Summary
The COPII vesicular coat forms on the endoplasmic reticulum from Sar1–GTP, Sec23/24 and Sec13/31 protein subunits. Here, we define the interaction between Sec23/24•Sar1 and Sec13/31, involving a forty-residue Sec31 fragment. In the crystal structure of the ternary complex, Sec31 binds as an extended polypeptide across a composite surface of the Sec23 and Sar1–GTP molecules, explaining the stepwise character of Sec23/24•Sar1 and Sec13/31 recruitment to the membrane. The Sec31 fragment stimulates GAP activity of Sec23/24, and a convergence of Sec31 and Sec23 residues at the Sar1 GTPase active site explains how GTP hydrolysis is triggered leading to COPII coat disassembly. The Sec31 active fragment is accommodated in a binding groove supported in part by Sec23 residue Phe380. Substitution of the corresponding residue F382L in human Sec23A causes cranio-lenticulo-sutural dysplasia, and we suggest that this mutation disrupts the nucleation of COPII coat proteins at endoplasmic reticulum exit sites.
Introduction
Vesicle transport pathways support the growth and maintenance of organelles. Vesicles are formed in a budding reaction involving the coordination of two coat protein complexes: an “inner shell” complex that binds to the membrane and captures cargo molecules, and an “outer shell” complex that polymerizes into a spherical cage to deform the membrane into a bud. A protein–protein link between the inner shell and the self-assembling outer shell—as observed for example between clathrin heavy chain and adaptors—is needed to couple the actions of these protein complexes, leading to coat protein clustering and cargo concentration into a membrane bud (Kirchhausen, 2000; McMahon and Mills, 2004).
COPII-coated vesicles bud from the endoplasmic reticulum (ER) to export newly synthesized proteins to the Golgi complex (Lee et al., 2004). The COPII coat forms through the sequential binding of three cytoplasmic proteins—Sar1, Sec23/24 and Sec13/31—to the ER membrane (Barlowe et al., 1994). Budding is initiated by the activation of the G protein Sar1 to its GTP-bound form, causing it to embed an N-terminal α helix in the bilayer (Antonny et al., 1997). Sar1-GTP recruits Sec23/24 to form the inner shell or “pre-budding” complex that binds directly to cargo molecules (Miller et al., 2003; Mossessova et al., 2003). Finally, the pre-budding complex recruits Sec13/31 to nucleate the polymerization of multiple Sec13/31 assembly units into an octahedral cage that constitutes the outer shell of the COPII coat (Barlowe et al., 1994; Stagg et al., 2006).
Structural analysis of Sec23/24•Sar1 reveals a bow-tie-shaped complex with a concave surface for binding the membrane vesicle; an extensive interface between Sec23 and Sar1 is stabilized by the bound GTP molecule (Bi et al., 2002). The Sec13/31 assembly unit is a heterotetramer—comprising Sec13/Sec31•Sec31/Sec13—the “architectural core” of which is organized as a linear array of α-solenoid and β-propeller domains to form a 28-nm long rod (Fath et al., 2007; Lederkremer et al., 2001; Matsuoka et al., 2001; Stagg et al., 2006). Twenty-four copies of the rod assemble to form the COPII cuboctahedron (Fath et al., 2007; Stagg et al., 2006).
The nature of the link between Sec23/24•Sar1 and Sec13/31 remains unclear. Studies of yeast and mammalian proteins suggest that an ~300-residue proline-rich region of Sec31 interacts with Sec23 (Shaywitz et al., 1997; Shugrue et al., 1999). This region is not part of the architectural core of the Sec13/31 assembly unit, and it seems not to fold into a structured domain (Fath et al., 2007). On this basis it has been suggested that the proline-rich region forms a somewhat flexible linker that projects from the outer shell cage in towards the membrane to engage Sec23/24 (Fath et al., 2007).
Defining the interactions between the inner and outer shell complexes is important not only for understanding COPII assembly, but also coat disassembly triggered by GTP hydrolysis. Rapid GTP hydrolysis on Sar1 requires Sec23/24, which is the GAP (GTPase-activating protein) for the reaction (Yoshihisa et al., 1993), and is accelerated an additional order of magnitude by Sec13/31 (Antonny et al., 2001). Thus, the GTP hydrolysis reaction on Sar1 is programmed in the COPII budding process, so as to couple coat assembly to disassembly.
In this paper, we define the active fragment of Sec31 that binds and stimulates the GAP activity of Sec23/24. We describe the 2.5 Å crystal structure of the ternary complex comprising the active fragment bound to Sec23•Sar1 stabilized with a non-hydrolyzable GTP analog. The structure reveals that Sec31 binds as an extended polypeptide across the Sec23 and Sar1 molecules, and inserts amino-acid side chains in the vicinity of the Sar1 active site to accelerate GTP hydrolysis in combination with Sec23. The F382L substitution in human Sec23A that causes cranio-lenticulo-sutural dysplasia maps close to the binding site for the Sec31 active fragment, implying that the mutation may disrupt nucleation of the COPII protein machinery. The functional consequences of the disease mutation are revealed in the accompanying manuscript by Fromme et al. (2007).
Results
Sec13/31 Stimulates the GAP Activity of Sec23/24 in Solution
The ability of Sec13/31 to accelerate GAP activity of Sec23/24 was discovered using S. cerevisiae COPII proteins bound to synthetic liposomes (Antonny et al., 2001). According to the results of that study, there are two general mechanisms by which Sec13/31 might alter the Sar1 active site to stimulate GAP activity: either a simple mechanism in which an element of Sec13/31 forms a stoichiometric contact with Sec23/24 or Sar1, or a more complex mechanism in which the polymerization of the coat induces changes at the active site—this might be triggered by inter-subunit contacts, perhaps in response to membrane curvature.
We favored the simple mechanism, which predicts that in vitro GAP stimulation by Sec13/31 should be retained in the absence of phospholipid membrane, and should require only a portion of the Sec13/31 molecule. We used the truncated, soluble form of S. cerevisiae Sar1 (residues 24–190, hereafter referred to as Sar1) in a fluorometric assay, and tested first whether GAP stimulation could be detected using full-length COPII subunits dispersed in solution. In preliminary experiments we used a substrate comprising Sar1 bound to the fluorescent GTP analog, mant-deoxyGTP (3′-O-[N-methyl-anthraniloyl]-deoxyGTP), because we were concerned that the fluorophore moiety attached to the 2′ position might interfere with the interaction between Sar1 and Sec23 residues that approach close to the ribose 2′-hydroxyl group (Bi et al., 2002). However, GTP hydrolysis experiments using Sar1 bound to mant-deoxyGTP and mant-GTP gave essentially identical results (data not shown), so we used mant-GTP in subsequent experiments (mant-GTP is a mixture of forms with the fluorescent mant moiety attached to either the 2′ or 3′ position).
Figure 1B shows a representative set of experiments in which the fluorescence output of a 1 μM solution of Sar1–mant-GTP was continuously monitored (λ = 438 nm) as COPII proteins were added. In curve (ii), the addition of Sec23/24 alone to the fluorescent Sar1 substrate caused a slow decrease in fluorescence output upon GTP hydrolysis, because mant-GDP has a decreased fluorescence at the Sar1 active site (see Ahmadian et al., 1997, for details of the mant fluorometric assay with Ras). The result mirrors earlier reports of Sec23-dependent GTP hydrolysis on Sar1 (Antonny et al., 2001; Yoshihisa et al., 1993). To confirm that the assay monitors GTP hydrolysis, we bound Sar1 to the non-hydrolyzable form of the fluorescent nucleotide, mant-GppNHp, and found that the addition of Sec23/24 caused no change in fluorescence (Figure 1B, curve i). We then tested the effect of full-length Sec13/31. The addition of 4 μM Sec13/31 to the solution containing Sec23/24 and Sar1–mant-GTP (curve iii) stimulated GTP hydrolysis beyond the rate due to Sec23/24 alone. Sec13/31 itself did not have GAP activity; rather it synergized with Sec23/24 to accelerate GTP hydrolysis on Sar1, as reported by Antonny et al. (2001).
Figure 1. Sec13/31 Stimulates the GAP Activity of Sec23/24 in Solution.
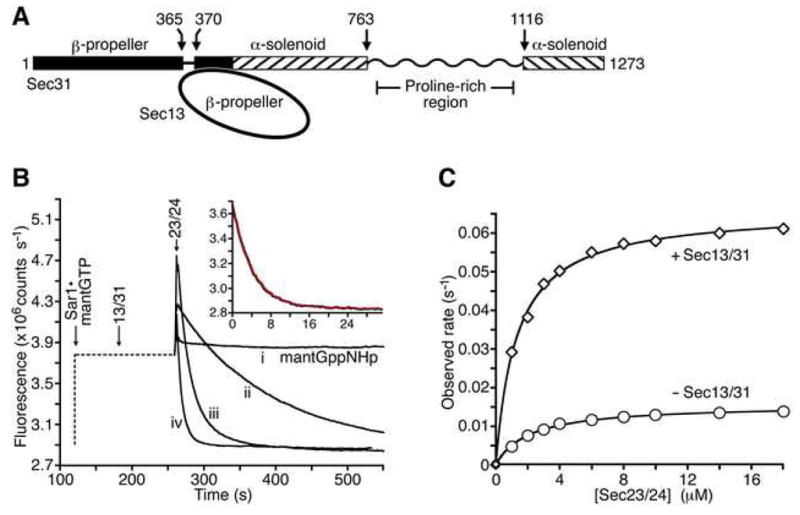
(A) Schematic diagram shows the domain structure of S. cerevisiae Sec13/31 defined previously (Dokudovskaya et al., 2006; Fath et al., 2007). The proline-rich region is drawn as a wavy line to convey that this polypeptide sequence seems not to contain a discrete domain, according to limited proteolysis experiments.
(B) The fluorometric GTPase assay. The graph shows time courses for four representative reactions. The flourescence output of a solution of 1 μM mant-GTP or mant-GppNHp was monitored upon addition of varying concentrations of Sec13/31, at a fixed concentration (3 μM) of Sec23/24. The left-hand portion of the curves, drawn as a dotted line, indicates the order of addition and incubation time of the various COPII components. Note the initial rapid increase in fluorescence intensity upon formation of the Sec23/24•Sar1-mantGTP complex. Curve (i) is a control experiment using mant-GppNHp. Curve (ii) shows GTP hydrolysis by Sec23/24 in the absence of Sec13/31. Curve (iii) shows the additional rate acceleration caused by 4 μM full-length Sec13/31. Curve (iv) shows rapid GTP hydrolysis by Sec23/24 plus 10 μM Sec31 active fragment (residues 899–947). The inset graph shows a least-squares fit to a first order exponential (red line), using data (black line) from an experiment containing Sec23/24 and 40μM Sec31 active fragment.
(C) Experiment to assess the ability of full-length Sec13/31 to stimulate the GAP activity of Sec23/24 in solution. GTP hydrolysis rates were measured across a range of concentrations of Sec23/24 in the absence (circles) or presence (diamonds) of 3 μM Sec13/31. Sar1–mantGTP was present at 1 μM in all experiments. The graph shows fits of the data to a simple hyperbolic equation. In the presence of Sec13/31, the maximal GTPase rate is 0.066 sec−1 and the apparent Km for Sec23/24 binding to Sar1 is 1.3 μM. In the absence of Sec13/31, the maximal rate is 0.015 sec−1 and the apparent Km is 2.1 μM.
Sec13/31 may stimulate GAP activity by increasing the intrinsic GTPase reaction rate of the Sec23/24•Sar1 complex or by increasing the affinity of Sec23/24 for Sar1. To delineate these two catalytic contributions, we measured GTP hydrolysis rates across a range of concentrations of Sec23/24, in the presence or absence of 3 μM Sec13/31 (Figure 1C). Reaction rates were obtained by fitting the time course data to a simple exponential function. In all the experiments there is a rapid increase in fluorescence upon formation of the Sec23/24•Sar1 complex (much like the fluorescent transient observed when neurofibromin binds to H-Ras–mant-GTP [Ahmadian et al., 1997]); the subsequent fluorescence decrease, which monitors GTP hydrolysis, fits well to the exponential function (inset in Figure 1B). The results of these preliminary experiments show that the presence of full-length Sec13/31 increases the GTPase rate roughly 4-fold, and increases the affinity between Sec23/24 and Sar1 roughly 2-fold (Figure 1C).
Thus, GAP stimulation by Sec13/31 does not depend on the accretion of COPII coat subunits on a phospholipid membrane. Stimulation can be measured in solution using the mant fluorometric assay and soluble coat subunits, and this provided a straightforward system to dissect the active fragment of Sec13/31.
A 50-Residue Fragment of Sec31 Binds to Sec23/24•Sar1 and Stimulates GAP Activity
The interaction between Sec13/31 and Sec23 maps to the proline-rich region of the Sec31 polypeptide (Figure 1A shows the domain structure of Sec31). Specifically, the S. cerevisiae Sec31 fragment comprising residues 850–1175 was shown to interact with full-length Sec23 in a two-hybrid analysis (Shaywitz et al., 1997). On this basis we tested whether the proline-rich region could stimulate GAP activity in the same manner as full-length Sec13/31. The proline-rich region has 20% proline content (between residues 770–1110) and seems not to contain a stably folded domain according to limited proteolysis experiments (Fath et al., 2007). Consistent with this, the majority of Sec31 fragments that we prepared were heavily degraded during expression in E. coli as fusions to glutathione S-transferase. We were able to express and purify a fragment encompassing most of the proline-rich region, residues 879–1114 (construct A in Figure 2B). Figure 2A shows the results of the experiment comparing the activities of this fragment and full-length Sec13/31, across a range of concentrations. The maximal GTPase rate for both proteins was 0.1 sec−1; the apparent Km for the proline-rich region binding to Sec23/24•Sar1 was 6.0 μM, just slightly weaker than the value of 4.8 μM measured for full-length Sec13/31. Thus, the proline-rich region retains the binding and catalytic capacity of full-length Sec13/31. This effect is exclusive to the proline-rich region, as other domains of Sec31 had no effect on GAP activity (data not shown).
Figure 2. Dissection of Sec13/31 and Identification of the Sec31 Active Fragment.

(A) The proline-rich region of Sec31 contains all of the binding and catalytic capacity of full-length Sec13/31. The graph shows the results of GTP hydrolysis experiments across a range of concentrations of full-length Sec13/31 (circles) or Sec31 proline-rich region, residues 879–1114 (diamonds). Sar1–mantGTP was present at 1 μM and Sec23/24 at 3 μM in all experiments. For full-length Sec13/31, the maximal GTPase rate is 0.10 sec−1, and the apparent Km for Sec13/31 binding to Sec23/24•Sar1 is 4.8 μM. For the Sec31 proline-rich region, the maximal GTPase rate is also 0.10 sec−1, and the apparent Km for the 236-residue region binding to Sec23/24•Sar1 is 6.0 μM.
(B) Diagram shows the polypeptide regions of Sec31 that were prepared in order to dissect the Sec31 active fragment.
(C) Bar graph shows the results of the Sec31 dissection experiment. The Sec31 polypeptide fragments A–K, as defined in (B), were tested for their ability to stimulate the GAP activity of Sec23/24. In all experiments the Sec31 polypeptide was present at 1.5 μM, Sar1–mantGTP at 1 μM and Sec23/24 at 3 μM. GAP stimulatory activity is expressed as a percentage of the activity of the active fragment (construct K, residues 899–947).
(D) Sec24 has no effect on the binding of Sec31 to the Sec23•Sar1 complex. GTP hydrolysis rates were measured across a range of concentrations of full-length Sec13/31 or Sec31 active fragment, in the presence of 3 μM Sec23/24 or 3 μM Sec23. For full-length Sec13/31 in the presence of Sec23/24 (circles), the maximal GTPase rate is 0.10 sec−1, and the apparent Km for Sec13/31 binding to Sec23/24•Sar1 is 4.8 μM. For full-length Sec13/31 in the presence of Sec23 (diamonds), the maximal GTPase rate is 0.10 sec−1, and the apparent Km for Sec13/31 binding to Sec23/24•Sar1 is 5.0 μM. For the Sec31 active fragment in the presence of Sec23/24 (crosses), the maximal GTPase rate is 0.31 sec−1, and the apparent Km for Sec13/31 binding to Sec23/24•Sar1 is 12.8 μM. Finally, for the Sec31 active fragment in the presence of Sec23 (triangles), the maximal GTPase rate is 0.32 sec−1, and the apparent Km for Sec13/31 binding to Sec23/24•Sar1 is 17.0 μM.
Next, we tested a series of Sec31 truncation constructs for the ability to stimulate GAP activity (Figure 2B and C). All the Sec31 protein fragments were added at 1.5 μM to an assay mix containing 1 μM Sar1–mant-GTP and 3 μM Sec23/24. Under these conditions, full stimulatory activity by Sec31 yielded an ~5-fold rate enhancement relative to the absence of Sec31 (compare, for example, the rate due to construct A with the control rate in Figure 2C). The results of the experiment show that an ~50-residue polypeptide, residues 899–947, retains full activity (construct K in Figure 2B and C). We refer to this as the active fragment of Sec31. Removal of additional residues from the N or C termini causes a reduction in activity (fragments D, E and J). However, there is not the dramatic loss of activity that one would predict if the terminal residues of a folded domain were removed. Consistent with this, the crystallographic analysis reveals that the Sec31 active fragment does not fold into a compact structure to interact with Sec23•Sar1, but binds as an extended polypeptide across the surface of the inner-shell complex (see below).
Surprisingly, when we tested the Sec31 active fragment across a range of concentrations, we found that it was more effective than full-length Sec13/31 at stimulating the GAP activity of Sec23/24. As shown in Figure 2D, the maximal rate of GTP hydrolysis measured with the active fragment was three times faster than for full-length Sec13/31 (0.3 sec−1 compared to 0.1 sec−1). Yet the active fragment lost a corresponding three-fold affinity for Sec23/24•Sar1 relative to the full-length protein. This subtle difference in stimulatory activity is perhaps not surprising given the very different forms of the two molecules—a 5 kDa peptide versus a ~340 kDa heterotetramer. Regardless, the data establish that GAP stimulatory activity is due to a circumscribed peptide element located in the middle of the Sec31 proline-rich region (Figure 2B).
Finally, the most rapid GTPase rate that we observed in this study—0.24 sec−1 measured using 40 μM active fragment—was thirty times faster than the rate due to Sec23/24 alone (Figure 2D). This ~30-fold stimulation of Sec23/24 GAP activity by the Sec31 active fragment compares to the 10-fold stimulation reported by Antonny et al. (2001) using liposomes and membrane-bound coat proteins.
Sec31 Does Not Contact Sec24 to Accelerate GTP Hydrolysis
The small size of the Sec31 active fragment suggests that it does not contact all three subunits of the Sec23/24•Sar1 complex. On the other hand, two-hybrid analysis identified two interactions involving the Sec31 proline-rich region—one with Sec23 and the other with a fragment of Sec24 (Shaywitz et al., 1997). We tested the contribution of Sec24 to the Sec31-stimulated GAP reaction (Figure 2D). First, we used the Sec31 active fragment, and found that Sec23 was almost as effective as Sec23/24 at catalyzing GTP hydrolysis on Sar1 (crosses and triangles in Figure 2D). There is a very slight loss of affinity of Sec31 for the inner shell proteins that may reflect experimental error (apparent Km of 17 μM for Sec23•Sar1 versus 12.8 μM for Sec23/24•Sar1). Next, we tested full-length Sec13/31 and found that Sec23 was as effective as Sec23/24 in the GAP reaction; Sec24 had no effect on the catalytic rate or the affinity of Sec13/31 for the inner-shell proteins (circles and diamonds in Figure 2D).
These results suggest that Sec24 is not involved in the contacts between the inner and outer shells. It is conceivable that an interaction between Sec13/31 and Sec24 occurs but is somehow not reported by our solution-based GTPase assay, though we consider this unlikely. The evidence for an interaction between Sec31 and Sec24 was obtained from a two-hybrid experiment employing a C-terminal region of Sec24, residues 666–926 (Shaywitz et al., 1997). According to the Sec24 crystal structure (Bi et al., 2002), the 666–926 fragment will have portions of the hydrophobic core of its helical domain exposed to solvent, and may not be a suitable interactor for two-hybrid analysis.
In summary, these data define a COPII core complex that stimulates rapid GTP hydrolysis, comprising Sec23•Sar1 and the active fragment of Sec31. We assembled this complex from purified S. cerevisae proteins and obtained crystals that diffract X-rays to 2.5 Å resolution (Table 1).
Table 1.
Data Collection and Refinement Statistics
| Space group | P21212 | |
| Cell dimensions (Å) | a = 94.7, b = 133.2, c = 82.0 | |
| PDB accession # | 2QTV | |
|
| ||
| Data processing | ||
| Overall | Outer shell | |
| Resolution (Å) | 30 – 2.50 | 2.59 – 2.50 |
| Measured reflections | 134,252 | 12,454 |
| Unique reflections | 35,291 | 3,417 |
| Completeness (%) | 96.2 | 95.0 |
| I/σ | 24.8 | 4.4 |
| Rmerge (%)¶ | 7.8 | 34.9 |
|
| ||
| Refinement statistics | ||
| Data range (Å) | 30 – 2.5 | |
| Reflections | 33,452 | |
| Non-hydrogen atoms | 7,388 | |
| Water molecules | 231 | |
| R.m.s. Δbonds (Å) § | 0.0065 | |
| R.m.s. Δangles (deg) § | 1.3 | |
| Mean B-factor (all atoms) | 35.9 | |
| Mean B-factor (Sec31 atoms) | 51.1 | |
| R-factor (%)† | 20.5 | |
| Rfree (%)†‡ | 26.2 | |
Rmerge = 100 × Σh Σi | Ih,i − <Ih> | / Σh Σi Ih,i.
Root-mean-squared deviation (R.m.s. Δ) from target geometry.
R-factor = 100 × Σ|FP − FP(calc) | / ΣFP; R values calculated for data with a 2σ cutoff.
Rfree was calculated with 5% of the data.
Crystal Structure Determination of Sec23•Sar1 Bound to the Active Fragment of Sec31
The crystals of the ternary complex of Sec23•Sar1 and the Sec31 active fragment were grown using the truncated form of Sar1 (residues 24–190) and were stabilized using the non-hydrolyzable GTP analog, GppNHp. The same strategy was used previously to crystallize the Sec23•Sar1 complex (Bi et al., 2002). Thus, the binary and ternary crystal structures can be compared straightforwardly to assess structural differences induced by Sec31.
The crystal structure of the ternary complex was solved by the molecular replacement method using yeast Sec23 and Sar1 as search models. The Sec31 active fragment was built into strong electron density for residues 907–942 of the peptide, and the structure was refined using data to 2.5 Å resolution (Figure 3 and Table 1).
Figure 3. Crystal Structure of Sec23•Sar1 Complexed with the Active Fragment of Sec31.

The ribbon representation is shown with the membrane-distal surface of the complex facing forward. Sec23 is orange and Sar1 is red. The Sec31 active fragment is in five colors: the N-terminal element that interacts solely with Sar1 (purple, residues 907–920); a short element that interacts with both Sec23 and Sar1 residues at the interface (white, residues 920–922); two elements that interact with Sec23 (blue, residues 923–927; green, residues 935–942); and the intervening stretch that interacts loosely with Sec23 (yellow, residues 928–934). The blue contour lines show difference electron density calculated prior to the inclusion of the Sec31 active fragment (at 2.5 Å resolution, contoured at 2.9 σ). Domains of the Sec23 protein are labeled, and the interface with Sec24 is indicated at the bottom of the picture. The switch 2 (labeled Sw2) and helix α3 elements of Sar1 to which Sec31 binds are indicated.
Discussion
Architecture of Sec23•Sar1 Bound to the Active Fragment of Sec31
The overall conformations of Sec23 and Sar1 observed in the crystal structure of the ternary complex (Figures 3 and 4) are essentially the same as in the Sec23•Sar1 binary complex (Bi et al., 2002), with only subtle alterations observed in the vicinity of the Sar1 active site. Sar1 is stabilized in the GTP conformation by the non-hydrolyzable analog, such that its β2–β3 switching element has shifted to eliminate the binding site for the Sar1 N-terminal “membrane anchor” (Bi et al., 2002).
Figure 4. Layered Appearance of COPII Coat Proteins in a Model of Sec23/24•Sar1 Bound to Sec31.

The ribbon representation on the left is a side view of Sec23/24•Sar1 complexed with the Sec31 active fragment. Sec23 is orange, Sar1 is red, GppNHp is blue, the Sec31 fragment is blue, and Sec24 is green. This is a composite model that includes Sec24 taken from a Sec23/24 crystal structure determined previously (Bi et al., 2002). The grey line indicates the curvature of membrane vesicle, and the dotted red line suggests the attachment of Sar1 to membrane via its N-terminal sequence. The view on the right is rotated 90° to show the membrane-distal surface in space-filling representation (same orientation as in Figure 3).
Thirty-six residues of Sec31, residues 907–942, are included in the molecular model, as no electron density is observed for the eight N-terminal and five C-terminal positions. This is consistent with the results of the biochemical dissection (see Figure 2B and C), and suggests that as few as 35–40 residues of Sec31 are required for GAP stimulation.
The Sec31 active fragment binds as an extended polypeptide across a composite surface of the Sec23 and Sar1 molecules. It does not adopt a folded tertiary structure; moreover, it has hardly any secondary structure other than a single turn of α helix, residues 915–919, that binds to the switch 2 element of Sar1 (Figure 3). The active fragment is oriented with its N terminus bound to Sar1 and C terminus bound to Sec23. A 16-residue segment (907–922) that includes the turn of α helix, extends ~15 Å across the Sar1 surface, interacting with switch 2 and the adjacent α3 helix (a common binding site for GAPs on G proteins [Vetter and Wittinghofer, 2001]). The 20 C-terminal residues 923–942 bind in a highly extended conformation, 47 Å long, across Sec23, and contact the gelsolin, trunk and β-barrel domains of the Sec23 molecule ((Bi et al., 2002)).
The observation that the Sec31 active fragment binds to a composite surface of Sec23 and Sar1 provides a molecular explanation for the ordered recruitment of the Sec23/24 and Sec13/31 complexes to ER membranes. Only upon GTP-dependent formation of Sec23/24•Sar1 is the receptor site for Sec13/31 formed on the membrane (Barlowe et al., 1994; Matsuoka et al., 1998). A structural corollary of this is the layered arrangement of the membrane and proteins in the coat, as highlighted in Figure 4. Thus, the Sec31 active fragment binds to the membrane-distal surface of Sec23•Sar1. All of its residues reside a uniform distance from the membrane surface, which we estimate to be ~45 Å (this assumes that the membrane-proximal surface of Sec23/24•Sar1 closely apposes the phospholipid membrane).
The sequence composition of the active fragment (Figure 5A) strongly suggests that the isolated peptide will adopt an unstructured conformation in solution. Only residue Trp922 of the ~40 amino acids is conserved as a large hydrophobic side chain. Moreover, a limited proteolysis analysis of Sec13/31 did not identify a stably-folded domain in this region or in the proline-rich region of Sec31 (residues 770–1110) as a whole. Whether the Sec31 active fragment is thoroughly unstructured in the context of full-length Sec13/31 is unclear. The 3-fold differences in catalytic rate and affinity on Sec23/24•Sar1 that we reported above may hint at regulatory conformational constraints imposed on the active fragment by adjacent domains of Sec13/31, but the development of this speculative idea must await further experimental work.
Figure 5. Sec31 Interactions at the Sec23•Sar1 Interface and the GTPase Active Site.
(A) Alignment of eight sequences of the Sec31 active fragment from seven species (including human forms A and B). The key at the top indicates the conservation of the predominant residue; two black bars corresponds to two common occurrences out of eight—the “noise level”—and red bars highlight the more highly conserved positions. Key tryptophan and asparagines residues are indicated with stars.
(B) Schematic drawing showing contacts at the protein–protein interfaces, colored as in Figures 3 and 4. Select contacts that are of structural interest are indicated with black lines. The contact between the arginine finger residue of Sec23—Arg722 (labeled R722)—and GTP phosphate groups is indicated with an arrow. The negative charge of the side chain of Sec31 residue D924 interacts with the electrostatic dipole of helix αI on Sec23. Residue Phe380 on helix αI is mutated to leucine in human Sec23A in individuals with cranio-lenticulo-sutural dysplasia (Boyadjiev et al., 2006).
(C) Bar graph shows the results of the mutagenesis experiment. The Sec31 fragment (residues 899–947) and mutants thereof were tested for the ability to stimulate the GAP activity of Sec23/24. In all experiments the Sec31 polypeptide was present at 1.5 μM, Sar1–mantGTP at 1 μM and Sec23/24 at 3 μM. (These conditions are the same as in the dissection experiment shown in Figure 2C). GAP stimulatory activity is expressed as a percentage of the activity of the active fragment.
(D) Closeup view showing Sec23 contacts at the Sar1 active site. This picture is generated from a previously determined crystal structure of Sec23 bound to Sar1 and GppNHp. Note the orientation of the arginine finger residue R722 and its bonds to the nucleotide.
(E) Closeup view of the ternary complex is oriented as in (C). The difference electron density map was calculated prior to the inclusion of the Sec31 active fragment (at 2.5 Å resolution, contoured at 2.9 σ). Note that the arginine finger residue R722 of Sec23 and a substrate water molecule in this ternary complex are in the same position as in the Sec23•Sar1 binary complex in (C). Note also the positions of key side chains W922, N923 and D924 of the Sec31 active fragment. The position of residue Phe380 on helix αI is also indicated.
Interfacial Contacts
In Figure 3, the Sec31 active fragment is colored to delineate five segments. The N-terminal residues, 907–919 (colored purple) interact solely with Sar1. These residues are not conserved in Sec31 sequences (Figure 5A), but we observe close interactions between this region and the switch 2 and α3 elements of Sar1. Next, residue Asp920 and the highly conserved residues Gly921 and Trp922 of the active fragment (colored white in Figure 3) contact both Sec23 and Sar1 to form a tripartite protein interface near the Sar1 active site (Figure 5D). The peptide geometry around residue Gly921 guides the insertion of the Trp922 side chain close to the seat of reaction (Figure 5D), and these interactions are key to the stimulation of GAP activity, as described in more detail below.
The remainder of the active fragment, residues 923–942, interacts solely with Sec23. A central stretch, residues 928–934 (yellow in Figure 3), appears to interact loosely, whereas the regions on either side form more intimate interactions with Sec23. Residues 923–927 (blue in Figure 3) are well conserved among Sec31 sequences (Figure 5A), and form a series of interactions with Sec23 that appear to be important for affinity and for buttressing Trp922 at the Sar1 active site (Fig 5B and D). Finally, the C-terminal stretch comprising residues 935–942 (green in Figure 3) is not well conserved, but several side chains—in particular Ala936, Val939 and Val941—form intimate contacts with residues of the trunk and β-barrel domains of Sec23 (Figure 5B).
In summary, the binding site for the Sec31 active fragment extends ~60 Å across the surface of Sec23 and Sar1, and involves three quasi-independent binding regions (colored purple, white/blue and green in Figure 3). Most residue positions in the Sec31 active fragment are not well conserved, in particular the N-terminal sequence that interacts with Sar1 and the C-terminal portion that interacts with Sec23. Nevertheless, these two terminal interaction regions are important for binding and stimulation of GAP activity, according to the results of the dissection experiment (Figure 2C). Sequence conservation is restricted to a central set of ~6 residues of the active fragment that is clustered around the tripartite interface near to the Sar1 active site.
Sec31 Residues Complete the Sar1 Active Site for Rapid GTP Hydrolysis
Sar1 has a very slow intrinsic rate of GTP hydrolysis because, like other Ras proteins, it lacks key catalytic residues (Bi et al., 2002; Vetter and Wittinghofer, 2001). The mechanism by which Sec23 acts as a GAP to accelerate the reaction involves the insertion of an arginine side chain, Arg722, into the active site to form bonds to the phosphates via its guanidinum group (Bi et al., 2002) (Figure 5C). This type of mechanism, involving an “arginine-finger” residue that neutralizes negative charge in the GTPase transition state is likewise a common feature of Ras proteins (Vetter and Wittinghofer, 2001).
The crystal structure of the Sec23•Sar1 binary complex revealed two catalytic features in addition to the identity of the arginine-finger residue. Firstly, an extensive interface between Sec23 and the switch 1 and 2 elements of Sar1 stabilizes these regions close to the active site. Secondly, a water molecule bridges the imidazole side chain of His77 and the γ-phosphate group of GppNHp, and is suitably located for nucleophilic attack (Bi et al., 2002, and Figure 5C). Taken together, the catalytic features of the Sec23•Sar1 active site are very similar to those seen in other GAP•G-protein complexes, so it is not immediately obvious from the Sec23•Sar1 crystal structure how Sec31 might stimulate GAP activity (discussed in Bi et al., 2002).
Inspection of the Sar1 active site and the tripartite interface in the ternary complex now reveals how Sec31 stimulates GAP activity of Sec23 (Figure 5D). The active fragment inserts two residues, Trp922 and Asn923, close to the active site, with the plane of the indole ring of Trp922 oriented almost parallel with the imidazole ring of His77 (Figure 5D). We propose that this interaction optimizes the geometry of the key histidine side chain for bonding to the nucleophilic water molecule (His77 is equivalent to Gln61 of H-Ras). The imidazole ring of His77 has rotated ~15° upon interaction with Trp922, to align with the plane of the tryptophan indole ring (the χ2 torsion angle of His 77 is −60° in the binary complex and −75° in the ternary complex). Other changes at the active site induced by Sec31 binding appear to support the role of Trp922. In particular, Sec23 residue Gln720, which is oriented away from the active site in the binary complex, is turned toward the active site and forms hydrogen bonds to residues Trp922 and Asn923 of the active fragment.
Thus, Sec31 side chains do not have a chemical catalytic role at the Sar1 active site like the arginine finger residue of Sec23—indeed, only residue Pro926 is invariant in the subset of sequences shown in Figure 5A. Rather, GAP stimulation is likely caused by Trp922 of Sec31 interacting with His77 to orient its imidazole ring at the Sar1 active site. In so doing, Trp922 and Asn923 plug a solvent-filled cavity that extends from bulk solvent to the vicinity of the active site, an unfavorable arrangement for catalysis in the binary complex that leaves one surface of the indole ring of His77 exposed to solvent (Figure 5C).
Structure-based mutagenesis of the Sec31 peptide confirmed the importance of these key residues. Mutation of residue Trp922 and Asn923 to alanine caused complete loss of GAP stimulatory activity (Figure 5C). Likewise, changes to Leu925 and residue Val939 in the C-terminal region were highly disruptive, even though these are not highly conserved positions (Figure 5A). Two very conservative residue changes in the N-terminal portion of the active fragment, Q910A and N915A, were tolerated with only modest loss of stimulatory activity; the asparagine residue does not in fact contact Sec23 or Sar1 directly.
In summary, the active-site configuration of the ternary complex explains how Sec31 synergizes with Sec23 to accelerate GTP hydrolysis. Sec31 cannot act alone on Sar1 because it only binds to the Sec23•Sar1 complex. In this way, the sequential binding reaction confers a two-gear mechanism for GTP hydrolysis on Sar1 (Antonny et al., 2001), whereby hydrolysis is initiated upon Sec23/24 binding and is accelerated further upon recruitment of Sec13/31. Thus GTP hydrolysis is programmed into the COPII system upon assembly—the slow rate of reaction on Sec23/24•Sar1 may provide the pre-budding complex the opportunity to gather cargo and SNARE molecules prior to Sec13/31 binding. Upon Sec13/31 recruitment, the rapid rate of hydrolysis—which we estimate could be as high as 0.5 sec−1—may compromise the stable attachment of ternary complexes to the membrane. But in the late stages of budding the dependence on Sar1–GTP for stabilization of COPII proteins on the ER probably diminishes as Sec13/31 polymerizes and as Sec23/24 collects cargo to provide additional bonds linking coat proteins laterally and to the membrane (discussed in Antonny et al., 2001).
The F382L Disease Mutation in Human Sec23A Maps Close to the Interface of Sec23 and Sec31
A substitution in human Sec23A, F382L, causes cranio-lenticulo-sutural dysplasia, a craniofacial and skeletal dysmorphic syndrome (Boyadjiev et al., 2006). Fibroblasts homozygous for the mutation have a disorganized endoplasmic reticulum. Molecular analysis of F382L Sec23A reveals that although it can combine with Sec24 and is recruited efficiently to membranes by Sar1, the mutant protein is deficient in vesicle formation. Moreover, Sec13/31 is mislocalized to the cytoplasm in the mutant cells, suggesting that the mutation affects the interaction of Sec23/24•Sar1 with Sec13/31 (Boyadjiev et al., 2006, and see accompanying manuscript by Fromme et al., 2007)
The corresponding phenylalanine residue in S. cerevisiae Sec23, Phe380, is located on helix αI of the trunk domain, which makes key interactions with the Sec31 active fragment (Figure 5). The phenylalanine residue is highly conserved (Boyadjiev et al., 2006), and its side chain resides on the internal face of helix αI, contributing to the hydrophobic core via contacts to adjacent hydrophobic side chains of Sec23—including Phe346, Phe375 and Tyr384. Although Phe380 does not contact Sec31 directly, residues 924–931 of the active fragment make contacts along the length of helix αI, the most notable of which is the electrostatic interaction between the side chain of Asp924 and the positive charge of the N terminus of the α helix (Figure 5D). As noted above, this set of residues of the active fragment—in particular 923–927—are important for buttressing Trp922 at the Sar1 active site. The disease mutation is a subtle change to leucine, but we predict that this will perturb the local structure of helix αI so as to weaken contacts to the active fragment. We infer that the Sec23 disease mutation impairs the recruitment and nucleation of Sec13/31 at sites of COPII budding.
Connection of Inner and Outer Shell Complexes in the COPII Coat
Figure 6 presents a model for the arrangement of Sec23/24 complexes in the COPII coat, and is based on recent structural data on the 60 nm cuboctahedron cage built from 24 assembly units (Fath et al., 2007; Stagg et al., 2006). Since each assembly unit is a Sec13/Sec31•Sec31/Sec13 heterotetramer, there are 48 copies of Sec31 per cage, and binding sites for 48 copies of Sec23/24•Sar1. Put differently, the cage has twelve vertices, each vertex formed from the convergence of four Sec13/31 rods, so there are binding sites for four Sec23/24•Sar1 molecules to nestle on the membrane underneath each vertex.
Figure 6. A Model for the Interactions and Organization of Proteins in the COPII Coat.
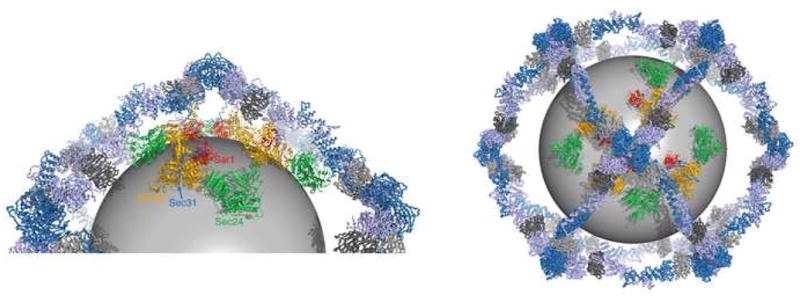
The picture shows a model of the COPII cage, a cuboctahedron built from 24 copies of the Sec13/Sec31-Sec31/Sec13 assembly unit (Fath et al., 2007; Stagg et al., 2006)). Each of the twelve vertices of the cage is formed by the convergence of four Sec13/31 subunits. Thus, there are binding sites for four copies of Sec23/24•Sar1 under each vertex. In the picture we have modeled four copies of Sec23/24•Sar1 complexed with the Sec31 active fragment. The zigzag lines represent the ~130-residue proline-rich segment (residues 764–898) that connects the α-solenoid to the active fragment of Sec31 (see Figures 1A and 2B). Since this linker is likely to be flexible, we positioned the four Sec23/24•Sar1 complexes such that they do not conform precisely to the 2-fold symmetry of the vertex. The sphere of diameter 38 nm represents the membrane vesicle. The closeup view on the left has a vertex at top. The view on the right is along the vertex 2-fold axis. Sec23 is orange, Sec24 is green, Sar1 is red, Sec31 is blue and lilac, and Sec13 is dark grey and light grey.
With respect to the present study, there are three salient features of the model. Firstly, the membrane vesicle is drawn as a 40 nm sphere inside the cage, to allow an ~5 nm space for Sec23/24•Sar1 (see Barlowe et al., 1994, and Fath et al., 2007). Secondly, the polypeptide linker (zigzag line) that connects the Sec31 active fragment to the upstream α-solenoid domain is a 130-residue portion of the proline-rich region, and is very likely unstructured. Thirdly, the C-terminus of the Sec31 α-solenoid domain, from where the linker projects down toward the membrane, is roughly equidistant (~70 Å) from the vertex dyad and the center of the assembly unit.
The flexible connection between the cage and Sec23/24•Sar1, via the 130-residue linker, suggests that Sec23/24•Sar1 complexes may be somewhat mobile on the membrane, such that the 432 symmetry of the cage is not imposed strictly on the inner shell proteins. Nevertheless, the mobility of Sec23/24•Sar1 is probably restricted by its dense packing on the membrane surface—we previously estimated that 48 copies of Sec23/24•Sar1 would cover as much as 80% of the surface area of a 40 nm membrane vesicle (Fath et al., 2007).
We have depicted this situation as four Sec23/24•Sar1 complexes under a vertex but not conforming closely to the cage symmetry (figure 6). The complexes are drawn nestled close to the vertex center, and paired in an approximate fashion with the Sec31 α-solenoid domain to which each is attached, rather than in a jumbled arrangement. But we predict that this order arises as much from the dense surface packing of Sec23/24•Sar1 as it does from the arrangement and symmetry of the outer shell assembly units.
A flexible connection between the inner and outer shells of the vesicular coat is highly reminiscent of the situation in clathrin cages, where the N-terminal β-propeller domain of the clathrin heavy chain interacts with adaptors via short peptide elements that are nested in flexible regions of polypeptide (ter Haar et al., 2000). Thus, a flexible connection between the architectural outer shell and the cargo-gathering inner shell complex may be a common feature of vesicle coat organization that facilitates the packaging of a range of cargo molecules of different shapes and sizes.
Experimental Procedures
Protein Preparation
For protein expression in insect cells, baculoviruses (Bac-to-Bac, Gibco) were prepared encoding yeast Sec13 and His6–Sec31. These proteins, as well as yeast His6–Sec23 and Sec23/His6–Sec24 complex, were expressed in insect cells and purified as described (Bi et al., 2002). Histidine tags were removed by cleavage with TEV protease. The soluble, truncated form of yeast Sar1, comprising residues 24–190, was complexed with the non-hydrolyzable GTP analog guanosine-5′-[β, γ-imido]-triphosphate (GppNHp) and Mg2+, and purified as described (Bi et al., 2002). Fragments of Sec31 were subcloned according to the domain analysis of (Fath et al., 2007), and were overexpressed in E. coli as C-terminal fusions to glutathione S-transferase using the pETGEXCT vector (Sharrocks, 1994). Proteins were purified by glutathione sepharose chromatography. Mutagenesis was performed using the Quickchange reagents (Stratagene).
Crystallization and Structure Determination
For crystallization, Sec23 was mixed with Sar1•GppNHp•Mg2+ (truncated Sar1) and the active fragment of Sec31 (residues 899–947); each protein was at a final concentration of 360 μM in 150 mM NaCl, 20 mM HEPES (pH 7.4) and 4 mM DTT. Crystals (space group P21212) were grown at 22°C by the hanging-drop method by mixing 1 μl of protein solution with 1 μl of a well solution comprising 15% PEG-1500 and 100 mM HEPES (pH 7.0). Crystals, which appeared after three days, were transferred to a solution containing 20% PEG-1500, 16% glycerol, and 100 mM HEPES (pH 6.8), and flash-frozen in liquid propane. Crystals treated in this manner diffracted X-rays to at least 2.5 Å resolution using a synchrotron X-ray source.
X-ray diffraction data to 2.5 Å resolution were measured at beamline X-25 of the National Synchrotron Light Source (NSLS). Data were processed with programs DENZO and SCALEPACK (Otwinowski and Minor, 1997). The structures were solved by molecular replacement with the program AMORE (CCP4, 1994) using yeast Sec23 as the search model (Bi et al., 2002). The initial model was improved by rigid-body and positional refinement with program CNS (Brunger et al., 1998). Sar1•GppNHp•Mg2+ was then placed into clear electron density. Finally, residues 907–942 of Sec31 were modeled into electron density, and the structure was refined to an R-factor of 20.5% (Rfree = 26.2%). The X-ray data and refinement statistics are summarized in Table 1. The model contains three residues of Sec23 that are outliers in the Ramachandran plot: Phe659 seems to be modeled correct; residues Val30 and Gln683 are located in regions of weak electron density. None of these residues is in the vicinity of the interface with Sar1 or Sec31. All Sec31 residues lie within allowed regions of the Ramachandran plot. The final model comprises 7,354 protein atoms, one molecule of GppNHp•Mg2+, one zinc atom (in the zinc-finger domain of Sec23) and 231 water molecules. No electron density is observed for the eight N terminal and five C terminal residues of the Sec31 polypeptide.
GTPase Assays
For GTPase assays, we prepared the substrate Sar1(24–190) bound to the fluorescent nucleotide derivatives mant-GTP, mant-dGTP or mant-GppNHp (mant nucleotides were purchased from Jena Bioscience). Sar1 was incubated with a 30-fold molar excess of mant nucleotide overnight at 4°C, and excess nucleotide was separated from protein by gel filtration on a superdex 75 column. The extent of mant nucleotide incorporation was estimated by absorbance measurements at 280 and 355 nm.
The decrease in fluorescence, due to hydrolysis of mant-GTP on Sar1, was monitored in a fluorimeter (Fluoromax-2, Horiba Jobin-Yvon). All reactions were performed at 25°C in 150 mM NaCl, 25 mM HEPES (pH 7.4), 2 mM DTT and 2 mM MgCl2 using an excitation wavelength of 360 nm and emission measured at 438 nm. In a typical experiment, the reaction was initiated by the addition of Sec23 (or Sec23/24) to a filtered, degassed solution containing Sar1•mant-nucleotide plus Sec31; the total reaction volume was 1.2 ml. Reaction rates were determined from exponential fits to the data.
Acknowledgments
We thank Anand Saxena for assistance at synchrotron beamline X25 at NSLS. This work was supported by grants from the National Institutes of Health and the Howard Hughes Medical Institute.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmadian MR, Hoffman U, Goody RS, Wittinghofer A. Individual rate constants for the interaction of Ras proteins with GTPase-activating proteins determined by fluorescence spectroscopy. Biochemistry. 1997;36:4535–4541. doi: 10.1021/bi962556y. [DOI] [PubMed] [Google Scholar]
- Antonny B, Beraud-Dufour S, Chardin P, Chabre M. N-terminal hydrophobic residues of the G-protein ADP-ribosylation factor-1 insert into membrane phospholipids upon GDP to GTP exchange. Biochemistry. 1997;36:4675–4684. doi: 10.1021/bi962252b. [DOI] [PubMed] [Google Scholar]
- Antonny B, Madden D, Hamamoto S, Orci L, Schekman R. Dynamics of the COPII coat with GTP and stable analogues. Nat Cell Biol. 2001;3:531–537. doi: 10.1038/35078500. [DOI] [PubMed] [Google Scholar]
- Barlowe C, Orci L, Yeung T, Hosobuchi M, Hamamoto S, Salama N, Rexach MF, Ravazzola M, Amherdt M, Schekman R. COPII: a membrane coat formed by Sec proteins that drive vesicle budding from the endoplasmic reticulum. Cell. 1994;77:895–907. doi: 10.1016/0092-8674(94)90138-4. [DOI] [PubMed] [Google Scholar]
- Bi X, Corpina RA, Goldberg J. Structure of the Sec23/24-Sar1 pre-budding complex of the COPII vesicle coat. Nature. 2002;419:271–277. doi: 10.1038/nature01040. [DOI] [PubMed] [Google Scholar]
- Boyadjiev SA, Fromme JC, Ben J, Chong SS, Nauta C, Hur DJ, Zhang G, Hamamoto S, Schekman R, Ravazzola M, et al. Cranio-lenticulo-sutural dysplasia is caused by a SEC23A mutation leading to abnormal endoplasmi-reticulum-to-Golgi trafficking. Nat Genet. 2006;38:1192–1197. doi: 10.1038/ng1876. [DOI] [PubMed] [Google Scholar]
- Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: A new software suite for macromolecular structure determination. Acta Crystallogr D Biol Crystallogr. 1998;54:905–21. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- CCP4. The CCP4 suite: programs for X-ray crystallography. Acta Crystallogr D. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- Dokudovskaya S, Williams R, Devos D, Sali A, Chait BT, Rout MP. Protease accessibility laddering: A proteomic tool for probing protein structure. Structure. 2006;14:653–660. doi: 10.1016/j.str.2006.02.006. [DOI] [PubMed] [Google Scholar]
- Fath S, Mancias JD, Bi X, Goldberg J. Structure and organization of coat proteins in the COPII cage. Cell. 2007;129:1325–1336. doi: 10.1016/j.cell.2007.05.036. [DOI] [PubMed] [Google Scholar]
- Fromme JC, Ravazzola M, Hamamoto S, Al-Balwi M, Eyaid W, Boyadjiev SA, Cosson P, Schekman R, Orci L. The genetic basis of a craniofacial disease provides insight into COPII coat assembly, Dev. Cell. 2007;X:YYY–ZZZ. doi: 10.1016/j.devcel.2007.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirchhausen T. Three ways to make a vesicle. Nat Rev Mol Cell Biol. 2000;1:187–98. doi: 10.1038/35043117. [DOI] [PubMed] [Google Scholar]
- Lederkremer GZ, Cheng Y, Petre BM, Vogan E, Springer S, Schekman R, Walz T, Kirchhausen T. Structure of the Sec23p/24p and Sec13p/31p complexes of COPII. Proc Natl Acad Sci U S A. 2001;98:10704–9. doi: 10.1073/pnas.191359398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee MC, Miller EA, Goldberg J, Orci L, Schekman R. Bidirectional protein transport between the ER and Golgi. Annu Rev Cell Dev Biol. 2004;20:87–123. doi: 10.1146/annurev.cellbio.20.010403.105307. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Orci L, Amherdt M, Bednarek SY, Hamamoto S, Schekman R, Yeung T. COPII-coated vesicle formation reconstituted with purified coat proteins and chemically defined liposomes. Cell. 1998;93:263–75. doi: 10.1016/s0092-8674(00)81577-9. [DOI] [PubMed] [Google Scholar]
- Matsuoka K, Schekman R, Orci L, Heuser JE. Surface structure of the COPII-coated vesicle. Proc Natl Acad Sci U S A. 2001;98:13705–9. doi: 10.1073/pnas.241522198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McMahon HT, Mills IG. COP and clathrin-coated vesicle budding: different pathways, common approaches. Curr Op Cell Biol. 2004;16:379–391. doi: 10.1016/j.ceb.2004.06.009. [DOI] [PubMed] [Google Scholar]
- Miller EA, Beilharz TH, Malkus PN, Lee MC, Hamamoto S, Orci L, Schekman R. Multiple cargo binding sites on the COPII subunit Se24p ensure capture of diverse membrane proteins into transport vesicles. Cell. 2003;114:497–509. doi: 10.1016/s0092-8674(03)00609-3. [DOI] [PubMed] [Google Scholar]
- Mossessova E, Bickford LC, Goldberg J. SNARE selectivity of the COPII coat. Cell. 2003;114:395–397. doi: 10.1016/s0092-8674(03)00608-1. [DOI] [PubMed] [Google Scholar]
- Otwinowski W, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- Sharrocks AD. A T7 expression vector for producing N- and C-terminal fusion proteins with glutathione S-transferase. Gene. 1994;138:105–108. doi: 10.1016/0378-1119(94)90789-7. [DOI] [PubMed] [Google Scholar]
- Shaywitz DA, Espenshade PJ, Gimeno RE, Kaiser CA. COPII subunit interactions in the assembly of the vesicle coat. J Biol Chem. 1997;272:25413–6. doi: 10.1074/jbc.272.41.25413. [DOI] [PubMed] [Google Scholar]
- Shugrue CA, Kolen ER, Peters H, Czernik A, Kaiser C, Matovcik L, Gorelick F. Identification of the putative mammalian orthologue of Sec31P, a component of the COPII coat. J Cell Sci. 1999;112:4546–4556. doi: 10.1242/jcs.112.24.4547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stagg SM, Gürkan C, Fowler DM, LaPointe P, Foss TR, Potter CS, Carragher B, Balch WE. Structure of the Sec13/31 COPII coat cage. Nature. 2006;439:234–238. doi: 10.1038/nature04339. [DOI] [PubMed] [Google Scholar]
- ter Haar E, Harrison SC, Kirchhausen T. Peptide-in-groove interactions link target protein to the beta-propeller of clathrin. Proc Natl Acad Sci U S A. 2000;97:1096–1100. doi: 10.1073/pnas.97.3.1096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vetter IR, Wittinghofer A. The guanine nucleotide-binding switch in three dimensions. Science. 2001;294:1299–304. doi: 10.1126/science.1062023. [DOI] [PubMed] [Google Scholar]
- Yoshihisa T, Barlowe C, Schekman R. Requirement for a GTPase-activating protein in vesicle budding from the endoplasmic reticulum. Science. 1993;259:1466–8. doi: 10.1126/science.8451644. [DOI] [PubMed] [Google Scholar]



