Abstract
The gene-dense chromosomes of archaea and bacteria were long thought to be devoid of pseudogenes, but with the massive increase in available genome sequences, whole genome comparisons between closely related species have identified mutations that have rendered numerous genes inactive. Comparative analyses of sequenced archaeal genomes revealed numerous pseudogenes, which can constitute up to 8.6% of the annotated coding sequences in some genomes. The largest proportion of pseudogenes is created by gene truncations, followed by frameshift mutations. Within archaeal genomes, large numbers of pseudogenes contain more than one inactivating mutation, suggesting that pseudogenes are deleted from the genome more slowly in archaea than in bacteria. Although archaea seem to retain pseudogenes longer than do bacteria, most archaeal genomes have unique repertoires of pseudogenes.
Keywords: comparative genomics, pseudogenes
Introduction
Genomic studies have allowed for the in-depth analysis of the genetic structure of organisms from all domains of life. Unfortunately, archaea are rather poorly represented among the more than 400 fully sequenced genomes, due in part to the difficulties associated with their cultivation and genetic manipulation (Schleper et al. 2005). Although the Eukarya and the Archaea are sister taxa (Ciccarelli et al. 2006), the overall organization of archaeal genes and genomes is more similar to that of the Bacteria. Like bacteria, archaea usually contain a single circular chromosome and have a high gene density, with genes organized in operons and lacking introns.
The elucidation of complete genome sequences has instigated large-scale experimental and computational analyses that have attempted to identify and annotate all genes encoded in a genome. Despite such efforts, up to 40% of the predicted coding sequences in many archaeal genomes lack a predicted function (Galperin and Koonin 2004, Fricke et al. 2006). It has been suggested that, within virtually all genomes, there are annotated genes that have been mutationally inactivated and can never be assigned a function (Ochman and Davalos 2006). Eukaryotic genomes have long been known to contain large numbers of non-functional genes (Vanin 1985), but the full extent of their pseudogene contents was not evident until whole genome sequences became available. Genome-wide analyses of the nematode (Harrison et al. 2001) and human (Torrents et al. 2003) genomes have detected massive amounts of now-defunct genes, whose numbers likely exceed the number of functional genes in each genome.
Because of the small size and high gene density of bacterial genomes, it was originally thought that prokaryotes would contain few, if any, pseudogenes (Lawrence et al. 2001). In addition, the majority of pseudogenes in higher eukaryotes are generated by retrotransposition (Vanin 1985), a process unknown in bacteria or archaea. Nevertheless, pseudogenes are now known to be a common feature of many bacterial genomes (Lerat and Ochman 2004, 2005) and may constitute nearly half of the annotated coding sequences (CDSs) in the genomes of some pathogens (Andersson et al. 1998, Cole et al. 2001, Toh et al. 2006).
A previous assessment of prokaryotic genomes estimated that up to 5% of the annotated genes in archaeal genomes may, in fact, be pseudogenes (Liu et al. 2004). This analysis, in which the contents of 53 bacterial and 11 archaeal genome sequences were compared, suggested that the pseudogenes originated predominantly from failed horizontal gene transfer events (as opposed to the mutational inactivation of resident genes). However, this analysis included comparisons of genes over broad phylogenetic distances, did not discriminate between orthologous and paralogous genes, and ignored genes without an assigned function. As such, this approach could lead to inaccurate appraisals of the pseudogene contents, particularly in the Archaea, which were not densely sampled and in which the functions of many genes have not been characterized. Moreover, if archaea have more split genes than bacteria, as suggested previously (Snel et al. 2000), the recognition of pseudogenes through inter-domain comparisons becomes less straightforward.
An alternative approach, one in which closely related genome sequences are compared, can enhance the resolution of pseudogenes because larger fractions of the genome are shared and there are fewer problems in assigning orthology. Most bacterial genomes have been found to contain largely unique sets of pseudogenes, suggesting that pseudogenes are constantly formed in, and rapidly eliminated from, the genome (Lerat and Ochman 2004); however, the frequencies with which pseudogenes are generated, maintained and removed from archaeal genomes are unknown. With the current availability of numerous archaeal genome sequences, including multiple members of particular genera, we sought to identify and enumerate the pseudogenes in archaea, and assessed their mechanisms of formation and erosion.
Materials and methods
Selection of phylogenetically related species
We used 16S rDNA sequences to determine the phylogenetic relationships of archaea for which genome sequences were available at NCBI as of April 1, 2006. Using the MEGA3.1 software package (Kumar et al. 2004), we applied a Minimum Evolution bootstrap test of phylogeny with pairwise deletion and 10,000 replicates. The relationships, accession numbers and genomic properties of the 15 selected species are presented in Figure 1.
Figure 1.
Relationships and features of archaeal genome sequences used in this study. Numbers at the nodes of the tree represent bootstrap values, with asterisks indicating 100% bootstrap support. The column labels indicating the numbers of pseudogenes represent total numbers, with the numbers in brackets indicating the positional pseudogenes.Horizontal lines separate different clades, and members within each clade were compared to detect pseudogenes.
Pseudogene identification
Pseudogenes were identified by comparing the genome contents of sequenced members of the same clade, as shown in Figure 1. Full genome sequences and sets of GenBank annotated proteins for the corresponding genomes were obtained from NCBI (http://www.ncbi.nih.gov). Inclusion thresholds for aligned sequences were as described previously (Lerat and Ochman 2005): E-values of < 1 0–15 and sequence identity of > 75% for clades containing species with 16S rDNA identity > 95% (i.e.,Pyrococcus spp., Methanosarcina spp., Thermoplasma spp.; highlighted in grey in Figure 1); and E-values of < 1 0–10and sequence identity of > 49% for clades containing species with 16S rDNA identity < 95% (i.e., Halobacterium/Haloarcula, Methanococcus/Methanocaldococcus, Sulfolobus spp.).
Within each clade of closely related organisms, the annotated proteins within the genome of one species were queried against the complete nucleotide sequence of another species from the same clade. This was done reciprocally for all crosswise comparisons within each clade using the TBLASTN search tool (Altschul et al. 1997). The Y –F program suite, developed to recognize truncated and otherwise mutationally altered CDSs (Lerat and Ochman 2004), was applied to the TBLASTN output, returning a list of candidate pseudogenes that was then curated manually. One way in which Y –F recognizes potential pseudogenes is by identifying internal stop codons in a query gene. For the comparisons of the three Methanosarcina spp., it was necessary to disable this feature for the amber stop codon TAG, which instead codes for pyrrolysine in this genus (James et al. 2001). In addition, all pseudogenes were curated manually for known recoding events (Cobucci-Ponzano et al. 2005).
Pseudogenes detected by the comparative analysis of full genome sequences can be either positional homologs or non-positional homologs of CDSs in the reciprocal genomes, based on gene context conservation: positional pseudogenes are those that share at least one neighboring gene with the corresponding functional copy in the related genome.
To identify gene-inactivating mutations, the putative pseudogenes were aligned with their counterparts using CLUSTALW 1.83 (Thompson et al. 1994). Gene-inactivating mutations were partitioned into five classes: frameshifts (insertions or deletions of 1 or 2 nucleotides in length), deletions (> 2 nucleotides in length), insertions (> 2 nucleotides in length), truncations (large deletions at either or both ends of a coding sequence), and nonsense mutations. In cases where more than one gene-inactivating mutation was identified in an alignment, the mutation was classified as a combination of two or more of these classes.
Results
Pseudogene content analyses in archaeal genomes
We assessed the pseudogene contents of 15 species, representing eight genera of archaea, by comparing the full genome sequences of the most closely related taxa. The fractions of predicted pseudogenes range from 0.3% to 8.6% of the total number of annotated protein coding sequences, including unannotated pseudogenes, i.e., intergenic regions that contain the eroded remnants of genes that have been annotated as CDS in a related genome (Figure 2). Applying identical methods, the range and the average pseudogene fractions are much lower in archaea than in pathogenic bacteria but similar to those of free-living bacteria (Lerat and Ochman 2004, 2005).
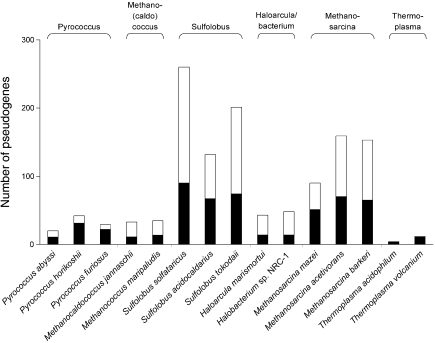
Figure 2.
Numbers of positional (filled bars) and non-positional pseudogenes (open bars) in archaeal genomes.
There are almost equal fractions of positional (i.e., sharing at least one neighboring gene with its counterpart in a related genome) and non-positional pseudogenes in most genomes, and within both classes, there are large numbers of unannotated pseudogenes (Table 1). Across taxa, the fraction of these unannotated pseudogenes increases with the total number of detected pseudogenes. As expected, large proportions of predicted pseudogenes are annotated as having hypothetical functions (Table 1). Inactivated mobile element-associated genes are more abundant among non-positional pseudogenes, especially in Sulfolobus spp. and Methanosarcina spp. The lists of the predicted pseudogenes are provided in Supplementary Tables S1 and S2.
Table 1.
Numbers, proportions and characteristics of pseudogenes in the Archaea. Numbers depicted are the total number of predicted pseudogenes, with bracketed numbers indicating the number of positional pseudogenes.
| Species | Total no. of pseudogenes | Percentage of pseudogenes | No. of annotated pseudogenes | No. of unannotated pseudogenes | Number of hypotheticals |
| Pyrococcus abyssi | 20 (9) | 1.1 (0.6) | 13 (9) | 7 (2) | 8 (4) |
| Pyrococcus horikoshii | 42 (11) | 1.9 (1.4) | 31 (23) | 11 (8) | 24 (16) |
| Pyrococcus furiosus | 29 (7) | 1.5 (1.1) | 25 (19) | 4 (3) | 19 (15) |
| Methanocaldococcus jannaschii | 31 (10) | 1.8 (0.6) | 31 (10) | 2 (0) | 11 (1) |
| Methanococcus maripaludis | 34 (13) | 2.0 (0.8) | 32 (12) | 2 (1) | 5 (0) |
| Sulfolobus solfataricus | 260 (90) | 8.6 (3.0) | 179 (58) | 81 (32) | 97 (21) |
| Sulfolobus acidocaldarius | 132 (67) | 5.7 (2.9) | 41 (20) | 93 (47) | 47 (13) |
| Sulfolobus tokodaii | 201 (74) | 6.9 (2.6) | 85 (37) | 116 (37) | 105 (22) |
| Haloarcula marismortui | 43 (14) | 1.2 (0.4) | 37 (10) | 6 (4) | 12 (5) |
| Halobacterium sp. NRC-1 | 48 (14) | 2.3 (0.7) | 33 (12) | 15 (2) | 26 (6) |
| Methanosarcina mazei | 90 (51) | 2.6 (1.5) | 63 (37) | 27 (14) | 37 (21) |
| Methanosarcina acetivorans | 159 (70) | 3.4 (1.5) | 101 (56) | 58 (14) | 59 (16) |
| Methanosarcina barkeri | 153 (65) | 4.2 (1.8) | 117 (50) | 36 (15) | 60 (18) |
| Thermoplasma acidophilum | 4 (4) | 0.3 (0.3) | 4 (4) | 0 (0) | 1 (1) |
| Thermoplasma volcanium | 12 (11) | 0.8 (0.7) | 12 (11) | 0 (0) | 3 (3) |
Mechanisms of gene inactivation
Whencompared to bacterial pseudogenes, archaeal pseudogenes are more highly decayed, with a larger fraction containing more than one inactivating mutation (Figure 3). In both archaea and bacteria, non-positional pseudogenes show greater gene decay than do positional pseudogenes. Truncations, frameshifts, and combinations thereof, are the most widespread mechanisms by which genes are inactivated in archaea (Supplementary Figure S1).
Figure 3.
Differences in the gene decay ratios (calculated as the ratio of pseudogenes eroded by multiple inactivating mutations to pseudogenes inactivated by a single mutation) for archaeal (filled bars) and bacterial (open bars) pseudogenes.
A total of six pseudogenes were inactivated by changes in the lengths of a homopolymeric stretch spanning more than six nucleotides, and all of these frameshifts occurred in A/T tracts (data not shown). Unlike the situation observed in bacterial pathogens, the interruption of genes by insertion sequences is relatively rare in archaea, with only 5 and 28 IS-inactivated genes from a total of 550 positional and 711 non-positional pseudogenes, respectively. Among the non-positional pseudogenes, 22 of the 28 IS-inactivated genes occur in S. solfataricus, which contains exceptionally large numbers of mobile elements (She et al. 2001).
Because pseudogenes are under no functional constraint and are evolutionarily neutral, such regions can divulge the underlying rate and pattern of mutations within a genome (Li et al. 1981, Mira et al. 2001). In general, deletions occur more frequently than insertions in archaeal pseudogenes (Figure 4). Thermoplasma volcanium is the only archaeal species in which the cumulative indel length is positive (i.e., more DNA is inserted than deleted in the detected pseudogenes), and this is due to a single 88-bp duplication in the NADH ubiquinone oxidoreductase gene. Although their overall indel lengths are negative, in the non-positional pseudogenes of species within the Methanosarcina clade, insertions outnumber deletions, with most cases involving the insertional inactivation of transposases within mobile elements.
Figure 4.
Cumulative size of insertion (open bars) and deletion (filled bars) events in positional (P) and non-positional (NP) pseudogenes of archaeal genomes. Numbers above the bars indicate numbers of pseudogenes on which the analyses are based.
GC content of archaeal genes with pseudogene counterparts
Previous analyses have reported a significant difference between the nucleotide composition of genes that have a pseudogene counterpart and those that do not (Lerat and Ochman 2004). We searched for this in archaea for the functional counterparts of both positional and non-positional pseudogenes (Supplementary Table S3) and found no consistent trend. Genes with positional or non-positional pseudogene counterparts have higher GC contents than genes without pseudogene counterparts in Methanocaldococcus jannaschii, Methanococcus maripaludis and the Sulfolobus spp. genomes. Genes with non-positional pseudogene counterparts have a significantly lower GC percentage in the Methanosarcina clade. In this case, the difference in GC contents is due to the many inactivated mobile elements, which typically have a different GC content than that of their hosts.
Shared pseudogenes between closely related archaeal genomes
The occurrence of only a single inactivating mutation in the vast majority of bacterial pseudogenes implies that pseudogenes are rapidly generated in, and removed from, these genomes. This process has resulted in genomes containing largely nonoverlapping pseudogene inventories, even among strains averaging only 1% in sequence divergence (Lerat and Ochman 2004). Although archaea seem to retain pseudogenes longer than do bacteria (as evident from the higher incidence of pseudogenes containing multiple inactivating mutations), most archaeal genomes have unique repertoires of pseudogenes. Among Pyrococcus spp., which average 30 positional pseudogenes, not more than three are shared between any two strains. Higher fractions of shared pseudogenes are observed in Sulfolobus and Methanosarcina, which can be ascribed, in part, to shared inactivated transposases that occur in multiple copies (Supplementary Tables S1 and S2).
Discussion
Comparative analyses of full genome sequences show that pseudogenes can occur at high frequencies and often outnumber the functional copies of genes. Even among bacteria, which have relatively small and streamlined genomes, up to half of the genome of some facultative pathogens, such as Rickettsia prowazekii and Mycobacterium tuberculosis (Andersson et al. 1998, Cole et al. 2001), is relegated to pseudogenes. In our analyses of 15 archaeal genomes representing eight genera, we found that the predicted fractions of pseudogenes range from 0.3% to 8.6%, corresponding to 4 to 260 pseudogenes per genome. These numbers are lower than the pseudogene contents found in most bacteria by the same approach; however, previous studies focused primarily on bacterial pathogens, which are known to have more severely degraded genomes than non-pathogenic bacteria. The pseudogene contents of archaea are likely to be more similar to those non-pathogenic, free-living bacteria, although relatively few bacterial genomes have been evaluated by these methods.
The available annotations of many of the completed archaeal genomes identify no pseudogenes. Although the original publications of the genomes of M. jannaschii (Bult et al. 1996), S. solfataricus (She et al. 2001), S. acidocaldarius (Chen et al. 2005), T. acidophilum (Ruepp et al. 2000) and T. volcanium (Kawashima et al. 2000) each describe certain genes as being inactivated, they are not classified as such in the most commonly used public databases (e.g., NCBI, www. ncbi.nlm.nih.gov). Among the genomes that we analyzed, only the annotations of M. mazei and M. barkeri contain pseudogenes (112 and 136 compared with the 90 and 153 that we identified in these genomes, respectively). And similar to our observations for S. solfataricus, She et al. (2001) identified 43 partial transposases.
In contrast to NCBI, the IMG database (img.jgi.doe.gov) (Markowitz et al. 2006) provides re-annotated archaeal genomes and lists somewhat different numbers of total genes as well as pseudogenes for these genomes. Because the specific annotation can impact the identification of pseudogenes (IMG predicts on average 2.5% more protein coding genes than does NCBI), it is not possible to compare directly the numbers of pseudogenes listed at IMG with those detected in the present study. Perrodou et al. (2006) have pointed out an association between different genome annotation approaches and pseudogene prediction, noting that, in some genomes, pseudogenes might be attributable to sequencing errors. Because a minority of the pseudogenes is formed by the most common sequencing errors, such artifacts have probably contributed little to the sets of pseudogenes that we recognized. Although it has been found that, in Pyrobaculum aerophilum, a putative deficiency in mismatch repair genes has created a variety of long and variable mononucleotide runs(Fitz-Gibbon et al. 2002), intraspecific variation in homopolymeric tract lengths are not a major source of gene inactivation in the archaeal genomes considered in this study.
Liu et al. (2004) also applied a comparative approach to identify the pseudogenes in 64 sequenced genomes, including 11 archaeal species. Their analyses compared species from different domains (e.g., the Archaea versus the Bacteria), which can make assignments of orthology difficult. To refine their analyses and to safeguard against the inclusion of annotation artifacts, they limited their comparisons to protein coding genes with assigned functions, thereby excluding all those annotated as “hypothetical.” Such restrictions, though valid, preclude accurate assessments of the full complement of pseudogenes in a genome. On one hand, the numbers of pseudogenes detected by such methods might be overestimated because orthologous genes from divergent taxa might remain functional despite extreme length or sequence differences. And this may be complicated by the occurence of more split genes in archaea than in bacteria (Snel et al. 2000). On the other hand, most pseudogenes detected through comparisons of closely related genomes are functionally annotated as “hypothetical,” which is not surprising since expendable genes are less likely to be among those whose functions have been assigned. Therefore, excluding these genes would result in underestimates of the actual number of pseudogenes. Despite these caveats, the pseudogene contents of archaea reported by Liu et al. (2004) are qualitatively similar to those identified with Y –F; both approaches indicate that archaeal genomes contain fewer pseudogenes than do pathogenic bacteria, and both indicate that archaeal pseudogenes are more decayed than bacterial pseudogenes. Also, in both our and Liu et al.’s analyses, S. solfataricus contains the largest number of pseudogenes of all surveyed archaeal genomes.
In addition to cataloging archaeal pseudogenes, our analyses provide insights into the mutational processes that occur within these genomes. In bacteria, strand slippage in mononucleotide repeats is considered a common mutagenic mechanism; however, few archaeal pseudogenes are attributable to variation in mononucleotide repeats. The association of strand slippage with immune evasion in bacterial pathogens (van der Woude and Baumler 2004) may explain the lower incidence of this type of frameshift in archaea. Strand slippage is evident in some of the mobile-element associated genes in S. acidocaldarius (IS1-family pseudogenes) and S. solfataricus (IS1048-family pseudogenes), many of which were modified by frameshifts in adenine mononucleotide repeats larger than six residues. Because mobile elements have recently been shown to create different functional proteins by strand slippage (Baranov et al. 2006), such events might not always indicate an inactivated gene. In addition, recent evidence has indicated that recoding may occur in Sulfolobus (Cobucci-Ponzano et al. 2003, 2005).
We find that the primary mechanism of gene inactivation in archaea is by truncation, which is responsible for the formation of over 30% of all pseudogenes. Among pseudogenes that have been inactivated by a single truncation event, unannotated pseudogenes are, on average, shorter than annotated pseudogenes, and non-positional pseudogenes are shorter than positional pseudogenes. Defunct transposable elements contain more inactivating mutations than other pseudogenes in S. tokodaii, S. solfataricus, M. acetivorans and M. barkeri. Whether the higher decay observed in transposable elements is caused by unsuccessful transposition events is unclear, although previous studies in Sulfolobus have shown that the rate of precise excision of mobile elements was low (Blount and Grogan 2005).
Pseudogene contents, as predicted by most comparative studies, still represent rather conservative estimates of the actual numbers of inactivated genes within a genome. Several classes of pseudogenes, such as those caused by missense mutations that abolish protein function as well as regulatory mutations that disrupt gene expression, will go undetected by this approach. Such comparative analyses also ignore strain-specific genes (i.e., ORFans) for which there are no homologous sequences available for comparison. Although many ORFans are thought to be functional (Daubin and Ochman 2004), they are unlikely to be essential to cell function and are prone to inactivation and loss.
Pseudogene detection by comparative analyses relies on the quality of the genome annotation, which can deviate substantially among different approaches (Brenner 1999). Increases in the availability of genome sequences from closely related species, which are still in short supply for archaea, have greatly facilitated genome annotation. As more genome sequences become available, we suspect that there will be less need to rely on experimental evidence to make accurate functional predictions about the majority of genes in a genome.
Supplementary material
Supplementary Table S1. List of archaeal positional pseudogenes.
Supplementary Table S2. List of archaeal non-positional pseudogenes.
Supplementary Figure S1. Numbers of pseudogenes and their mechanisms of inactivation in each of the archaeal genomes considered.
Supplementary Table S3. The GC percentages of gene sets that do and do not contain a pseudogene counterpart.
Acknowledgments
The authors thank Emmanuelle Lerat for assistance with the computational analyses and Becky Nankivell for assistance in the preparation of this manuscript. This work is supported by a research grant No. GM56120 from the National Institutes of Health to H.O.
References
- R1.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Andersson S.G., Zomorodipour A., Andersson J.O., et al. The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature. 1998;396:133–140. doi: 10.1038/24094. [DOI] [PubMed] [Google Scholar]
- R3.Baranov P.V., Fayet O., Hendrix R.W., Atkins J.F. Recoding in bacteriophages and bacterial IS elements. Trends Genet. 2006;22:174–181. doi: 10.1016/j.tig.2006.01.005. [DOI] [PubMed] [Google Scholar]
- R4.Blount Z.D., Grogan D.W. New insertion sequences of Sulfolobus: functional properties and implications for genome evolution in hyperthermophilic archaea. Mol. Microbiol. 2005;55:312–325. doi: 10.1111/j.1365-2958.2004.04391.x. [DOI] [PubMed] [Google Scholar]
- R5.Brenner S.E. Errors in genome annotation. Trends Genet. 1999;15:132–133. doi: 10.1016/s0168-9525(99)01706-0. [DOI] [PubMed] [Google Scholar]
- R6.Bult C.J., White O., Olsen G.J., et al. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii . Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- R7.Chen L., Skovgaard M., et al. The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R8.Ciccarelli F.D., Doerks T., Von mering C., Creevey C.J., Snel B., Bork P. Toward automatic reconstruction of a highly resolved tree of life. Science. 2006;311:1283–1287. doi: 10.1126/science.1123061. [DOI] [PubMed] [Google Scholar]
- R9.Cobucci-Ponzano B., Trincone A., Giordano A., Rossi M., Moracci M. Identification of an archaeal a-L-fucosidase encoded by an interrupted gene. Production of a functional enzyme by mutations mimicking programmed –1 frameshifting. J. Biol. Chem. 2003;278:14622–14631. doi: 10.1074/jbc.M211834200. [DOI] [PubMed] [Google Scholar]
- R10.Cobucci-Ponzano B., Rossi M., Moracci M. Recoding in archaea. Mol. Microbiol. 2005;55:339–348. doi: 10.1111/j.1365-2958.2004.04400.x. [DOI] [PubMed] [Google Scholar]
- R11.Cole S.T., Eiglmeier K., Parkhill J., et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. doi: 10.1038/35059006. [DOI] [PubMed] [Google Scholar]
- R12.Daubin V., Ochman H. Bacterial genomes as new gene homes: the genealogy of ORFans in E. coli. Genome Res. 2004;14:1036–1042. doi: 10.1101/gr.2231904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R13.Fitz-Gibbon S.T., Ladner H., Kim U.J., Stetter K.O., Simon M.I., Miller J.H. Genome sequence of the hyperthermophilic crenarchaeon Pyrobaculum aerophilum . Proc. Natl. Acad. Sci. USA. 2002;99:984–989. doi: 10.1073/pnas.241636498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Fricke W.F., Seedorf H., Henne A., Kruer M., Liesegang H., Hedderich R., Gottschalk G., Thauer R.K. The genome sequence of Methanosphaera stadtmanae reveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J. Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R15.Galperin M.Y., Koonin E.V. ‘Conserved hypothetical’ proteins: prioritization of targets for experimental study. Nucleic Acids Res. 2004;32:5452–5463. doi: 10.1093/nar/gkh885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Harrison P.M., Echols N., Gerstein M.B. Digging for dead genes: an analysis of the characteristics of the pseudogene population in the Caenorhabditis elegans genome. Nucleic Acids Res. 2001;29:818–830. doi: 10.1093/nar/29.3.818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.James C.M., Ferguson T.K., Leykam J.F., Krzycki J.A. The amber codon in the gene encoding the monomethylamine methyltransferase isolated from Methanosarcina barkeri is translated as a sense codon. J. Biol. Chem. 2001;276:34252–34258. doi: 10.1074/jbc.M102929200. [DOI] [PubMed] [Google Scholar]
- R18.Kawashima T., Amano N., Koike H., et al. Archaeal adaptation to higher temperatures revealed by genomic sequence of Thermoplasma volcanium . Proc. Natl. Acad. Sci. USA. 2000;97:14257–14262. doi: 10.1073/pnas.97.26.14257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Kumar S., Tamura K., Nei M. MEGA3: integrated software for Molecular Evolutionary Genetics Analysis and sequence alignment. Brief. Bioinform. 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- R20.Lawrence J.G., Hendrix R.W., Casjens S. Where are the pseudogenes in bacterial genomes? Trends Microbiol. 2001;9:535–540. doi: 10.1016/s0966-842x(01)02198-9. [DOI] [PubMed] [Google Scholar]
- R21.Lerat E., Ochman H. Y –F: exploring the outer limits of bacterial pseudogenes. Genome Res. 2004;14:2273–2278. doi: 10.1101/gr.2925604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R22.Lerat E., Ochman H. Recognizing the pseudogenes in bacterial genomes. Nucleic Acids Res. 2005;33:3125–3132. doi: 10.1093/nar/gki631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R23.Li W.H., Gojobori T., Nei M. Pseudogenes as a paradigm of neutral evolution. Nature. 1981;292:237–239. doi: 10.1038/292237a0. [DOI] [PubMed] [Google Scholar]
- R24.Liu Y., Harrison P.M., Kunin V., Gerstein M. Comprehensive analysis of pseudogenes in prokaryotes: widespread gene decay and failure of putative horizontally transferred genes. Genome Biol. 2004;5:R64. doi: 10.1186/gb-2004-5-9-r64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R25.Markowitz V.M., Korzeniewski F., Palaniappan K., et al. The integrated microbial genomes (IMG) system. Nucleic Acids Res. 2006;34:D344–348. doi: 10.1093/nar/gkj024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R26.Mira A., Ochman H., Moran N.A. Deletional bias and the evolution of bacterial genomes. Trends Genet. 2001;17:589–596. doi: 10.1016/s0168-9525(01)02447-7. [DOI] [PubMed] [Google Scholar]
- R27.Ochman H., Davalos L.M. The nature and dynamics of bacterial genomes. Science. 2006;311:1730–1733. doi: 10.1126/science.1119966. [DOI] [PubMed] [Google Scholar]
- R28.Perrodou E., Deshayes C., Muller J., Schaeffer C., Van dorsselaer A., Ripp R., Poch O., Reyrat J.M., Lecompte O. ICDS database: interrupted CoDing sequences in prokaryotic genomes. Nucleic Acids Res. 2006;34:D338–343. doi: 10.1093/nar/gkj060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Ruepp A., Graml W., Santos-martinez M.L., et al. The genome sequence of the thermoacidophilic scavenger Thermoplasma acidophilum . Nature. 2000;407:508–513. doi: 10.1038/35035069. [DOI] [PubMed] [Google Scholar]
- R30.Schleper C., Jurgens G., Jonuscheit M. Genomic studies of uncultivated archaea. Nat. Rev. Microbiol. 2005;3:479–488. doi: 10.1038/nrmicro1159. [DOI] [PubMed] [Google Scholar]
- R31.She Q., Singh R.K., Confalonieri F., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R32.Snel B., Bork P., Huynen M. Genome evolution. Gene fusion versus gene fission. Trends Genet. 2000;16:9–11. doi: 10.1016/s0168-9525(99)01924-1. [DOI] [PubMed] [Google Scholar]
- R33.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.Toh H., Weiss B.L., Perkin S.A., Yamashita A., Oshima K., Hattori M., Aksoy S. Massive genome erosion and functional adaptations provide insights into the symbiotic lifestyle of Sodalis glossinidius in the tsetse host. Genome Res. 2006;16:149–156. doi: 10.1101/gr.4106106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R35.Torrents D., Suyama M., Zdobnov E., Bork P. A genome-wide survey of human pseudogenes. Genome Res. 2003;13:2559–2567. doi: 10.1101/gr.1455503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R36.van der Woude M.W., Baumler A.J. Phase and antigenic variation in bacteria. Clin. Microbiol. Rev. 2004;17:581–611. doi: 10.1128/CMR.17.3.581-611.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R37.Vanin E.F. Processed pseudogenes: characteristics and evolution. Annu. Rev. Genet. 1985;19:253–272. doi: 10.1146/annurev.ge.19.120185.001345. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1. List of archaeal positional pseudogenes.
Supplementary Table S2. List of archaeal non-positional pseudogenes.
Supplementary Figure S1. Numbers of pseudogenes and their mechanisms of inactivation in each of the archaeal genomes considered.
Supplementary Table S3. The GC percentages of gene sets that do and do not contain a pseudogene counterpart.