Abstract
Despite having provided the first example of a prokaryal glycoprotein, little is known of the rules governing the N-glycosylation process in Archaea. As in Eukarya and Bacteria, archaeal N-glycosylation takes place at the Asn residues of Asn-X-Ser/Thr sequons. Since not all sequons are utilized, it is clear that other factors, including the context in which a sequon exists, affect glycosylation efficiency. As yet, the contribution to N-glycosylation made by sequon-bordering residues and other related factors in Archaea remains unaddressed. In the following, the surroundings of Asn residues confirmed by experiment as modified were analyzed in an attempt to define sequence rules and requirements for archaeal N-glycosylation.
Keywords: Archaea, N-glycosylation, oligosaccharide transferase, post-translational modification
Introduction
The covalent attachment of polysaccharides represents one of the most prevalent post-translational modifications proteins experience. In N-glycosylation, glycan moieties are linked to Asn residues found as part of NXS or NXT motifs referred to as sequons, where X is any residue apart from proline, although rare exceptions to these rules exist (Gavel and von Heijne 1990, Kaji et al. 2003). Although essential for N-glycosylation, the presence of a sequon is not sufficient for modification, given the large number of unmodified sequons in both glycosylated and non-glycosylated proteins (Ben-Dor et al. 2004, Petrescu et al. 2004).
To delineate the rules determining whether modification of a given sequon occurs, researchers have considered numerous and varied properties of the target protein. These include the amino acid composition and secondary structure of the sequon and its surroundings, the number of sequons in the protein, their position(s) relative to the protein termini or to other sequons in cases where multiple potential N-glycosylation sites exist. Accordingly, studies on eukaryal glycoproteins have pointed to the importance of the character of the amino acids occupying several positions both upstream and downstream of Asn residues experimentally verified to be glycan-bearing. Based on consideration of the residues neighoring those eukaryal glycoprotein sequons experimentally shown to be modified, Ben-Dor et al. (2004) proposed a series of rules designed to predict whether a given sequon is modified. Similarly, a survey of some 500 nonredundant glycoproteins listed in the PDB database revealed an increased occurrence of aromatic residues at the +2 position following sequon Ser/Thr residues and that N-glycosylation often transpires at sites where changes in secondary structure occur (Petrescu et al. 2004). Other studies reported that the presence of a Pro residue immediately following a sequon most often hinders glycosylation (Mellquist et al. 1998). In addition, distinctions can be made between the rules governing the glycosylation of NXS and NXT sequons. For example, glycosylation occurs less frequently at NXS than at NXT sequons (Bause and Legler 1981, Kasturi et al. 1995), Furthermore, NXS glycosylation efficiency is affected by the character of the residues at the X position and at the +1 positions following the sequon serine (Shakin-Eshleman et al. 1996, Mellquist et al. 1998).
Once thought to be exclusively restricted to Eukarya, it is now clear that prokaryotes are also capable of N-glycosylation (Lechner and Wieland 1989, Szymanski and Wren 2005). In addressing N-glycosylated proteins in Campylobacter jejuni, the only bacterium for which a full N-glycosylation pathway has been described (Szymanski et al. 1999), it was observed that modification occurs at NXS/T sequons, where X is any residue apart from proline, as in Eukarya (Nita-Lazar et al. 2005). Although essential for bacterial N-glycosylation, the presence of a sequon is not sufficient for Asn modification. In a recent detailed analysis of C. jejuni glycoproteins (Kowarik et al. 2006), it was noted that only those Asn residues found as part of a D/EYNXS/T motif, where X and Y are not proline, experience glycosylation. By contrast, glycosylation of a given sequon in yeast did not require a negatively charged reside at the –2 position. Moreover, earlier analysis of eukaryal N-glycosylation sites revealed that Asp and Glu are disfavored at this position (Ben-Dor et al. 2004). Such differences in the sequence requirements of eukaryal and bacterial N-glycosylation likely reflect differences between eukaryal and bacterial oligosaccharide transferases (OST), responsible for transferring the lipid-linked glycan moieties to sequons in the target protein (Kelleher and Gilmore 2006, Weerapana and Imperiali 2006). In Eukarya, OST comprises a multisubunit complex at the heart of which lies the Stt3 protein in yeast and its homologues in other species. In Bacteria, the Stt3 homologue PglB is thought to be solely responsible for OST activity (Wacker et al. 2002).
Although prokaryal N-glycosylation was first described in Archaea (Mescher and Strominger 1976, Lechner and Sumper 1987), still little is known of either the extent of such modification or the rules governing the process in this form of life. Indeed, while the ability to glycosylate proteins has been demonstrated in numerous species by a variety of experimental methods, the modification of glycoprotein Asn residues has been directly demonstrated in only a limited number of cases (Eichler and Adams 2005).
In this study, archaeal glycoprotein sequences were considered in an attempt to unmask any putative contribution to Asn glycosylation made by residues found in the positions bordering modified sequons. Defining such N-glycosylation sequence requirements in Archaea carries significant implications for understanding how the enzymes involved in the process (in particular the archaeal Stt3 homologue) operate. In addition, selected archaeal genomes were analyzed in an effort to predict the extent of N-glycosylation in Archaea because such information could provide insight into the relationship between post-translational modification and the ability of archaeal polypeptides to remain properly folded in the face of extreme physical conditions that would normally lead to protein denaturation, loss of solubility and aggregation.
Materials and methods
Predicted N-glycosylated archaeal secretory proteins
A database of proteins secreted by ten archaeal species (Aeropyrum pernix, Archaeoglobus fulgidus, Halobacterium sp. NRC-1, Methanococcus jannaschii, Methanothermobacter thermoautotrophicus, Pyrococcus abyssi, Pyrococcus horikoshii, Sulfolobus solfataricus, Thermoplasma acidophilum and Thermoplasma volcanium) was created as described previously (Bardy et al. 2003). Briefly, predicted protein-encoding genes from each organism were analysed using SignalP v.2.0 (www.cbs.dtu.dk/services/SignalP-2.0), designed to detect the presence of signal peptides. In these analyses, protein sequences were examined using the hidden Markov model with truncation set to 70 amino acids, trained on the different data sets available (Eukarya, Gram-positive or Gram-negative Bacteria). Proteins predicted as containing signal peptides by at least one data set were then subcellularly localized by Psort (Nakai and Horton 1999; www.psort.nibb.ac.jp). Only those proteins considered to be secreted in Gram-positive organisms or localized to the periplasm or outer membrane of Gram-negative organisms (listed in Bardy et al. 2003) were scanned for the presence of sequons.
Results and discussion
Sequence profile of regions bordering modified sequons in archaeal glycoproteins
Amino acid residues near sequons could influence N-glycosylation by modulating access to the oligosaccharide transferase active site or the affinity of such interactions, by affecting sequon conformation or by providing sequon residues with partners for hydrogen bonding or other associations that could interfere with N-glycosylation. Toward determining whether any of these potential scenarios are applicable to archaeal N-glycosylation, the ten amino acids both proceeding and following archaeal sequons experimentally verified as glycan-bearing (Table 1) were considered, as were the amino acids surrounding unmodified and proposed, but as yet uncharacterized, sequons in the same proteins. At each amino acid position, the relative frequency of a representative of a particular amino acid group was noted. In this study, amino acids were first clustered based on their being non-polar (AGVLIPFMWC), uncharged and polar (QNSTY), positively charged (RKH) or negatively charged (DE). Where warranted, the amino acid clusterings adopted by Ben-Dor et al. (2004), designed to further divide residues along chemical property lines, were also considered. In these instances, the groups of the first clustering set were MLIV, TSC, FYW, AG, KR, DE, QN, P and H, whereas the groups comprising the second clustering set were DE, KRH, QNST, AMLIVFWY, C, G and P. In the latter sets of clusterings, the nonpolar and the uncharged, polar amino acids were further subdivided according to their chemical properties (e.g., hydrophobic, aromatic, small in size).
Table 1.
Neighboring residues of experimentally verified N-glycosylated sequons in Archaea. References: 1 = Paul et al. 1986; 2 = Lechner and Sumper 1987; 3 = Sumper et al. 1990; 4 = Mengele and Sumper 1992; 5 = Brockl et al. 1991; 6. Voisin et al. 2005; 7 = Zahringer et al. 2000; and 8 = Lin and Tang 1990. Abbreviation: SLG = S-layer glycoprotein. N-glycosylated sequons are shown in bold.
| Species | Protein | No. of N- glycosylation sites1 | Sequence surrounding verified N-glycosylated sequons | Ref. |
| H. salinarum | SLGa | 12 | VAFTGGAAAANASDLNDYQRFNE | 1 |
| LGIIAKSDAVNSSGGVKDNIDTS | ||||
| SISDLEDEVGNYTSGPPTGDQIR | 2 | |||
| H. volcanii | SLG | 7 | GNLDADSESFNKTIQSGDRVFLG | 3 |
| IGDTSEVGIANSSATNTSGSSTG | 4 | |||
| EVGIANSSATNTSGSSTGPTVET | ||||
| PVVETGTVDINGTASGANSVLVI | ||||
| M. fervidus | SLG | 20 | IAGAAEVKNLNTSKTFTLIQEAI | 5 |
| M. voltae | FlaA | 2 | GKGDMAVITINCTNLDLAPRTRL | 6 |
| FlaB1 | 5 | VAAVAASVLINTSGFLQQKASTT | 6 | |
| GLQVTGVTGVNSTDSENITHIVA | ||||
| VTGVNSTDSENITHIVAYITPKA | ||||
| YNGVSAVLKANESAIDGTSGTPD | ||||
| PDVLSATLLSNTSATEYQVVSLQ | ||||
| FlaB2 | 6 | VAAVAASVLINTSGFLQQKASTT | 6 | |
| LQISQVMGMHNNSNINKTAIYIS | ||||
| VMGMHNNSNINKTAIYISPNAGS | ||||
| DGSNKRVYKYNESSYKDLTNGGD | ||||
| NKGDLVAITLNTTSFSTTPRTSI | ||||
| ISFTTPATYLNDSKVVQLQ | ||||
| FlaB3 | 2 | RKMSLLYDSSNASNGGKVRLSTA | 6 | |
| NGGKVRLSTANGTSDIFKYDDVY | ||||
| SLG | 2 | DDANLKSLLTNGTNDFTELDAGK | 6 | |
| S. acidocaldarius | CbsA | 16 | TPGTTYSLERNYTNYVSIINGKE | 7 |
| PNNTEITVMSNGTILLWHSPRPV | ||||
| Thermopsin | 11 | PAGIASFGLYNYSGNVTPYVITT | 8 | |
| ASFGLYNYSGNVTPYVITTNEML | ||||
1 Both putative and verified N-glycosylation sites.
A search of the literature (June, 2006) uncovered 27 experimentally verified archaeal sequons distributed between ten proteins from five species, including halophiles (Halobacterium salinarum, Haloferax volcanii), methanogens (Methanothermus fervidus, Methanococcus voltae) and a thermoacidophile (Sulfolobus acidocaldarius). The evidence for Asn glycosylation in these cases came either from amino acid sequencing (Lechner and Sumper 1987, Lin and Tang 1990, Brockl et al. 1991, Mengele and Sumper 1992) or from mass spectrometry approaches (Paul et al. 1986, Zahringer et al. 2000, Voisin et al. 2005). Examination of the sequons themselves revealed that, as in Eukarya and Bacteria (Gavel and von Heijne 1990, Nita-Lazar et al. 2005), the X position immediately downstream of every glycosylated Asn residue was never a Pro. In one third of the sequons, however, Ser or Thr were found at the X position. By contrast, the same residues account for only 17% of the amino acid content of these glycoproteins, apart from the sequon and bordering 10 residues. When the residues bordering glycosylated sequons were considered, several general traits were observed. In contrast to what has been reported for C. jejuni, where the sequon motif was recently expanded to include a necessary Asp or Glu at the –2 position upstream of the sequon Asn residue (Kowarik et al. 2006), negatively charged residues were detected only at the –2 position in the archaeal sequences in one of the 27 examples considered. By contrast, nonpolar residues were found in this position 48% of the time, whereas uncharged polar residues were detected in 37% of the cases. The remaining examples presented positively charged residues at this position. At the +1 position following the sequon, either Ala or Gly were detected in 37% of the instances, although the same residues account for only 18% of the amino acid content outside the sequon and bordering residues, thus suggesting a preference for a small amino acid at the +1 site. The presence of Pro at the +1 position did not prevent glycosylation, as demonstrated by thermopsin peptide 2, although it should be noted that this is the only example where Pro is detected at this position. Further from the sequon, possible amino acid preferences were noted at the –4 position, where charged residues were detected only once but Ser and Thr were detected 41% of the time, and at the –8, –9 and –10 positions, where nonpolar residues were noted in 81%, 56% and 67% of the sequences, respectively. Specifically, Ala and Gly were observed at position –8 in 48% of the cases and at position –9 in 33% of the cases. Again, since Ser and Thr together and Ala and Gly together account for 17% and 18% of the amino acid content outside the sequon and its up- and downstream flanking 10 residues, respectively, the character of the –4, –8, –9 and –10 positions appears to be an enrichment in a particular type of residue.
Insight into archaeal N-glycosylation can be derived not only from analysis of the environment of modified sequons but also by addressing the surroundings of sequons shown to contain unmodified Asn residues. In the set of sequences examined in the present study, only Asn-17 of the H. salinarum S-layer glycoprotein was verified as unmodified despite being part of an NXT sequon (Lechner and Sumper 1987). In contrast to the 27 glycosylated archaeal sequons, where aromatic residues were never observed at the +1 position, the position immediately after 17NYT19 is occupied by a Tyr residue. Given the apparent preference of the glycosylation machinery for smaller residues at this site (see above), it is thus possible that the presence of a bulky Tyr at this site hinders glycan attachment. When the amino acids proceeding (NDYQRFNENT) and following (YSTASEDGKT) this H. salinarum S-layer glycoprotein sequon were considered, no marked differences from the corresponding residues surrounding modified sequons were evident. Similarly, no obvious patterns were apparent when the amino acid composition of the upstream and downstream regions surrounding the remaining 56 proposed, but as yet unverified, sequons in the same proteins was considered (Table A1). Nonetheless, no Pro residues were detected at the X position in any of these sequons, whereas at the +1 position, only nine sequences presented an aromatic residue and just one sequon offered a Pro at this site. Thus, it is likely that many of the additional sequons are glycosylated.
In addition to addressing questions of amino acid sequence, examining the distibution of modified sequons may also provide details on the N-glycosylation process in Archaea. Such information could offer insight into the ability of the archaeal oligosaccharide transferase to process sequons lying in close proximity to each other, thereby reflecting spatial considerations of the enzyme’s activity. In the present study, such analysis revealed that, despite their proximity, both Asn residues within the 308NSSATNTS315 section of the H. volcanii S-layer glycoprotein are modified (Mengele and Sumper 1992). Similary, Asn-72 and Asn-77 of M. voltae FlaB2, again separated by four residues, are both modified (Voisin et al. 2005). In Eukarya, the closest glycosylated pair of naturally occurring Asn residues, found in haptoglobin-1, are three residues apart (Kurosky et al. 1980, Gavel and von Heijne 1990). At present, no information on the acceptable proximity of Asn for dual glycosylation tolerated by the C. jejuni N-glycosylation machinery is available.
In terms of the distribution of modified sequons within the glycoprotein, only the M. voltae flagellins were considered, since 14 of the 15 putative N-glycosylation sites have been characterized (Voisin et al. 2005). In the four proteins considered, i.e., FlaA, FlaB1, FlaB2 and FlaB3, N-glycosylation sites were distributed throughout the proteins and showed no preference for any particular secondary structural region.
The environments of occupied archaeal NXS and NXT sequons are distinct
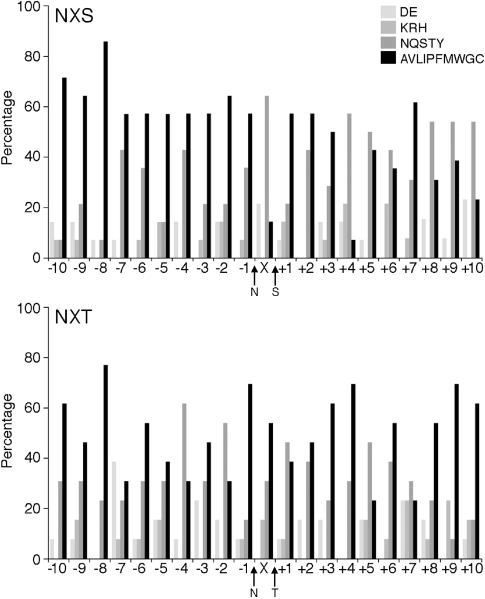
Several studies have pointed to the differential processing of NXS and NXT sequons in eukaryal glycoproteins (Kasturi et al. 1995, Shakin-Eshleman et al. 1996, Mellquist et al. 1998, Ben-Dor et al. 2004). When the two classes of sequons found in the archaeal glycoprotein sequences considered in the present study were examined, it appeared that the residues surrounding experimentally verified NXS sequons (n = 14) revealed traits distinct from the residues surrounding experimentally verified NXT sequons (n = 13). As reflected in Figure 1, the two sequon classes could be distinguished at the X position, with Ser or Thr being detected in 50% of the NXS sequons, but in only 15% of the NXT sequons. By contrast, the X position of NXT sequons corresponded to Gly in 31% of the sequences, but in only 14% of the NXS sequons. At the –2 position, Ser or Thr were detected in 46% of the NXT sequons, but in only 7% of the NXS sequons. At the +1 position, NXS sequons incorporated Gly at the X position 36% of the time, whereas Gly was not detected in the same position of the NXT sequons. Distinctions between NXS and NXT were also noted at positions further upstream or downstream of the sequon. Acidic residues were observed in 39% of the –7 positions of those sequences including the NXS sequon, but were absent from the same position in NXT-containing sequences. At the +8, +9 and +10 positions, NXS-containing sequences presented Ser or Thr 46% of the time, whereas the same residues were only employed at these positions in 8% of the NXT-containing sequences. On the other hand, the hydrophobic residues Ile, Leu, Met and Val were observed in positions +9 and +10 in 46% and 39% of those sequences including NXT, respectively, but only 15% and 8% of the time at the same positions in NXS-containing sequences.
Figure 1.
Analysis of the characteristics of amino acids surrounding modified Asn residues in archaeal glycoproteins. The relative proportions of acidic (DE), basic (KRH), polar uncharged (NQSTY) and polar (AVLIPFMWGC) residues were considered at 10 positions up- and downstream of modified archaeal NXS (upper panel, based on 14 sequences) and NXT (lower panel, based on 13 sequences) sequons. In each case, the character of the residue at the X position was also considered. In each panel, the positions of the modified Asn as well as the sequon Ser (upper panel) or Thr (lower panel) are indicated.
Putative N-glycosylation of predicted secretory proteins in Archaea
To get a general sense of the extent of N-glycosylation in Archaea, a previously assembled database of predicted secretory proteins from 10 archaeal genomes (Bardy et al. 2003) was scanned for the presence of NXS and NXT sequons (where X was not a proline), assuming these to be potential N-glycosylation sites (Table A2). Such analysis predicted major differences in the levels of putative N-glycosylated secreted proteins expressed by the various genomes examined (Table 2). Whereas only 39% of the 121 proteins supposedly secreted by A. pernix are predicted to be N-glycosylated, 30 of the 32 predicted M. jannaschii secretory proteins (i.e., 94%) contain NXS or NXT sequons. The other genomes considered are predicted to N-glycosylate 50–88% of their putatively secreted proteins. Moreover, not only do the different species potentially N-glycosylate their predicted secretomes to differing extents, but they can also often be distinguished by the unbalanced presence of either the NXS or NXT motif. In A. pernix, a total of 37 proteins contain either the NXS or NXT sequon alone yet only 10 proteins contain both. In Halobacterium sp. NRC-1, 11 proteins contain the NXS sequon, 13 contain the NXT sequon, whereas 10 proteins contain both. By contrast, the NXS and NXT sequons are found in only three and five M. jannaschii proteins, respectively, whereas 22 polypeptides present both motifs. In M. thermoautotrophicus, most of the 45 predicted N-glycosylated secretory proteins contain comparable amounts of the NXS and NXT sequons. This number includes, however, 13 proteins containing the NXT motif alone and only two sequences that include just the NXS motif. In A. pernix, the NXS motif is found in 23 proteins, whereas only 14 proteins present the NXT sequence.
Table 2.
Tally of predicted N-glycosylated secretory proteins in selected archaeal species
| Species | No. of predicted secretory proteins | No. of predicted N-glycosylated secretory protein1 | No. of sequons per predicted secreted glycoprotein | ||||||
| NXS + XST | NXS alone | NXT alone | Total | 1 | 2–4 | 5–6 | > 6 | ||
| A. pernix | 121 | 10 | 23 | 14 | 47 | 26 | 18 | 3 | 0 |
| A. fulgidus | 79 | 28 | 9 | 13 | 50 | 17 | 25 | 3 | 5 |
| Halobacterium sp. NRC-1 | 68 | 10 | 11 | 13 | 34 | 19 | 12 | 3 | 0 |
| M. jannaschii | 32 | 22 | 3 | 5 | 30 | 2 | 13 | 4 | 11 |
| M. thermoautrophicum | 68 | 30 | 2 | 13 | 45 | 7 | 17 | 9 | 12 |
| P. abyssi | 28 | 13 | 4 | 7 | 24 | 5 | 13 | 3 | 3 |
| P. horikoshii | 63 | 21 | 10 | 11 | 42 | 12 | 18 | 4 | 8 |
| S. solfataricus | 46 | 25 | 6 | 5 | 36 | 3 | 18 | 4 | 11 |
| T. acidophilum | 26 | 17 | 5 | 1 | 23 | 3 | 5 | 5 | 10 |
| T. volcanium | 24 | 12 | 2 | 3 | 17 | 5 | 5 | 0 | 7 |
| Total | 555 | 188 | 75 | 85 | 348 | 99 | 144 | 38 | 67 |
1 The motifs NPS (40 instances) and NPT (20 instances) were omitted from the tally.
The putative glycoprotein populations in the different species also differed in the number of proposed N-glycosylation sites per protein. In A. fulgidus, 42 proteins contained 1–4 putative N-glycosylation sites, whereas only eight proteins contained five or more such sites. In Halobacterium sp. NRC-1, 31 proteins are predicted to contain 1–4 N-glycosylation motifs, but only three proteins are thought to contain five or six such sites. No proteins are predicted to contain more than six sequons in this species. Likewise, A. pernix is thought to include 44 predicted glycoproteins that contain four or less possible N-glycosylation motifs, but only three proteins presenting more. By contrast, 43% of the putative secreted proteins that may be N-glycosylated in S. solfataricus contain more than six sequons. In M. jannaschii, only two proposed secetory proteins contain single N-glycosylation sites, whereas 28 sequences present multiple sequons for possible modification.
Comparing sequences bordering modified archaeal, bacterial and eukaryal sequons
Analysis of experimentally confirmed glycan-modified sequons and their surrounding residues in archaeal glycoprotein, albeit limited in number, reveals that although the archaeal sequences share much in common with their eukaryal and bacterial counterparts, differences from the rules and tendencies governing eukaryal and bacterial sequon glycosylation also exist.
Based on available data, the most obvious difference between sequon glycosylation in Archaea and Bacteria (as exemplified by C. jejuni) is the neccessity for an acidic residue in the –2 position in the latter but not the former. Analysis of the putative N-glycosylation sites of the proposed secretory protein populations of 10 archaeal genomes failed to detect an enhanced presence of Asp or Glu at this position. Although further characterization of N-glycosylated proteins from other Bacteria will confirm whether such a requirement for negatively charged residues at the –2 position is unique to C. jejuni, the independence of archaeal sequon modification from this rule points to the archaeal Stt3, believed to be responsible for the transfer of a lipid-linked glycan moiety to target sequons (Abu-Qarn and Eichler 2006, Chaban et al. 2006), being closer to its eukaryal than to its C. jejuni homologue, PglB (Wacker et al. 2002). This relationship was previously suggested on the basis of sequence comparison, even though the archaeal OST appears to be composed of a single Stt3-like component as in Bacteria (Abu-Qarn and Eichler 2006, Chaban et al. 2006). However, the amino acid composition surrounding modified sequons in archaeal and eukaryal glycoproteins also show differences. In eukaryal glycoproteins, there is a high probability of finding aromatic residues at positions –2 and –1, a small hydrophobic residue (Gly or Val) at the X position and a larger hydrophobic residue (Ile, Leu, Met, Phe, Trp or Tyr) at the +1 position (Ben-Dor et al. 2004, Petrescu et al. 2004). By contrast, Phe, Trp and Tyr were detected at archaeal positions –2 and –1 in only 4% and 11% of the archaeal sequences, respectively, Ser or Thr were readily found at the X position and Ala and Gly predominated at the archaeal +1 position. Aromatic residues were never detected at this site. The only time a Tyr was detected at the +1 position was in the only sequon experimentally shown as being unmodified (see above). Nonetheless, as is the case in archaeal glycoproteins, Asp and Glu are also rarely found at the –2 and –1 positions adjacent to modified sequons in eukaryal N-glycosylated proteins (Petrescu et al. 2004). Moreover, in both archaeal and eukaryal glycoproteins, basic residues are poorly represented at position +1.
Insights into the archaeal N-glycosylation process from sequence analysis
The present study not only offers insight into the relationship between the environment in which archaeal sequons lie and their likelihood of being modified but also sheds light onto the interplay between protein translation, protein translocation and N-glycosylation in Archaea. Earlier results have shown that the Asn residues of sequons found < 12–14 positions downstream from a transmembrane domain are not modified by the vertebrate oligosaccharide transferase, suggesting the active site of the complex lies 30–40 Å from the ER membrane (Nilsson and von Heijne 1993). If the active site of archaeal Stt3 is similarly positioned, then the modification of the Asn residue 9 positions from the C-terminus of M. voltae FlaB2 (Voisin et al. 2005) would require glycosylation at this position to transpire post-translationally, following translocation of the protein across the membrane. However, topological analysis of the yeast and mouse Stt3 proteins (Kim et al. 2005) predicts that the WWDYG motif, thought to be responsible for the oligosaccharide transferase activity of the protein (Yan and Lennarz 2002, Wacker et al. 2002), is located 42 residues from the last transmembrane domain of the polypeptide. By contrast, predictive software (HMMTOP; Tusnady and Simon 2001) puts the same motif only 25 residues from the final transmembrane domain of the H. volcanii protein (Abu-Qarn and Eichler 2006). Thus, the spatial dimensions of co-translational/translocational modification by archaeal Stt3 may differ from those employed by its eukaryal homologue. Similar spatial considerations also apply when considering that Asn residues in the H. salinarum, M. fervidus and H. volcanii S-layer glycoproteins found one, six and 12 positions from the N-terminus, respectively, are glycan-modified. These observations suggest that glycosylation occurs following signal peptide cleavage since these residues would otherwise lie too close to the plasma membrane to be accessible to the archaeal Stt3 active site. A similar scenario has been proposed for the processing of eukaryal signal peptide-bearing glycoprotein precursors (Chen et al. 2001).
Conclusions
The finding that the characteristics of the amino acids surrounding glycosylated sequons in Archaea differ from those in Bacteria or Eukarya is in accord with earlier observations pointing to unique aspects of the archaeal process. Although NXS sequons are clearly modified in archaeal glycoproteins, exchanging the Ser residue of a particular Asn-Ala-Ser sequon within the H. salinarum S-layer glycoprotein sequence with a Val, Leu or Asn did not prevent N-glycosylation (Zeitler et al. 1998). Moreover, whereas glycans are linked to Asn residues of eukaryal glycoproteins almost exclusively through N-acetylglucosamine, Archaea can use other linking sugars (Lechner and Wieland 1989, Voisin et al. 2005). Together, these observations suggest that the archaeal OST works differently from its eukaryal and bacterial counterparts or that it is less stringent in its target selection criteria. Alternatively, Archaea could express a second version of the OST that functions in a novel manner. To date, no candidates for this putative second OST have been proposed, although A. fulgidus is thought to encode two Stt3 proteins (Burda and Aebi 1999) that could be imagined to function distinctly. Accordingly, different catalytic activities have been reported for the two Stt3 homologues detected in vertebrates (Kelleher et al. 2003).
Analysis of the regions bordering the glycan-bearing Asn residues of archaeal glycoproteins may provide insight into various aspects of the N-glycosylation process in Archaea. With the recent development of experimental systems for deciphering the steps involved in the archaeal version of this post-translational modification (Abu-Qarn and Eichler 2006, Chaban et al. 2006), the rules governing sequon occupation, including those proposed here, can be tested.
Acknowledgments
This work was supported by the Israel Science Foundation–The Charles H. Revson Foundation (Grant 433/03).
Appendix
Table A1.
Neighboring residues of putative sequons in archaeal glycoproteins containing experimentally verified N-glycosylated sequons. References: 1 = Lechner and Sumper 1987; 2 = Sumper et al. 1990; 3 = Brockl et al. 1991; 4 = Voisin et al. 2005; 5 = Zahringer et al. 2000; and 6 = Lin and Tang 1990. Abbreviation: SLG = S-layer glycoprotein. Putative sequons are shown in bold.
| Species | Protein | Sequence surrounding putative sequons | Ref. |
| H. salinarum | SLG | DTVDTGLVVDNDSTTEIVDDYEN | 1 |
| STTEIVDDYENTSISDVDYAYAI | |||
| DYVNSNEELTNGSALDGVSTDDD | |||
| AYVVGSEVDINGTANEGTDDVVL | |||
| LLQGDASIEINSTDEWNSDGQWS | |||
| DVPLSNVEPGNYTVEADDGDNTD | |||
| DQTTVDQPENNQTMTTTMTETTT | |||
| ETTTEMTTQENTTENGSEGTSDG | |||
| MTTTQENTTENGSEGTSDGESGG | |||
| H. volcanii | SLG | DVDSGSATTPNVTLLAPRITDSE | 2 |
| QYVVGSEVDINGTATSSDSVAIY | |||
| EREDTTTSSDNATDTTTTTDGPT | |||
| M. fervidus | SLG | DGNIIIVGPGNYTENILVNKSLT | 3 |
| PGNYTENILVNKSLTLLKSNGSA | |||
| SLTLKSTLKSNGSAIINAVSSEK | |||
| IGGKNGIYMENVTGCTITNNTIQ | |||
| MENVTGCTITNNTIQNAFVSGWE | |||
| YYGGNGICLVNSTNNTITNNIIR | |||
| GNGICLVNSTNNTITNNIIRNNT | |||
| NNTITNNIIRNNTWNGINVCESK | |||
| VCESKGNIIKNNTIMYSGGIGIY | |||
| NNIIENNRIINATYGGIYLFRPS | |||
| NNKICRNYIANVSSGGGGMSGAI | |||
| PKGELVDNIWNTTEGGNYWSNYT | |||
| NTTEGGNYWSNYTGNNTGDGTGN | |||
| GGNYWSNYTGNNTGDGTGNIPYY | |||
| VRWDKIVPEGNHTIKAEIPYSAE | |||
| FSKIVQGFVQNKTFTITLTNLGK | |||
| YYISIYTNPVNGTKVSYRELTIT | |||
| YRELTITLKPNETKTIELGKYPF | |||
| M. voltae | FlaA | VAAVAASVLINTSGFLQQKASST | 4 |
| SLG | ADSDYSDALINATTGFANIADNV | 4 | |
| S. acidocaldarius | CbsA | FLLSIIFTLENVSLAQTSPQISV | 5 |
| AGYWSQIPWTNISLTANIPMAPT | |||
| THYLLVKAAWNGSWIFILEEWQA | |||
| WSTAVAGIYPNASGPGLFRMIEL | |||
| GKEETGRIVFNYSGITLPAPNNT | |||
| NYSGITLPAPNNTEITVMSNGTI | |||
| NDGMFYGYYVNSTWYYPDRAAIM | |||
| IWMWVSGATWNNSTYDPAFKSNI | |||
| WMWVSGATWNNSTYDPAFKSNIW | |||
| DPAFKSNIWQNESLTGLSYVDSG | |||
| NHGFAVPLYTNNTMYEVDTAGIW | |||
| WYTPVASEGLNGSLFFIWTGAKY | |||
| FFIWTGAKYENGSWVVEFARPLS | |||
| VPLDYQPFMPNITVGKTYYVAFA | |||
| Thermopsin | PQNIYFFPHRNTTGATISSGLYV | 6 | |
| ITTNEMLGYVNITSLLAYNREAL | |||
| TLQFNIVLSVNTSNGVYAYWLQD | |||
| NSLTFIDNVWNLTGSLSTLSSSA | |||
| SFPLSYITIINMSYTSNAVYVWI | |||
| ISASLMINGNNYTPNGLYYDAEL | |||
| NGAPTSFNSLNCTLGLYYISNGS | |||
| NCTLGLYYISNGSITPVPSLYTF | |||
| GVPIAYNGIENLTILTNNFSVIL | |||
| GIENLTILTNNFSVILI | |||
Table A2.
Putative N-glycosylated predicted secretory proteins.
| Species | Proteins |
| A.fulgidus | AF0078, 0101, 0141, 0142, 0254, 0274, 0378, 0487, 0637, 0663, 0681, 0711, 0736, 0740, 0759, 0816, 0817, 0912, 0924, 0992, 1025, 1054, 1055, 1096, 1118, 1177, 1231, 1276, 1354, 1411, 1464, 1574, 1658, 1659, 1660, 1661, 1675, 1763, 1884, 1945, 1948, 2017, 2048, 2079, 2080, 2081, 2090, 2119, 2163, 2188 |
| A. pernix | APE0016, 0018, 0174, 0268, 0419, 0592, 0595, 0620, 0622, 0654, 0725, 0814, 0825, 0858, 0876, 0916, 0958, 1013, 1143, 1171, 1303, 1334, 1458, 1463, 1521, 1574, 1575, 1604, 1625, 1675, 1676, 1820, 1901, 1905, 1944, 2014, 2073, 2095, 2175, 2186, 2204, 2422, 2434, 2545, 2594, 2610, |
| Halobacterium sp. NRC-1 | VNG0200, 0204, 0219, 0220, 0262, 0281, 0399, 0539, 0620, 0681, 0816, 0818, 0819, 0931, 0953, 1009, 1036, 1100, 1181, 1337, 1372, 1659, 1675, 1951, 2148, 2516, 2567, 2573, 2589, 6064, 6178, 6323 |
| M. jannaschii | MJ0418, 0492, 0505, 0753, 0755, 0756, 0784, 0815, 0822, 0832.1, 0833, 0835.1, 0872, 0891, 0893, 0903, 0954, 1128, 1284, 1289, 1291, 1292, 1396, 1400, 1402, 1472, 1488, 1502 |
| M. thermoautotrophicus | MTH57, 60, 61, 97, 117, 247, 302, 318, 330, 353, 354, 412, 424, 479, 508, 513, 523, 567, 570, 596, 597, 604, 612, 635, 670, 715, 719, 795, 806, 845, 910, 911, 924, 968, 977, 995, 1038, 1216, 1290, 1330, 1400, 1513, 1566, 1653, 1772, 1777, 1782 |
| P. abyssi | PAB0061, 0133, 0275, 0337, 0378, 0485, 0602, 0741, 0847, 0936, 0982, 0984, 1279, 1379, 1380, 1384, 1456, 1651, 1687, 1790, 1823, 1825, 2366 |
| P. horikoshii | PH004, 0036, 0066, 0188, 0244, 0298, 0310, 0399, 0462, 0480, 0548, 0549, 0550, 0551, 0555, 0564, 0572, 0602, 0649, 0733, 0777, 0875, 0954, 0996, 1070, 1130, 1184, 1190, 1191, 1192, 1289, 1303, 1336, 1341, 1495, 1569, 1668, 1690, 1740, 1790, 1929, 1964 |
| S. solfararicus | SSO0037, 0117, 0147, 0330, 0483, 0489, 0519, 0522, 0541, 0650, 0999, 1053, 1093, 1131, 1167, 1320, 1872, 1981, 2088, 2187, 2420, 2570, 2611, 2681, 2684, 2712, 2776, 2795, 2819, 2846, 2969, 2971, 2972, 3104, 3138, 3177 |
| T. acidophilum | Ta0129, 0261, 0297, 0308, 0367, 0380, 0496, 0510, 0553, 0556, 0591, 0721, 0741, 0851, 0951, 1029, 1052, 1171, 1236, 1403, 1421, 1462, 1499 |
| T. volcanium | TVN0085, 0128, 0205, 0231, 0366, 0371, 0544, 0610, 0611, 0711, 0840, 0948, 0969, 1143, 1200, 1290, 1452, 1469 |
References
- R1.Abu-Qarn M., Eichler J. Protein N-glycosylation in Archaea: defining Haloferax volcanii genes involved in S-layer glycoprotein glycosylation. Mol. Microbiol. 2006;61:511–525. doi: 10.1111/j.1365-2958.2006.05252.x. [DOI] [PubMed] [Google Scholar]
- R2.Bardy S.L., Eichler J., Jarrell K.F. Archaeal signal peptides—a comparative survey at the genome level. Protein Sci. 2003;12:1833–1843. doi: 10.1110/ps.03148703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Bause E., Legler G. The role of the hydroxy amino acid in the triplet sequence Asn-Xaa-Thr(Ser) for the N-glycosylation step during glycoprotein biosynthesis. Biochem. J. 1981;195:639–644. doi: 10.1042/bj1950639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Ben-Dor S., Esterman N., Rubin E., Sharon N. Biases and complex patterns in the residues flanking protein N-glycosylation sites. Glycobiology. 2004;14:95–101. doi: 10.1093/glycob/cwh004. [DOI] [PubMed] [Google Scholar]
- R5.Brockl G., Behr M., Fabry S., Hensel R., Kaudewitz H., Biendl E., Konig H. Analysis and nucleotide sequence of the genes encoding the surface-layer glycoproteins of the hyperthermophilic methanogens Methanothermus fervidus and Methanothermus sociabilis . Eur. J. Biochem. 1991;199:147–152. doi: 10.1111/j.1432-1033.1991.tb16102.x. [DOI] [PubMed] [Google Scholar]
- R6.Burda P., Aebi M. The dolichol pathway of N-linked glycosylation. Biochim. Biophys. Acta. 1999;1426:239–257. doi: 10.1016/s0304-4165(98)00127-5. [DOI] [PubMed] [Google Scholar]
- R7.Chaban B., Voisin S., Kelly J., Logan S.M., Jarrell K.F. Identification of genes involved in the biosynthesis and attachment of Methanococcus voltaeN-linked glycans: insight into N-linked glycosylation pathways in Archaea. Mol. Microbiol. 2006;61:259–268. doi: 10.1111/j.1365-2958.2006.05226.x. [DOI] [PubMed] [Google Scholar]
- R8.Chen X., Vanvalkenburgh C., Liang H., Fang H., Green N. Signal peptidase and oligosaccharyltransferase interact in a sequential and dependent manner within the endoplasmic reticulum. J. Biol. Chem. 2001;276:2411–2416. doi: 10.1074/jbc.M007723200. [DOI] [PubMed] [Google Scholar]
- R9.Eichler J., Adams M.W. Posttranslational protein modification in Archaea. Microbiol. Mol. Biol. Rev. 2005;69:393–425. doi: 10.1128/MMBR.69.3.393-425.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Gavel Y., von Heijne G. Sequence differences between glycosylated and non-glycosylated Asn-X-Thr/Ser acceptor sites: implications for protein engineering. Protein Eng. 1990;3:433–442. doi: 10.1093/protein/3.5.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R11.Kaji H., Saito H., Yamauchi Y., Shinkawa T., Taoka M., Kasai K., Takahashi N., Isobe T. Lectin affinity capture, isotope-coded tagging and mass spectrometry to identify N-linked glycoproteins. Nat. Biotechnol. 2003;21:667–672. doi: 10.1038/nbt829. [DOI] [PubMed] [Google Scholar]
- R12.Kasturi L., Eshleman J.R., Wunner W.H., Shakin-eshleman S.H. The hydroxy amino acid in an Asn-X-Ser/Thr sequon can influence N-linked core glycosylation efficiency and the level of expression of a cell surface glycoprotein. J. Biol. Chem. 1995;270:14756–14761. doi: 10.1074/jbc.270.24.14756. [DOI] [PubMed] [Google Scholar]
- R13.Kelleher D.J., Gilmore R. An evolving view of the eukaryotic oligosaccharyl transferase. Glycobiology. 2006;16:47R–62R. doi: 10.1093/glycob/cwj066. [DOI] [PubMed] [Google Scholar]
- R14.Kelleher D.J., Karaoglu D., Mandon E.C., Gilmore R. Oligosaccharyltransferase isoforms that contain different catalytic STT3 subunits have distinct enzymatic properties. Mol. Cell. 2003;12:101–111. doi: 10.1016/s1097-2765(03)00243-0. [DOI] [PubMed] [Google Scholar]
- R15.Kim H., Von heijne G., Nilsson I.M. Membrane topology of the STT3 subunit of the oligosaccharyl transferase complex. J. Biol. Chem. 2005;280:20261–20267. doi: 10.1074/jbc.M412213200. [DOI] [PubMed] [Google Scholar]
- R16.Kowarik M., Young N.M., Numao S., et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006;25:1957–1966. doi: 10.1038/sj.emboj.7601087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.Kurosky A., Barnett D.R., Lee T.H., Touchstone B., Hay R.E., Arnott M.S., Bowman B.H., Fitch W.M. Covalent structure of human haptoglobin: a serine protease homolog. Proc. Natl. Acad. Sci. USA. 1980;77:3388–3392. doi: 10.1073/pnas.77.6.3388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.Lechner J., Sumper M. The primary structure of a procaryotic glycoprotein. Cloning and sequencing of the cell surface glycoprotein gene of halobacteria. J. Biol. Chem. 1987;262:9724–9729. [PubMed] [Google Scholar]
- R19.Lechner J., Wieland F. Structure and biosynthesis of prokaryotic glycoproteins. Annu. Rev. Biochem. 1989;58:173–194. doi: 10.1146/annurev.bi.58.070189.001133. [DOI] [PubMed] [Google Scholar]
- R20.Lin X., Tang J. Purification, characterization, and gene cloning of thermopsin, a thermostable acid protease from Sulfolobus acidocaldarius . J. Biol. Chem. 1990;265:1490–1495. [PubMed] [Google Scholar]
- R21.Mellquist J.L., Kasturi L., Spitalnik S.L., Shakin-eshleman S.H. The amino acid following an asn-X-Ser/Thr sequon is an important determinant of N-linked core glycosylation efficiency. Biochemistry. 1998;37:6833–6837. doi: 10.1021/bi972217k. [DOI] [PubMed] [Google Scholar]
- R22.Mengele R., Sumper M. Drastic differences in glycosylation of related S-layer glycoproteins from moderate and extreme halophiles. J. Biol. Chem. 1992;267:8182–8185. [PubMed] [Google Scholar]
- R23.Mescher M.F., Strominger J.L. Purification and characterization of a prokaryotic glycoprotein from the cell envelope of Halobacterium salinarium . J. Biol. Chem. 1976;251:2005–2014. [PubMed] [Google Scholar]
- R24.Nakai K., Horton P. PSORT: a program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 1999;24:34–36. doi: 10.1016/s0968-0004(98)01336-x. [DOI] [PubMed] [Google Scholar]
- R25.Nilsson I.M., von Heijne G. Determination of the distance between the oligosaccharyltransferase active site and the endoplasmic reticulum membrane. J. Biol. Chem. 1993;268:5798–5801. [PubMed] [Google Scholar]
- R26.Nita-Lazar M., Wacker M., Schegg B., Amber S., Aebi M. The N-X-S/T consensus sequence is required but not sufficient for bacterial N-linked protein glycosylation. Glycobiology. 2005;15:361–367. doi: 10.1093/glycob/cwi019. [DOI] [PubMed] [Google Scholar]
- R27.Paul G., Lottspeich F., Wieland F. Asparaginyl-N-acetylgalactosamine. Linkage unit of halobacterial glycosaminoglycan. J. Biol. Chem. 1986;261:1020–1024. [PubMed] [Google Scholar]
- R28.Petrescu A.J., Milac A.L., Petrescu S.M., Dwek R.A. Statistical analysis of the protein environment of N-glycosylation sites: implications for occupancy, structure, and folding. Glycobiology. 2004;14:103–114. doi: 10.1093/glycob/cwh008. [DOI] [PubMed] [Google Scholar]
- R29.Shakin-Eshleman S.H., Spitalnik S.L., Kasturi L. The amino acid at the X position of an Asn-X-Ser sequon is an important determinant of N-linked core-glycosylation efficiency. J. Biol. Chem. 1996;271:6363–6366. doi: 10.1074/jbc.271.11.6363. [DOI] [PubMed] [Google Scholar]
- R30.Szymanski C.M., Wren B.W. Protein glycosylation in bacterial mucosal pathogens. Nat. Rev. Microbiol. 2005;3:225–237. doi: 10.1038/nrmicro1100. [DOI] [PubMed] [Google Scholar]
- R31.Szymanski C.M., Yao R., Ewing C.P., Trust T.J., Guerry P. Evidence for a system of general protein glycosylation in Campylobacter jejuni . Mol. Microbiol. 1999;32:1022–1030. doi: 10.1046/j.1365-2958.1999.01415.x. [DOI] [PubMed] [Google Scholar]
- R32.Tusnady G.E., Simon I. The HMMTOP transmembrane topology prediction server. Bioinformatics. 2001;17:849–850. doi: 10.1093/bioinformatics/17.9.849. [DOI] [PubMed] [Google Scholar]
- R33.Voisin S., Houliston R.S., Kelly J., Brisson J.R., Watson D., Bardy S.L., Jarrell K.F., Logan S.M. Identification and characterization of the unique N-linked glycan common to the flagellins and S-layer glycoprotein of Methanococcus voltae . J. Biol. Chem. 2005;280:16586–16593. doi: 10.1074/jbc.M500329200. [DOI] [PubMed] [Google Scholar]
- R34.Wacker M., Linton D., Hitchen P.G., et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. Science. 2002;298:1790–1793. doi: 10.1126/science.298.5599.1790. [DOI] [PubMed] [Google Scholar]
- R35.Weerapana E., Imperiali B. Asparagine-linked protein glycosylation: from eukaryotic to prokaryotic systems. Glycobiology. 2006;16:91R–101R. doi: 10.1093/glycob/cwj099. [DOI] [PubMed] [Google Scholar]
- R36.Yan Q., Lennarz W.J. Studies on the function of oligosaccharyl transferase subunits. Stt3p is directly involved in the glycosylation process. J. Biol. Chem. 2002;277:47692–47700. doi: 10.1074/jbc.M208136200. [DOI] [PubMed] [Google Scholar]
- R37.Zahringer U., Moll H., Hettmann T., Knirel Y.A., Schafer G. Cytochrome b558/566 from the archaeon Sulfolobus acidocaldarius has a unique Asn-linked highly branched hexasaccharide chain containing 6-sulfoquinovose. Eur. J. Biochem. 2000;267:4144–4149. doi: 10.1046/j.1432-1327.2000.01446.x. [DOI] [PubMed] [Google Scholar]
- R38.Zeitler R., Hochmuth E., Deutzmann R., Sumper M. Exchange of Ser-4 for Val, Leu or Asn in the sequon Asn-Ala-Ser does not prevent N-glycosylation of the cell surface glycoprotein from Halobacterium halobium . Glycobiology. 1998;8:1157–1164. doi: 10.1093/glycob/8.12.1157. [DOI] [PubMed] [Google Scholar]