [Google Scholar]
- 22.Dice JF. Chaperone-mediated autophagy. Autophagy. 2007;3:295–299. doi: 10.4161/auto.4144. [DOI] [PubMed] [Google Scholar]
- 23.Kiffin R, Christian C, Knecht E, Cuervo AM. Activation of chaperone-mediated autophagy during oxidative stress. Mol Biol Cell. 2004;15:4829–4840. doi: 10.1091/mbc.E04-06-0477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cuervo AM, Dice JF. Age-related decline in chaperone-mediated autophagy. J Biol Chem. 2000;275:31505–31513. doi: 10.1074/jbc.M002102200. [DOI] [PubMed] [Google Scholar]
- 25.Finn PF, Dice JF. Ketone bodies stimulate chaperone-mediated autophagy. J Biol Chem. 2005;280:25864–25870. doi: 10.1074/jbc.M502456200. [DOI] [PubMed] [Google Scholar]
- 26.Bryant NJ, Stevens TH. Vacuole biogenesis in Saccharomyces cerevisiae: protein transport pathways to the yeast vacuole. Microbiol Mol Biol Rev. 1998;62:230–247. doi: 10.1128/mmbr.62.1.230-247.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones EW. Three proteolytic systems in the yeast Saccharomyces cerevisiae. J Biol Chem. 1991;266:7963–7966. [PubMed] [Google Scholar]
- 28.Klionsky DJ, Herman PK, Emr SD. The fungal vacuole: composition, function and biogenesis. Microbiol Rev. 1990;54:266–292. doi: 10.1128/mr.54.3.266-292.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Raymond CK, Howald-Stevenson I, Vater CA, Stevens TH. Morphological classification of the yeast vacuolar protein sorting mutants: evidence for a prevacuolar compartment in class E vps mutants. Mol Biol Cell. 1992;3:1389–1402. doi: 10.1091/mbc.3.12.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson JS, Klionsky DJ, Banta LM, Emr SD. Protein sorting in Saccharomyces cerevisiae: isolation of mutants defective in the delivery and processing of multiple vacuolar hydrolases. Mol Cell Biol. 1988;8:4936–4948. doi: 10.1128/mcb.8.11.4936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rothman J, Stevens TH. Protein sorting in yeast: mutants defective in vacuolar biogenesis mislocalize vacuolar proteins into the late secretory pathway. Cell. 1986;47:1041–1051. doi: 10.1016/0092-8674(86)90819-6. [DOI] [PubMed] [Google Scholar]
- 32.Klionsky DJ. The molecular machinery of autophagy: unanswered questions. J Cell Sci. 2005;118:7–18. doi: 10.1242/jcs.01620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Huang WP, Klionsky DJ. Autophagy in yeast: a review of the molecular machinery. Cell Struct Funct. 2002;27:409–420. doi: 10.1247/csf.27.409. [DOI] [PubMed] [Google Scholar]
- 34.Reggiori F, Klionsky DJ. Autophagosomes: biogenesis from scratch? Curr Opin Cell Biol. 2005;17:415–422. doi: 10.1016/j.ceb.2005.06.007. [DOI] [PubMed] [Google Scholar]
- 35.Wang CW, Klionsky DJ. The molecular mechanism of autophagy. Mol Med. 2003;9:65–76. [PMC free article] [PubMed] [Google Scholar]
- 36.Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, et al. A ubiquitin-like system mediates protein lipidation. Nature. 2000;408:488–492. doi: 10.1038/35044114. [DOI] [PubMed] [Google Scholar]
- 37.Mizushima N, Noda T, Yoshimori T, Tanaka Y, Ishii T, George MD, et al. A protein conjugation system essential for autophagy. Nature. 1998;395:395–398. doi: 10.1038/26506. [DOI] [PubMed] [Google Scholar]
- 38.Dunn WA., Jr Studies on the mechanisms of autophagy: formation of the autophagic vacuole. J Cell Biol. 1990;110:1923–1933. doi: 10.1083/jcb.110.6.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tuttle DL, Dunn WA. Divergent modes of autophagy in the methylotrophic yeast Pichia pastoris. J Cell Sci. 1995;108:25–35. doi: 10.1242/jcs.108.1.25. [DOI] [PubMed] [Google Scholar]
- 40.Yuan W, Stromhaug PE, Dunn WA. Glucose-induced autophagy of peroxisomes in Pichia pastoris requires a unique E1-like protein. Mol Biol Cell. 1999;10:1353–1366. doi: 10.1091/mbc.10.5.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gancedo C. Inactivation of fructose-1,6-bisphosphatase by glucose in yeast. J Bacteriol. 1971;107:401–405. doi: 10.1128/jb.107.2.401-405.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gancedo JM. Yeast carbon catabolite repression. Microbiol Mol Biol. 1998;62:334–361. doi: 10.1128/mmbr.62.2.334-361.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Toyoda Y, Fuji H, Miwa I, Okuda J, Sy J. Anomeric specificity of glucose effect on cAMP, fructose-1,6-bisphosphatase and trehalase in yeast. Biochem Biophys Res Comm. 1987;43:212–217. doi: 10.1016/0006-291x(87)90652-8. [DOI] [PubMed] [Google Scholar]
- 44.Carlson M. Glucose repression in yeast. Curr Opin Microbiol. 1999;2:202–207. doi: 10.1016/S1369-5274(99)80035-6. [DOI] [PubMed] [Google Scholar]
- 45.Holzer H. Proteolytic catabolite inactivation in Saccharomyces cerevisiae. Revis Biol Cellular. 1989;21:305–319. [PubMed] [Google Scholar]
- 46.Gamo FJ, Navas MA, Blazquez MA, Gancedo C, Gancedo JM. Catabolite inactivation of heterologous fructose-1,6-bisphosphatases and fructose-1,6-bisphosphatase-beta-galactosidase fusion proteins in Saccharomyces cerevisiae. Eur J Biochem. 1994;222:879–884. doi: 10.1111/j.1432-1033.1994.tb18935.x. [DOI] [PubMed] [Google Scholar]
- 47.Horak J, Wolf DH. Two distinct proteolytic systems responsible for glucose induced degradation of fructose-1,6-bisphosphatase and the Gal2p transporter in the yeast Saccharomyces cerevisiae share the same protein components of the glucose signaling pathway. J Biol Chem. 2002;77:8248–8254. doi: 10.1074/jbc.M107255200. [DOI] [PubMed] [Google Scholar]
- 48.Riballo EM, Herwerjer D, Wolf D, Lagunas R. Catabolite inactivation of the yeast maltose transporter occurs in the vacuole after internalization by endocytosis. J Bacteriol. 1995;177:5622–5627. doi: 10.1128/jb.177.19.5622-5627.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Horak J, Wolf DH. The ubiquitin ligase SCF(Grr1) is required for Gal2p degradation in the yeast Saccharomyces cerevisiae. Biochem Biophys Res Commun. 2005;335:1185–1190. doi: 10.1016/j.bbrc.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 50.Gadura N, Michels CA. Sequences in the N-terminal cytoplasmic domain of Saccharomyces cerevisiae maltose permease are required for vacuolar degradation but not glucose-induced internalization. Curr Genet. 2006;50:101–114. doi: 10.1007/s00294-006-0080-3. [DOI] [PubMed] [Google Scholar]
- 51.Mazon MJ, Gancedo JM, Gancedo C. Inactivation of yeast fructose-1,6-bisphosphatase. In vivo phosphorylation of the enzyme. J Bio Chem. 1982;257:1128–1130. [PubMed] [Google Scholar]
- 52.Rittenhouse J, Moberly L, Marcus F. Phosphorylation in vivo of yeast (Saccharomyces cerevisiae) fructose-1,6-bisphosphatase at the cyclic AMP-dependent site. J Biol Chem. 1987;262:10114–10119. [PubMed] [Google Scholar]
- 53.Lamponi SP, Galassi P, Tortora, Guerritore A. Glucose induced degradation of yeast fructose-1,6-bisphosphatase requires additional triggering events besides protein phosphorylation. FEBS. 1987;216:265–269. doi: 10.1016/0014-5793(87)80703-2. [DOI] [PubMed] [Google Scholar]
- 54.Jiang Y, Davis C, Broach JR. Efficient transition to growth on fermentable carbon sources in Saccharomyces cerevisiae requires signaling through the Ras pathway. EMBO J. 1998;17:6942–6951. doi: 10.1093/emboj/17.23.6942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Cui DY, Brown CR, Chiang HL. The type 1 phosphatase Reg1p-Glc7p is required for the glucose-induced degradation of fructose-1,6-bisphosphatase in the vacuole. J Biol Chem. 2004;279:9713–9724. doi: 10.1074/jbc.M310793200. [DOI] [PubMed] [Google Scholar]
- 56.Hammerle M, Bauer J, Rose M, Szallies A, Thumm M, Dusterhus S, et al. Proteins of newly isolated mutants and the amino-terminal proline are essential for ubiquitin-proteasome catalyzed catabolite degradation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. J Biol Chem. 1998;273:25000–25005. doi: 10.1074/jbc.273.39.25000. [DOI] [PubMed] [Google Scholar]
- 57.Regelmann J, Schule T, Josupeit FS, Horak J, Rose M, Entian KD, et al. Catabolite degradation of fructose-1,6-bisphosphatase in the yeast Saccharomyces cerevisiae: a genome-wide screen identifies eight novel GID genes and indicates the existence of two degradation pathways. Mol Biol Cell. 2003;14:1652–1663. doi: 10.1091/mbc.E02-08-0456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Schork SM, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase of Saccharomyces cerevisiae. Degradation via the ubiquitin pathway. J Biol Chem. 1995;270:26446–26450. doi: 10.1074/jbc.270.44.26446. [DOI] [PubMed] [Google Scholar]
- 59.Schork SM, Bee G, Thumm M, Wolf DH. Catabolite inactivation of fructose-1,6-bisphosphatase in yeast is mediated by the proteasome. FEBS Lett. 1994;349:270–274. doi: 10.1016/0014-5793(94)00668-7. [DOI] [PubMed] [Google Scholar]
- 60.Chiang HL, Schekman R. Regulated import and degradation of a cytosolic protein in the yeast vacuole. Nature. 1991;350:313–318. doi: 10.1038/350313a0. [DOI] [PubMed] [Google Scholar]
- 61.Chiang HL, Schekman R, Hamamoto S. Selective uptake of cytosolic, peroxisomal and plasma membrane proteins by the yeast vacuole. J Biol Chem. 1996;271:9934–9941. doi: 10.1074/jbc.271.17.9934. [DOI] [PubMed] [Google Scholar]
- 62.Chiang H-L, Schekman R. Site of catabolite inactivation. Nature. 1994;369:284. doi: 10.1038/369283a0. [DOI] [PubMed] [Google Scholar]
- 63.Hung GC, Brown CR, Wolfe AB, Liu J, Chiang HL. Degradation of the gluconeogenic enzymes fructose-1,6-bisphosphatase and malate dehydrogenase is mediated by distinct proteolytic pathways and signaling events. J Biol Chem. 2004;279:49138–49150. doi: 10.1074/jbc.M404544200. [DOI] [PubMed] [Google Scholar]
- 64.Shieh HL, Chiang HL. In vitro reconstitution of glucose induced targeting of fructose-1,6-bisphosphatase into the vacuole in semi-intact yeast cells. J Biol Chem. 1998;273:3381–3387. doi: 10.1074/jbc.273.6.3381. [DOI] [PubMed] [Google Scholar]
- 65.Minard KI, McAlister-Henn L. Glucose-induced degradation of the MDH2 isozyme of malate dehydrogenase in yeast. J Biol Chem. 1992;267:17458–17464. [PubMed] [Google Scholar]
- 66.Minard KI, McAlister-Henn L. Glucose-induced phosphorylation of the MDH2 isozyme of malate dehydrogenase in Saccharomyces cerevisiae. Arch Biochem Biophys. 1994;315:302–309. doi: 10.1006/abbi.1994.1504. [DOI] [PubMed] [Google Scholar]
- 67.Gibson N, McAlister-Henn L. Physical and genetic interactions of cytosolic malate dehydrogenase with other gluconeogenic enzymes. J Biol Chem. 2003;278:25628–25636. doi: 10.1074/jbc.M213231200. [DOI] [PubMed] [Google Scholar]
- 68.Hoffman M, Chiang HL. Isolation of degradation-deficient mutants defective in the targeting of fructose-1,6-bisphosphatase into the vacuole for degradation in Saccharomyces cerevisiae. Genetics. 1996;143:1555–1566. doi: 10.1093/genetics/143.4.1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Brown CR, McCann JA, Hung G, Elco C, Chiang HL. Vid22p, a novel plasma membrane is required for the fructose-1,6-bisphosphatase degradation pathway. J Cell Sci. 2001;115:655–666. doi: 10.1242/jcs.115.3.655. [DOI] [PubMed] [Google Scholar]
- 70.Brown CR, Wolfe AB, Cui D, Chiang HL. The vacuolar import and degradation pathway merges with the endocytic pathway to deliver fructose-1,6-bisphosphatase to the vacuole for degradation. J Biol Chem. 2008;283:26116–26127. doi: 10.1074/jbc.M709922200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Huang PH, Chiang HL. Identification of novel vesicles in the cytosol to vacuole protein degradation pathway. J Cell Biol. 1997;136:803–810. doi: 10.1083/jcb.136.4.803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brown CR, McCann JA, Chiang HL. The heat shock protein Ssa2p is required for import of fructose 1-6-bisphosphatase into Vid vesicles. J Cell Biol. 2000;150:65–76. doi: 10.1083/jcb.150.1.65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Brown CR, Cui DY, Hung GG, Chiang HL. Cyclophilin A mediates Vid22p function in the import of fructose-1,6-bisphosphatase into Vid vesicles. J Biol Chem. 2001;276:48017–48026. doi: 10.1074/jbc.M109222200. [DOI] [PubMed] [Google Scholar]
- 74.Shieh HL, Chen Y, Brown CR, Chiang HL. Biochemical analysis of fructose-1,6-bisphosphatase import into vacuole import and degradation vesicles reveals a role for UBC1 in vesicle biogenesis. J Biol Chem. 2001;276:10398–10406. doi: 10.1074/jbc.M001767200. [DOI] [PubMed] [Google Scholar]
- 75.Chiang MC, Chiang HL. Vid24p, a novel protein localized to the fructose-1,6-bisphosphatase containing vesicles, regulates targeting of fructose-1,6-bisphosphatase from the vesicles to the vacuole for degradation. J Cell Biol. 1998;140:1347–1356. doi: 10.1083/jcb.140.6.1347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Liu JJ, Brown CR, Chiang HL. The vacuolar ATPase is required for the degradation of fructose-1,6-bisphoaphatase in the vacuole. Autophagy. 2005;3:146–156. doi: 10.4161/auto.1.3.2036. [DOI] [PubMed] [Google Scholar]
- 77.Wieland F, Harter C. Mechanisms of vesicles formation: insights from the COP system. Curr Opin Cell Biol. 1999;11:440–446. doi: 10.1016/s0955-0674(99)80063-5. [DOI] [PubMed] [Google Scholar]
- 78.Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bi-directional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- 79.McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Opin Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- 80.Duden R. ER-to-Golgi transport: COP I and COP II function. Mol Membr Biol. 2003;20:197–207. doi: 10.1080/0968768031000122548. [DOI] [PubMed] [Google Scholar]
- 81.Whitney JA, Gomez M, Sheff D, Kreis T, Mellman I. cytoplasmic coat proteins involved in endosome function. Cell. 1995;67:239–253. doi: 10.1016/0092-8674(95)90183-3. [DOI] [PubMed] [Google Scholar]
- 82.Aniento F, Gu F, Parton RG, Gruenberg J. An endosomal beta COP is involved in the pH-dependent formation of transport vesicles destined for late endosomes. J Cell Biol. 1996;133:29–41. doi: 10.1083/jcb.133.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Gu F, Aniento F, Parton RG, Gruenberg J. Functional dissection of COP-I subunits in the biogenesis of multivesicular endosomes. J Cell Biol. 1997;139:1183–1195. doi: 10.1083/jcb.139.5.1183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Piguet V, Gu F, Foti M, Demaurex N, Gruenberg J, Carpentier JL, et al. Nef-induced CD4 degradation: a diacidic-based motif in Nef functions as a lysosomal targeting signal through the binding of beta-COP in endosomes. Cell. 1999;97:63–73. doi: 10.1016/s0092-8674(00)80715-1. [DOI] [PubMed] [Google Scholar]
- 85.Daro E, Sheff D, Gomez M, Kreis T, Mellman I. Inhibition of endosome function in CHO cells bearing a temperature-sensitive defect in the coatomer (COPI) component epsilon-COP. J Cell Biol. 1997;139:1747–1759. doi: 10.1083/jcb.139.7.1747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gabriely G, Kama R, Gerst J. Involvement of specific COPI subunits in protein sorting from the late endosome to the vacuole in yeast. Mol Cell Biol. 2007;27:526–540. doi: 10.1128/MCB.00577-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Vida TA, Emr SD. A new vital stain for visualizing vacuolar membrane dynamics and endocytosis in yeast. J Cell Biol. 1995;128:790–792. doi: 10.1083/jcb.128.5.779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Toret CP, Drubin DG. The budding yeast endocytic pathway. J Cell Sci. 2006;119:4585–4587. doi: 10.1242/jcs.03251. [DOI] [PubMed] [Google Scholar]
- 89.Engqvust-Goldstein A and Drubin D. Actin assembly and endocytosis:from yeast to mammals. Annu Rev Cell Dev Biol. 2003;19:287–332. doi: 10.1146/annurev.cellbio.19.111401.093127. [DOI] [PubMed] [Google Scholar]
- 90.Dewar H, Warren DT, Gardiner FC, Gourlay CG, Satish N, Richardson MR, Andrews PD, Ayscough KR. Novel proteins linking the actin cytoskeleton to the endocytic machinery in Saccharomyces cerevisiae. Mol Biol Cell. 2002;13:3646–3661. doi: 10.1091/mbc.E02-05-0262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Warren DT, Andrews PD, Gourlay CW, Ayscough KR. Sla1p couples the yeast endocytic machinery to proteins regulating actin dynamics. J Cell Sci. 2002;115:1703–1715. doi: 10.1242/jcs.115.8.1703. [DOI] [PubMed] [Google Scholar]
- 92.Howard JP, Hutton JL, Olson JM, Payne GS. Sla1p serves as the targeting signal recognition factor for NPFX(1,2)D-mediated endocytosis. J Cell Biol. 2002;157:315–326. doi: 10.1083/jcb.200110027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Kaksonen M, Sun Y, Drubin DG. A pathway for association of receptors, adaptors, and actin during endocytic internalization. Cell. 2003;115:475–487. doi: 10.1016/s0092-8674(03)00883-3. [DOI] [PubMed] [Google Scholar]
- 94.Santt O, Pfirrmann T, Braun B, Juretschke J, Kimmig P, Scheel H, et al. The Yeast GID Complex, a novel ubiquitin ligase (E3) involved in the regulation of carbohydrate metabolism. Mol Biol Cell. 1998;19:3323–3333. doi: 10.1091/mbc.E08-03-0328. [DOI] [PMC free article] [PubMed] [Google Scholar]