Abstract
Reverse gyrase, an enzyme of uncertain funtion, is present in all hyperthermophilic archaea and bacteria. Previous phylogenetic studies have suggested that the gene for reverse gyrase has an archaeal origin and was transferred laterally (LGT) to the ancestors of the two bacterial hyperthermophilic phyla, Thermotogales and Aquificales. Here, we performed an in-depth analysis of the evolutionary history of reverse gyrase in light of genomic progress. We found genes coding for reverse gyrase in the genomes of several thermophilic bacteria that belong to phyla other than Aquificales and Thermotogales. Several of these bacteria are not, strictly speaking, hyperthermophiles because their reported optimal growth temperatures are below 80 °C. Furthermore, we detected a reverse gyrase gene in the sequence of the large plasmid of Thermus thermophilus strain HB8, suggesting a possible mechanism of transfer to the T. thermophilus strain HB8 involving plasmids and transposases. The archaeal part of the reverse gyrase tree is congruent with recent phylogenies of the archaeal domain based on ribosomal proteins or RNA polymerase subunits. Although poorly resolved, the complete reverse gyrase phylogeny suggests an ancient acquisition of the gene by bacteria via one or two LGT events, followed by its secondary distribution by LGT within bacteria. Finally, several genes of archaeal origin located in proximity to the reverse gyrase gene in bacterial genomes have bacterial homologues mostly in thermophiles or hyperthermophiles, raising the possibility that they were co-transferred with the reverse gyrase gene. Our new analysis of the reverse gyrase history strengthens the hypothesis that the acquisition of reverse gyrase may have been a crucial evolutionary step in the adaptation of bacteria to high-temperature environments. However, it also questions the role of this enzyme in thermophilic bacteria and the selective advantage its presence could provide.
Keywords: adaptation, Archaea, evolution, genome context, HGT, hyperthermophily, plasmid, thermophily, Thermus thermophilus
Introduction
Reverse gyrase, an enzyme first discovered in the hyperthermophilic archaeon Sulfolobus acidocaldarius, is formed by the combination of an N-terminal helicase module and a C-terminal topoisomerase module (see Declais et al. 2000 for a review). Comparative genomic analysis performed 4 years ago showed that reverse gyrase was at that time the only protein specific for hyperthermophiles (i.e., present in all hyperthermophiles and absent in all non-hyperthermophiles) (Forterre 2002). In this study, the threshold between thermophiles (which lack the reverse gyrase gene) and hyperthermophiles (harboring a reverse gyrase gene) was set to 80 °C, in agreement with the original definition given by Carl Stetter (1989) for hyperthermophiles. Recently, it has been claimed that reverse gyrase is not essential for hyperthermophilic life since a reverse gyrase knock-out mutant of Thermococcus kodakarensis grows (although poorly) at 90 °C (Atomi et al. 2004). However, growth of the mutant is significantly reduced compared with the wild type, and the difference increases with increasing temperature. Furthermore, unlike the wild type, the reverse gyrase mutant cannot grow above 90 °C (Atomi et al. 2004).
The exact role of reverse gyrase in vivo remains obscure. It is usually suggested that it protects DNA against thermodenaturation (unwinding of the double helix), since it introduces positive supercoils in vitro (Kikuchi and Asai 1984). However, topologically closed DNA is extremely resistant to denaturation at high temperature (at least up to 107 °C) (Marguet and Forterre 1994) and some hyperthermophiles have negatively supercoiled plasmids, indicating that DNA positive supercoiling is not essential for hyperthermophily (Guipaud et al. 1997, Lopez-Garcia and Forterre 2000). Recent in vivodata suggest that reverse gyrase is more likely involved in the protection of DNA against chemical degradation (such as depurination) or other forms of DNA damage (such as DNA breakage) occurring at high temperature (Kampmann and Stock 2004, Napoli et al. 2004).
The systematic presence of reverse gyrase in hyperthermophiles and its critical role for optimal growth at high temperatures suggest that the origin of this enzyme may have coincided with the appearance of modern hyperthermophilic organisms. The basal position of hyperthermophilic archaea and bacteria in the classical rRNA tree of life has led to the hypothesis that hyperthermophily is an ancestral feature and that the Last Universal Common Ancestor (LUCA) itself was hyperthermophilic (Stetter 1996, Nisbet and Fowler 1996). However, our previous work on reverse gyrase does not support this idea, suggesting instead that reverse gyrase originated in the archaeal domain and was subsequently transferred to the ancestors of Thermotogales and Aquificales (the two bacterial phyla containing hyperthermophiles) by two distinct lateral gene transfer (LGT) events (Forterre et al. 2000, Forterre 2002). This conclusion was inferred from the mixing of archaeal and bacterial lineages in an unrooted reverse gyrase phylogeny, and by the genomic context of bacterial reverse gyrase genes that appeared to be surrounded by genes of potential archaeal origin (Forterre et al. 2000). Other genomic studies have strengthened the hypothesis that the ability of bacterial hyperthemophiles to grow at high temperatures could be a derived trait, whose acquisition was facilitated by extensive LGTs from archaea (Aravind et al. 1998, Nelson et al. 1999, Koonin et al. 2001). The idea that reverse gyrase originated in archaea agrees with the suggestion that all modern archaea derive from a common hyperthermophilic ancestor (Forterre et al. 2002), but that LUCA was either a mesophile or a moderate thermophile (Galtier et al. 1999, Brochier and Philippe 2002).
The analyses reported in our 2002 paper were performed at a time when the number of completely sequenced archaeal and bacterial genomes was still limited (Forterre 2002). In recent years, this number has increased enormously, especially in the bacterial domain (i.e., 27 archaeal and over 327 bacterial genomes are completely sequenced, while 56 archaeal and 987 bacterial genomes are being sequenced, according to the Genomes Online Database http://www.genomesonline.org/). We, thus, performed a new systematic search for reverse gyrase genes in public databases to gain further insight into the evolutionary history of this enzyme and its implication for the origin of hyperthermophiles. This updated analysis revealed the presence of reverse gyrase in several thermophilic bacteria that are not hyperthermophiles, stricto sensu. Most of these bacteria belong to phyla other than Aquificales and Thermotogales. Our phylogenetic analysis suggests a complex history for the reverse gyrase gene, with mostly vertical inheritance in the archaeal domain followed by different LGT events from archaea to bacteria and between different bacterial lineages, or simply between different bacterial lineages.
Materials and methods
Construction of the datasets
Twenty-two reverse gyrase sequences were retrieved from the National Center for Biotechnology Information (NCBI) by BLASTP searches in the nonredundant (nr) database at the NCBI (http://www.ncbi.nlm.nih.gov). The sequences of Sulfurihydrogenibium azorense, Caldicellulosiruptor saccharolyticus, Dictyoglomus thermophilum, Thermodesulfobacterium commune and Thermotoga neapolitana were obtained by TBLASTN searches (Altschul et al. 1990) from the ongoing NCBI genome project database and the Thermococcus gammatolerans reverse gyrase sequence was obtained from Yvan Zivanovic and Fabrice Confalonieri (Université Paris-Sud, personal communication). No reverse gyrase sequence from the environmental database was found by TBLASTN searches (Altschul et al. 1990). The 32 sequences were aligned with the CLUSTAL W program (Thompson et al. 1994). The alignment was manually refined using the MUST package (Philippe 1993). Regions where alignment was ambiguous were removed and a total of 651 positions were finally kept for phylogenetic reconstruction.
The same approach was used to reconstruct the other datasets.
Phylogenetic analyses
Maximum likelihood (ML) phylogenetic analyses were performed by PHYML (Guindon and Gascuel 2003) using a JTT model with a gamma correction (eight discrete classes, an estimated alpha parameter and an estimated proportion of invariant sites). Bootstrap analyses (1000 replicates) were performed using PHYML. Bayesian analyses were performed using MrBayes v.3.0b4 (Ronquist and Huelsenbeck 2003) with a mixed model of amino acid substitution and a gamma correction (eight discrete categories plus a proportion of invariant sites) to take into account among-site rate variations. MrBayes was run with four chains for 1 million generations and trees were sampled every 100 generations. To construct the consensus tree, the first 1500 trees were discarded as ‘‘burnin.’’
All the datasets and the phylogenetic trees are available on request from C. Brochier-Armanet.
Genomic context analysis
We retrieved the list of the ten open reading frames (ORFs) surrounding the reverse gyrase genes from the complete genomes at the NCBI. For each ORF, we performed BLASTP searches (Altschul et al. 1990) to determine the nature of the closest BLAST hit (archaeal, eukaryal or bacterial). Next, the taxonomic distribution of the BLASTP output homologues was manually examined to classify the ORF into one of three categories: ORF having (1) archaeal homologues only; (2) bacterial homologues only; and (3) both archaeal and bacterial homologues.
Results and discussion
New homologues of reverse gyrase
Our screening for reverse gyrase genes by BLASTP in the nr database and by TBLASTN in the microbial genomes database at the NCBI, revealed reverse gyrase genes in the genomes of three recently sequenced hyperthermophilic archaea, Nanoarchaeum equitans, Thermococcus gammatolerans and Thermococcus kodakarensis and in 11 bacteria, Thermoanaerobacter tengcongensis, Thermodesulfobacterium commune, five Thermotogales (T. neapolitana, Thermotoga petrophila, Thermotoga naphthophila and two Thermotoga sp.), Thermus thermophilus strain HB8, S. azorense, Dictyoglomus thermophilum and Caldicellulosiruptor saccharolyticus. Some of these bacteria are borderline hyperthermophilic, because their optimal growth temperature is close to 80 °C (Table 1) and they can grow above this temperature. However, several others (C. saccharolyticus, S. azorense, T. commune and T. thermophilus HB8) have maximum growing temperatures well below the 80 °C theoretical threshold (Table 1). This indicates that reverse gyrase can no longer be considered as a specific hyperthermophilic protein stricto sensu. However, it remains present in all hyperthermophiles without exception, whereas it is dispensable in thermophilic bacteria and archaea, e.g., T. thermophilus strain HB27 and Methanothermobacter thermoautotrophicum lack reverse gyrase. The presence of reverse gyrase genes in several thermophilic bacterial genomes was not surprising because reverse gyrase activity has been detected previously in crude extracts of several thermophilic (but not hyperthermophilic) bacteria (Bouthier de la tour et al. 1991). However, the phylogenomic distribution of reverse gyrase still strongly suggests a critical role for this enzyme at high temperatures because we detected no reverse gyrase gene in any mesophilic bacteria or archaea, even though several hundred mesophilic prokaryotic genomes are now available. The present analysis indicates that bacterial reverse gyrases (previously known in only two phyla, Thermotogales and Aquificales, that include mostly thermophilic or hyperthermophilic species) are more widespread in the bacterial domain than previously thought, occurring in four other bacterial phyla; namely, the Firmicutes (Thermoanaerobacter tengcongensis and Caldicellulosiruptor saccharolyticus),the Thermodesulfobacteria (T. commune), the Dictyoglomales (Dictyoglomus thermophilum) and the Thermus/Deinococcus group (T. thermophilus strain HB8) (Table 1). Unlike the Thermotogales and Aquificales, the Firmicutes appear to be almost entirely mesophilic: Thermoanaerobacter tengcongensis and Caldicellulosiruptor saccharolyticus being the only members of the phylum containing reverse gyrase out of 198 strains whose genomes have been sequenced.
Table 1.
Prokaryotes containing a gene coding for the reverse gyrase. The asterisk (*) indicates a new reverse gyrase found since our last study in 2002. Abbreviations: T = thermophilic; and H = hyperthermophilic.
| Organism | Taxonomic affiliation | Top | Range | Locus | Length | Remarks |
| Aeropyrum pernix K1 | Archaea; Crenarchaeota; Thermoprotei; Desulfurococcales; Desulfurococcaceae | 90–95 | H | APE1376/APE1340 | 1222/1376 | |
| Aquifex aeolicus VF5 | Bacteria; Aquificae; Aquificales; Aquificaceae | 96 | H | AQ886/AQ1159 | 1146/1177 | |
| Archaeoglobus fulgidus DSM 4304 | Archaea; Euryarchaeota; Archaeoglobi; Archaeoglobales; Archaeoglobaceae | 83 | H | AF1024 | 1054 | |
| Caldicellulosiruptor saccharolyticus | Bacteria; Firmicutes; Clostridia; Syntrophomonadaceae; Caldicellulosiruptor | 65 | T | Incomplete | 1121 | |
| Dictyoglomus thermo-philum H-6–12* | Bacteria; Dictyoglomi; Dictyoglomales; Dictyoglomaceae | 78 | T | Incomplete | 1210 | |
| Methanocaldococcus jannaschii DSM 2661 | Archaea; Euryarchaeota; Methanococci; Methanococcales; Methanocaldococcaceae | 85 | H | MJ1512 | 1613 | Intein-containing |
| Methanopyrus kandleri AV19 | Archaea; Euryarchaeota; Methanopyri; Methanopyrales; Methanopyraceae | 98 | H | MK0049/MK0289 | 1221/358 | B/A subunit |
| Nanoarchaeum equitans Kin4-M* | Archaea; Nanoarchaeota | 90 | H | NEQ318/NEQ434 | 701/575 | Topoisomerase/helicase domain |
| Pyrobaculum aerophilum str. IM2 | Archaea; Crenarchaeota; Thermoprotei; Thermoproteales; Thermoproteaceae | 100 | H | PAE1108 | 1228 | |
| Pyrococcus abyssi GE5 | Archaea; Euryarchaeota; Thermococci; Thermococcales; Thermococcaceae | 103 | H | PAB2423 | 1214 | |
| Pyrococcus furiosus DSM 3638 | Archaea; Euryarchaeota; Thermococci; Thermococcales; Thermococcaceae | 100 | H | PF0495 | 1214 | |
| Pyrococcus horikoshii OT3 | Archaea; Euryarchaeota; Thermococci; Thermococcales; Thermococcaceae | 98 | H | PH0800 | 1624 | Intein-containing |
| Sulfolobus acidocaldarius | Archaea; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae | 70–75 | T | Incomplete | 1248 | |
| Sulfolobus shibatae | Archaea; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae | 75–80 | H | Incomplete | 1166 | |
| Sulfolobus solfataricus P2 | Archaea; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae | 85 | H | SSO0963/SSO0420 | 1166/1242 | |
| Sulfolobus tokodaii str. 7 | Archaea; Crenarchaeota; Thermoprotei; Sulfolobales; Sulfolobaceae | 80 | H | ST0374/ST1290 | 1233/1156 | |
| Sulfurihydrogenibium azorense Az-Fu1* | Bacteria; Aquificae; Aquificales; Hydrogenothermaceae | 68 | T | Incomplete | 1200 | |
| Thermoanaerobacter tengcongensis MB4* | Bacteria; Firmicutes; Clostridia; Thermoanaerobacteriales; Thermoanaerobacteriaceae | 75 | T | TTE1745 | 1117 | |
| Thermococcus gammatolerans* | Archaea; Euryarchaeota; Thermococci; Thermococcales; Thermococcaceae | 88 | H | Incomplete | 1226 | |
| Thermococcuskodakaraensis KOD1* | Archaea; Euryarchaeota; Thermococci; Thermococcales; Thermococcaceae | 85 | H | Incomplete | 1711 | Intein-containing |
| Thermodesulfobacterium commune DSM 2178* | Bacteria; Thermodesulfobacteria; Thermodesulfobacteriales; Thermodesulfobacteriaceae | 70 | T | Incomplete | 1200 | |
| Thermotoga maritima MSB8 | Bacteria; Thermotogae; Thermotogales; Thermotogaceae | 80 | H | TM0173 | 1104 | |
| Thermotoga naphthoplia | Bacteria; Thermotogae; Thermotogales; Thermotogaceae | 80 | H | Incomplete | 1104 | |
| Thermotoga neapolitana DSM 4359* | Bacteria; Thermotogae; Thermotogales; Thermotogaceae | 70–75 | T | Incomplete | 1209 | |
| Thermotoga petrophila | Bacteria; Thermotogae; Thermotogales; Thermotogaceae | 80 | H | Incomplete | 1104 | |
| Thermus thermophilus HB8* | Bacteria; Deinococcus-Thermus; Deino cocci; Thermales; Thermaceae | 68 | T | TTHB172 | 1116 | Gene localized on a plasmid |
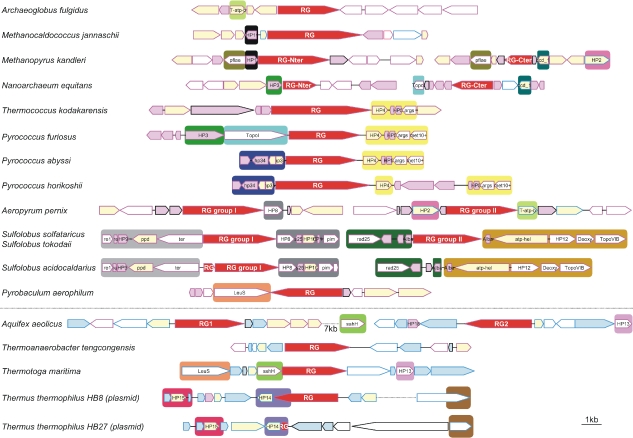
The discovery of a gene coding for a reverse gyrase in T. thermophilus strain HB8 was unexpected, because no reverse gyrase gene was found in the genome of its close relative T. thermophilus strain HB27 (Henne et al. 2004). The reverse gyrase of the T. thermophilus strain HB8 is encoded by a large plasmid closely related to the plasmid, pTT27, of Strain HB27. To our knowledge, this is the first report of a plasmid carrying a gene encoding reverse gyrase. Although T. thermophilus HB8 harbors a reverse gyrase, it was shown previously that cell-free extracts of this strain do not exhibit reverse gyrase activity, and the small plasmid pTT8 present in T. thermophilus HB8 is negatively supercoiled (Charbonnier and Forterre 1994). This indicates that, if the reverse gyrase is functional, the regular gyrase present predominates in determining the intracellular topological state of DNA in T. thermophilus HB8, as previously shown in Thermotoga maritima and Archaeoglobus fulgidus (Guipaud et al. 1997, Lopez-Garcia and Forterre 2000). TBLASTN analyses against the pTT27 plasmid of strain HB27 revealed a 126-amino-acid-long hypothetical protein, located at the TTP0124 locus, almost identical to the C-terminal end of reverse gyrase encoded by the large plasmid of strain HB8 (Figure 1; Appendix 1). This indicates that reverse gyrase was probably encoded in the ancestor of the two plasmids and then partially deleted in the large plasmid present in strain HB27. Despite accurate searches, we found no region in the large plasmid (or in the chromosome) of strain HB27 corresponding to the N-terminus of reverse gyrase. The flanking regions around these reverse gyrase gene sequences and the TTP0124 loci are poorly conserved between the two plasmids (Figure 1), despite the close relationship between their host strains. This suggests that these regions are prone to recombination/rearrangement events that could have been facilitated by the large number of transposases encoded by these plasmids (not shown). These transposases belong to a few families with representatives in both bacterial and archaeal genomes (not shown), indicating that they could have been involved in the transfer of the reverse gyrase gene, between either archaea and bacteria or different bacterial lineages. It should be interesting to compare the two strains of T. thermophilus, HB8 and HB27, to study the exact function of reverse gyrase in vivo, because only one of these closely related strains contains a potentially functional reverse gyrase.
Figure 1.
Genomic organization of the regions surrounding reverse gyrase genes. Blue boxes indicate genes having exclusively bacterial or bacterial and eukaryotic homologues; pink boxes indicate genes having exclusively archaeal or archaeal and eukaryotic homologues; yellow boxes indicate genes having both archaeal and bacterial homologues; and empty boxes indicate genes with homologues in the three domains of life. The color of the box frames indicates the nature of each gene's closest homologue by BLASTP searches (blue = bacterial; and pink = archaeal). Large boxes with the same color surrounding genomic fragments indicate regions sharing homologous genes. The scale bar represents approximately 1000 nucleotides. Gene name abbreviations: T-atp-b = ABC transporter, ATP-binding protein (COG1131); HP1 = hypothetical protein (COG1590); pflae = pyruvate-formate lyase-activating enzyme (COG1180); dcd_1 = deoxycytidine deaminase (COG0717); HP2 = uncharacterized membrane protein (COG3174); HP3 = hypothetical protein (COG2433); TopoI = DNA topoisomerase I (COG0550); HP4 = predicted transcriptional regulators (COG1378); HP5 = uncharacterized Zn-finger containing protein (COG1645); args = arginase 1-like protein (COG0010); met10+ = met-10+ protein (COG2520); HP6 = uncharacterized membrane-associated protein/domain (COG2512); HP7 = predicted DNA-binding proteins with PD1-like DNA-binding motif (COG1661); HP8 = predicted amidohydrolase (COG0388); re1 = DNA-directed RNA polymerase, subunit E (COG1095); re = DNA-directed RNA polymerase, subunit E (COG2093); HP9 = hypothetical protein (COG1909); ppd = phosphonopyruvate decarboxylase (COG3635); ter = transitional endoplasmic reticulum ATPase (COG0464); s25 = SSU ribosomal protein S25E (COG4901); HP10 = membrane protein implicated in regulation of membrane protease activity (COG1585); HP11 = predicted transcriptional regulators containing the CopG/Arc/MetJ DNA-binding domain (COG3609); pim = protein L-isoaspartate methyltransferase (COG2519); rad25 = DNA repair protein rad56 (COG1061); Alba = archaeal DNA-binding protein (COG1581); atp-heli = ATP-dependent helicase (COG1201); HP12 = hypothetical protein (COG1690); Deoxy = deoxyhypusine synthase (COG1899); TopoVIB = DNA topoisomerase VI subunit B (COG1389); LeuS = leucyl-tRNA synthetase (COG0495); sahH = adenosylhomocysteinase (COG0499); HP13 = hypothetical protein (COG1578); HP14 = predicted metal-dependent membrane protease (COG1266); HP15 = hypothetical protein (COG1604); and HP16 = hypothetical protein (COG1458).
Phylogeny of the reverse gyrase
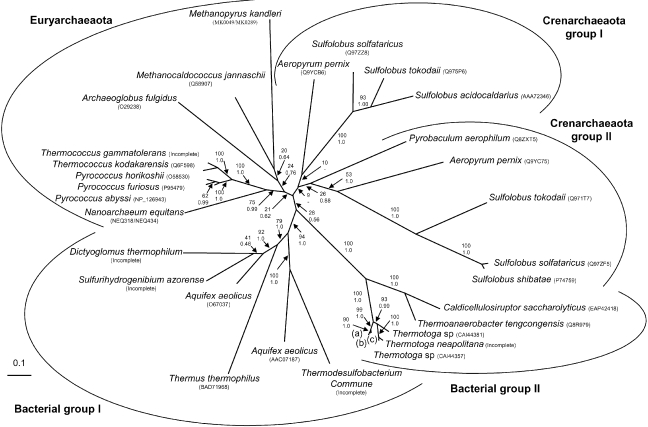
Archaeal and bacterial reverse gyrases form two groups (albeit weakly supported, bootstrap value (BV) = 28% and posterior probability (PP) = 0.56) separated by a very short branch in our ML (Figure 2) and Bayesian (not shown) phylogenetic trees of the 32 reverse gyrase sequences. A previous phylogenetic analysis suggested that reverse gyrase genes were independently acquired twice by bacteria, because T. maritima and Aquifex aeolicus sequences emerged within the Crenarchaeota and Euryarchaeota, respectively (Forterre et al. 2000). In our new phylogeny (Figure 2), bacterial and archaeal reverse gyrases are now both monophyletic. The relationships between archaeal reverse gyrases, albeit poorly supported, appeared to be in agreement with those observed in ribosomal and transcription protein concatenation trees, or in transcription protein concatenation trees alone (Brochier et al. 2005a, 2005b). The Crenarchaeota/Euryarchaeota dichotomy is recovered as well as the emerging order of various lineages within the two phyla: Pyrobaculum aerophilum emerges first in the crenarchaeotal part of the tree, followed by the separation of Aeropyrum pernix and Sulfolobales, whereas Thermococcales plus Nanoarchaeum equitans are the first emerging Euryarchaeota, followed by the separation of the A. fulgidus and the methanogens Class I (Bapteste et al. 2005). This tree suggests also that one duplication of the reverse gyrase gene occurred in the ancestor of hyperthermophilic Crenarchaea and that one of the two copies was subsequently lost in both P. aerophilum and S. acidocaldarius, because only one copy of the reverse gyrase gene is found in the completely sequenced genomes of these archaea (Figure 2).
Figure 2.
Maximum likelihood unrooted phylogenetic tree of reverse gyrase. The tree was calculated by PHYML (JTT model, gamma correction (eight discrete classes), an estimated alpha parameter and an estimation of the proportion of invariant positions). Numbers > 0 at nodes are bootstrap values calculated from 1000 replicates by PHYML and numbers < 0 are posterior probabilities computed by MrBayes. The scale bar represents the number of substitutions per site, and (a) Thermotoga naphthophila (CA144289); (b) Thermotoga maritima (O51934); and (c) Thermotoga petrophila (CA144306).
The reverse gyrase of the archaeon Nanoarchaeum equitans is atypical because its two modules (helicase and topoisomerase) are encoded by distinct, non-contiguous genes (Waters et al. 2003). It was suggested that this organization may correspond to the ancestral form of reverse gyrase, according to the early emergence of N. equitans at the base of the archaeal domain in both SSU rRNA (Huber et al. 2002) and concatenated ribosomal protein trees (Waters et al. 2003). However, this archaeon emerged as a sister group of Thermococcales in the reverse gyrase tree (BV = 75% and PP = 0.99, Figure 2) in agreement with a revised position of N. equitans based on a recent and more refined phylogenetic analysis of a concatenation of ribosomal proteins (Brochier et al. 2005b). The same position was observed in the phylogenies restricted to the helicase domain or to the topoisomerase domain (not shown). This result suggests that the presence of two distinct genes coding the reverse gyrase in N. equitans is not a plesiomorphy (ancestral character state) but rather an apomorphy (derived character state) that likely resulted in a secondary split event specific to the N. equitans lineages (as observed for M. kandleri).
As in our previous analysis, the bacterial reverse gyrase sequences fall into two clusters, Group I and Group II, supported by high bootstrap values and posterior probabilities (BV = 94% and PP = 1.0 and BV = 100% and PP = 1.0, respectively). Group I is composed of the two copies of A. aeolicus reverse gyrase and those of T. commune, T. thermophilus, D. thermophilum and S. azorense, whereas Group II clusters the reverse gyrases of the six Thermotogales (i.e., T. maritima, T. neapolitana, T. petrophila, T. naphthophila and the two Thermotaga sp.) and the two Clostridiales (T. tengcongensis and C. saccharolyticus).
The monophyly of archaeal and bacterial reverse gyrases in our new tree is consistent with the idea of a thermophilic or hyperthermophilic last common ancestor of bacteria and archaea containing a reverse gyrase-coding gene; the loss of the reverse gyrase-coding gene occurring secondarily in some thermophilic species and in all mesophilic species. However, in contradiction with this hypothesis, it should be noticed that the distances between archaeal and bacterial reverse gyrase sequences are not significantly higher on the average than those between archaeal reverse gyrases or between bacterial reverse gyrases (average distances of 1.3; 1.1 and 0.9 substitutions per site, respectively). In other words, there are not two canonical versions of reverse gyrase, one bacterial and one archaeal, strictosensu (Woese et al. 2000), but only a single version. Indeed, the monophyly of each domain is only weakly supported (BV = 28% and PP = 0.56). In our opinion, given the short size of the branch separating the two domains, it is unlikely that bacterial and archaeal reverse gyrases originated by speciation from an ancestral reverse gyrase present in the last common ancestor of archaea and bacteria, and we still favor the idea that reverse gyrase originated first in one of the two prokaryotic domains and was transferred later on to the other.
The hypothesis of a transfer from archaea to bacteria is supported by the contrast between the classical phylogeny observed for the archaeal part of the tree and the unusual aspect of the bacterial part. Whereas the archaeal reverse gyrase tree is similar to the canonical 16S rRNA, ribosomal protein or RNA polymerase subunit trees, the bacterial reverse gyrase tree is atypical by virtue of the unusual and well supported bacterial relationships inside the two reverse gyrase clusters (especially inside Group I). In our opinion, these relationships are better explained if we suppose that one of a small number of LGTs from archaea was followed by redistribution as a consequence of a few LGT events between bacterial lineages, rather than a single acquisition from the bacterial ancestor followed by strict vertical inheritance and numerous secondary losses. In particular, it is striking that the two copies of A. aeolicus reverse gyrase are no longer clustered together in our new phylogeny, but are instead grouped with reverse gyrases from species belonging to different bacterial phyla (i.e., D. thermophilum, T. commune and T. thermophilus). This can be explained by either an early duplication of the reverse gyrase gene in an Aquificales ancestor, followed by independent LGTs of each copy in different bacteria, or by two independent acquisitions.
Genomic context analysis of reverse gyrase genes
Based on best BLAST hit analyses, it was previously reported that the genomes of T. maritima and A. aeolicus contained genes coding for reverse gyrases, which were surrounded by genes of possible archaeal origin, supporting the hypothesis that they were recruited via LGTs from the Archaea (Forterre et al. 2000). In this study, we performed a more accurate and systematic analysis of the genome context in both archaea and bacteria for all the reverse gyrase genes detected (Figure 1). A rapid screening revealed that the genome context around the archaeal reverse gyrase genes is not conserved, except for closely related species. However, whereas the genomic context is well conserved in Sulfolobales (except for the loss of the reverse gyrase gene (Group II) in S. acidocaldarius), important rearrangements can already be observed between the four Thermococcales. The non-conservation of the reverse gyrase gene context suggests the absence of selective pressures for functional or regulatory purposes. However, despite the rapid genome shuffling around reverse gyrase genes, in several species we found a few conserved genes whose presence fits well with the reverse gyrase phylogeny. This is the case for the presence of the hypothetical protein 1 (HP1, COG1590, Figure 1) around the Methanopyrus kandleri and Methanocaldococcus jannaschii reverse gyrase genes that support the clustering of those two archaea in the phylogeny (Figure 2). The presence of hypothetical protein 8 (HP8, COG0388, Figure 1) supports the clustering of the Group I reverse gyrase genes found in A. pernix and Sulfolobus species (Figure 2), whereas the presence of the Type IA DNA topoisomerase and the hypothetical proteins 3 (TopoI, COG 0550 and HP3, COG2433, Figure 1) supports the grouping together of N. equitans and Thermococcales (Figure 2). Finally, the presence of two other genes, T-atp-b (COG 1131) and dcd1 (COG0717), in the reverse gyrase genomic contexts of species that do not form a clade in our tree (A. pernix, M. kandleri and N. equitans) can be easily explained if these genes were in the genomic context of reverse gyrase gene in the common ancestor of the three species. We found one gene (leuS) present both in the genome context of a crenarchaeon, P. aerophilum, and of a bacterium, T. maritima. This could indicate that reverse gyrase from the bacterial Group II originated from a crenarchaeon, as suggested in our previous work (Forterre et al. 2000). We are aware that these conjectures from genome context analysis are weak, especially if the contexts evolve rapidly, as is likely the case here. However, we feel that they are worth mentioning because their existence suggests that the sequencing of more archaeal genomes should permit us to test their validity by making links between the genomic context of species whose genomes are presently available.
The eight genes of possible archaeal origin identified by BLAST analyses in our previous analysis around the T. maritima and A. aeolicus reverse gyrase genes were again recovered because, despite the much higher number of genomic sequences in current databases, they still have an archaeal homologue as closest hit in BLASTP searches (Figure 1). We also identified three genes of possible archaeal origin surrounding the reverse gyrase gene of T. tengcongensis. Although we could detect some genes with a closest bacterial hit around reverse gyrase genes in archaeal genomes, they were less abundant. In particular, we found no genes with a bacterial close BLAST hit in the proximity of archaeal reverse gyrases genes from Thermococcales, Sulfolobales and A. fulgidus. However, such an approach is prone to error since close BLAST hits do not always correspond to phylogenetic neighbors (Koski and Golding 2001) and do not permit us to identify possible LGT from bacteria to archaea. Then, we investigated the phylogenies of all genes with an archaeal best blast hit located in proximity to reverse gyrase in bacterial genomes. The resulting phylogenetic trees were often poorly resolved or difficult to interpret because of numerous duplications, losses and LGTs, or because of LGTs alone (not shown). However, some of them not only confirmed an archaeal origin, but also revealed interesting patterns (Appendices 2–4). This is the case of the gene encoding the hypothetical protein 13 (HP13, COG1578, Figure 1) that is close to the reverse gyrase gene both in A. aeolicus and Thermotogales. This gene is also present in D. thermophilum, but the genomic context cannot be analyzed in this case because the genome of this species has not yet been completely sequenced.Both the genomic distribution and the phylogeny of HP13 support the hypothesis of an archaeal origin for this protein and its subsequent transfer to a few bacteria (Appendix 2). Indeed, HP13 is present in most Euryarchaeota and in the crenarchaeon, P. aerophilum, whereas it is only present sporadically in bacteria (i.e., in a subset of Proteobacteria, a cyanobacterium, and a low GC Gram positive, in addition to the three thermophilic/hyperthermophilic bacteria D. thermophilum, Thermotogales and A. aeolicus). We observed a similar pattern for the protein HP16 (COG1458, Figure 1) encoded by a gene located near one of the copies of the A. aeolicus reverse gyrase gene. In addition to A. aeolicus, this protein is present in all Euryarchaeota, but in only two thermophilic and hyperthermophilic bacteria (Thermodesulfovibrio yellowstonii and Thermodesulfobacterium commune, respectively) (Appendix 3). A third protein is SahH (COG 0499), an adenosylhomocysteinase, encoded by a gene located directly at the 5′ -end of the T. maritima reverse gyrase gene. A recent phylogenetic analysis has shown that SahH sequences can be divided into two major groups: one of them (Group I) comprised all of the archaea and a few bacteria, and the second (Group II) comprised all of the eukaryotes and the great majority of the bacteria (Stepkowski et al. 2005). Our more complete Group I SahH phylogeny confirms this result (Appendix 4), but it also indicates that Group I SahH are present in all bacteria that contain a chromosomal reverse gyrase, and that 23 of the 34 (68%) bacterial species harboring a Group I SahH are either thermophiles or hyperthermophiles (underlined species in Appendix 4). Conversely, bacterial Group II SahH (several hundreds) are all from non-hyperthermophilic species. Accordingly, we suggest that bacterial Group I SahH genes were acquired via an LGT event, possibly concomitantly with reverse gyrase, from an ancestral hyperthermophilic-adapted archaeal protein.
It is tempting to speculate that the presence of genes of archaeal origin in the vicinity of the reverse gyrase genes in the genomes of thermophilic and hyperthermophilic bacteria identifies a trace of the initial (and possibly massive) co-transfer of genes coding for hyperthermophilic-adapted proteins (including reverse gyrase) from archaea to bacteria when the latter started to adapt to hyperthermophilic environments (Aravind et al. 1998, Nelson et al. 1999, Koonin et al. 2001). A second and more prosaic hypothesis remains that these observations simply reflect more recent LGT events occurring in yet to be identified organisms coexisting in similar environments. However, our phylogenetic analyses did not reveal obvious recent LGT events of archaeal genes to bacteria (i.e., no short bacterial branch emerges within an archaeal assemblage). The apparent absence of recent LGT events of reverse gyrase and surrounding genes between the prokaryotic domains could be explained by gaps in our knowledge of hyperthermophile organisms diversity and in available data or by the fact that gene transfer occurs more easily between bacteria (intra-domain) than between archaea and bacteria (inter-domain). In that case, the presence of reverse gyrase and other archaeal genes in thermophilic and hyperthermophilic bacteria would testify to a limited number of massive ancient transfers that occurred under high selective pressure when the first bacteria ventured into high temperature environments already occupied by hyperthermophilic archaea.
In conclusion, our updated phylogenomic analyses of all reverse gyrase genes in archaea and bacteria confirm that a reverse gyrase is systematically present in all hyperthermophiles, in agreement with the importance of this enzyme for survival at high temperatures. Because we found a reverse gyrase gene in some thermophilic bacteria, we also confirm previous biochemical results that detected reverse gyrase activity in several thermophilic (but non-hyperthermophilic) organisms. In contrast to hyperthermophiles, the presence of a reverse gyrase in thermophiles appears to be optional (as exemplified by the presence of reverse gyrase in T. thermophilus strain HB8 and its absence in the strain HB27). The congruence between the phylogeny of archaeal reverse gyrases and that of archaeal species history suggests that the gene coding for reverse gyrase was probably not affected by LGTs during archaeal evolution. Conversely, a late acquisition of reverse gyrase in bacteria via LGT from archaeal donors followed by spreading individual LGT is supported by the complex relationships between bacterial reverse gyrase sequences, and by the presence of genes of archaeal origin around reverse gyrase genes in bacterial genomes. The phylogenetic analysis of some of these genes suggests that they may have been co-transferred with reverse gyrase and may have contributed to thermoadaptation in bacteria. In our opinion, it is likely that the first bacteria that fixed the reverse gyrase gene were already thermophilic, and that the presence of this enzyme helped some of them to become hyperthermophiles. This suggests that the presence of reverse gyrase should bring some selective advantage even in thermophilic species (explaining why some thermophilic bacteria contain a reverse gyrase coding gene), despite the fact that a reverse gyrase deleted hyperthermophilic mutant can grow at a temperature of 90 °C in the laboratory. It would be useful to perform competition experiments between wild type and reverse gyrase mutants at different temperatures to gain better insight into the role of this enzyme in vivo. In particular, if reverse gyrase is involved in DNA repair, it is possible that the advantage to a thermophile or a hyperthermophile of possessing a reverse gyrase gene can be fully appreciated only under stressful conditions that challenge DNA integrity. Finally, it should be interesting to study in more detail the physiology of all bacteria described here in which reverse gyrase has been discovered in order to verify their optimal and maximal growth temperatures, and to compare them systematically with thermophilic bacteria lacking this unique enzyme.
Acknowledgments
The authors are grateful to Simonetta Gribaldo for her comments and careful reading of the manuscript.
Appendix
Figure A1.
Alignment between the reverse gyrase sequence from Thermus thermophilus HB8 strain (green, bold) and the hypothetical protein sequence located at the TTP0124 locus from T. thermophilus HB27 strain (orange). Symbol: “-” = missing amino acids.
Figure A2.
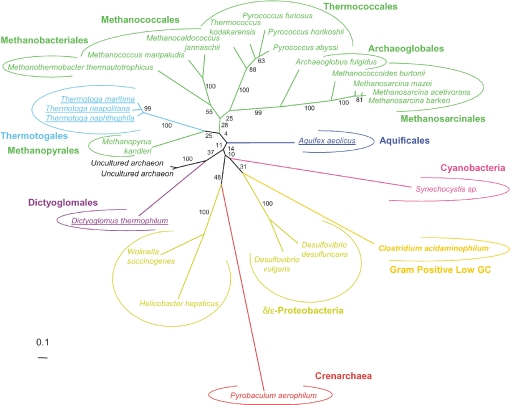
Maximum likelihood unrooted phylogenetic tree of the hypothetical protein HP13 (Figure 1). The tree was calculated by PHYML (JTT model, gamma correction (eight discrete classes), an estimated alpha parameter and an estimation of the proportion of invariant positions). Numbers at nodes are bootstrap values calculated from 1000 replicates by PHYML. The scale bar represents the percentage of substitutions per site. Underlined species are hyperthermophilic or thermophilic bacteria.
Figure A3.
Maximum likelihood unrooted phylogenetic tree of the hypothetical protein HP16 (Figure 1). The tree was calculated by PHYML (JTT model, gamma correction (eight discrete classes), an estimated alpha parameter and an estimation of the proportion of invariant positions). Numbers at nodes are bootstrap values calculated from 1000 replicates by PHYML. The scale bar represents the percentage of substitutions per site. Underlined species are hyperthermophilic or thermophilic bacteria.
Figure A4.
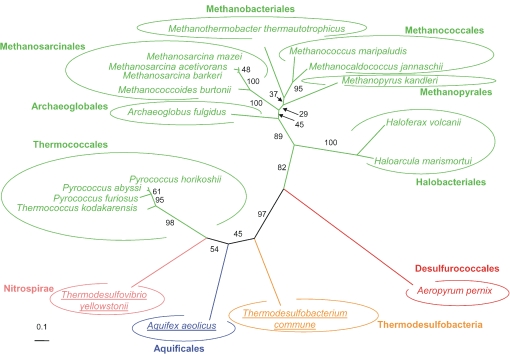
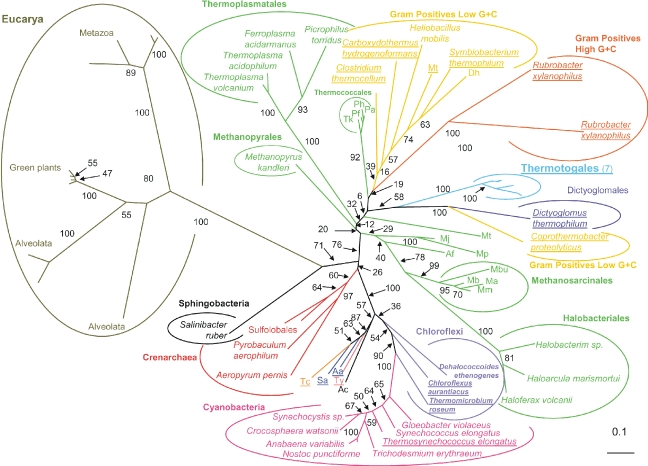
Maximum likelihood unrooted phylogenetic tree of the adenosylhomocysteinase sahH (Figure 1). The tree was calculated by PHYML (JTT model, gamma correction (eight discrete classes), an estimated alpha parameter and an estimation of the proportion of invariant positions). Numbers at nodes are bootstrap values calculated from 1000 replicates by PHYML. The scale bar represents the percentage of substitutions per site. Underlined species are hyperthermophilic or thermophilic bacteria. Abbreviations: Pf = Pyrococcus furiosus (Thermococcales); Ph = Pyrococcus horikoshii (Thermococcales); Pa = Pyrococcus abyssi (Thermococcales); Tk = Thermococcus kodakarensis (Thermococcales); Mt = Methanothermobacter thermautotrophicus; Mj = Methanococcus jannaschii; Mp = Methanococcus maripaludis; Af = Archaeoglobus fulgidus; Mbu = Methanococcoides burtonii; Mm = Methanosarcina mazei; Mb = Methanosarcina barkeri; Ma = Methanosarcina acetivorans; Ac = Acidobacterium capsulatum (Acidobacteria); Tc = Thermodesulfobacterium commune (Thermodesulfobacteria); Ty = Thermodesulfoyibrio yellowstonii (Nitrospirae); Sa = Sulfurihydrogenibium azorense (Aquificales); Aa = Aquifex aeolicus (Aquificales); Mt = Moorella thermoaceticat (Gram Positives Low G + C); and Dh = Desulfitobacterium hafniense (Gram Positives Low G + C).
References
- R1.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- R2.Aravind L., Tatusov R.L., Wolf Y.I., Walker D.R., Koonin E.V. Evidence for massive gene exchange between archaeal and bacterial hyperthermophiles. Trends Genet. 1998;14:442–444. doi: 10.1016/s0168-9525(98)01553-4. [DOI] [PubMed] [Google Scholar]
- R3.Atomi H., Matsumi R., Imanaka T. Reverse gyrase is not a prerequisite for hyperthermophilic life. J. Bacteriol. 2004;186:4829–4833. doi: 10.1128/JB.186.14.4829-4833.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Bapteste E., Brochier C., Boucher Y. Higher-level classification of the Archaea: evolution of methanogenesis and methanogens. Archaea. 2005;1:353–363. doi: 10.1155/2005/859728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R5.Bouthier de la tour C., Portemer C., Huber R., Forterre P., Duguet M. Reverse gyrase in thermophilic eubacteria. J. Bacteriol. 1991;173:3921–3923. doi: 10.1128/jb.173.12.3921-3923.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R6.Brochier C., Philippe H. A non-hyperthermophilic ancestor for bacteria. Nature. 2002;417:244. doi: 10.1038/417244a. [DOI] [PubMed] [Google Scholar]
- R7.Brochier C., Forterre P., Gribaldo S. An emerging phylogenetic core of Archaea: phylogenies of transcription and translation machineries converge following addition of new genome sequences. BMC Evol. Biol. 2005;5:36. doi: 10.1186/1471-2148-5-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R8.Brochier C., Gribaldo S., Zivanovic Y., Confalonieri F. Nanoarchaea: representatives of a novel archaeal phylum or a fast-evolving euryarchaeal lineage related to Thermococcales? Genome Biol. 2005;6:R42. doi: 10.1186/gb-2005-6-5-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.Charbonnier F., Forterre P. Comparison of plasmid DNA topology among mesophilic and thermophilic eubacteria and archaebacteria. J. Bacteriol. 1994;176:1251–1259. doi: 10.1128/jb.176.5.1251-1259.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Declais A.C., Marsault J., Confalonieri F., Bouthier de la tour C., Duguet M. Reverse gyrase, the two domains intimately cooperate to promote positive supercoiling. J. Biol. Chem. 2000;275:19498–19504. doi: 10.1074/jbc.M910091199. [DOI] [PubMed] [Google Scholar]
- R11.Forterre P. A hot story from comparative genomics: reverse gyrase is the only hyperthermophile-specific protein. Trends Genet. 2002;18:236–237. doi: 10.1016/s0168-9525(02)02650-1. [DOI] [PubMed] [Google Scholar]
- R12.Forterre P., Bouthier de la tour C., Philippe H., Duguet M. Reverse gyrase from hyperthermophiles: probable transfer of a thermoadaptation trait from archaea to bacteria. Trends Genet. 2000;16:152–154. doi: 10.1016/s0168-9525(00)01980-6. [DOI] [PubMed] [Google Scholar]
- R13.Forterre P., Brochier C., Philippe H. Evolution of the Archaea. Theor. Popul. Biol. 2002;6:409–422. doi: 10.1006/tpbi.2002.1592. [DOI] [PubMed] [Google Scholar]
- R14.Galtier N., Tourasse N., Gouy M. A nonhyperthermophilic common ancestor to extant life forms. Science. 1999;283:220–221. doi: 10.1126/science.283.5399.220. [DOI] [PubMed] [Google Scholar]
- R15.Guindon S., Gascuel O. A simple, fast, and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 2003;52:696–704. doi: 10.1080/10635150390235520. [DOI] [PubMed] [Google Scholar]
- R16.Guipaud O., Marguet E., Noll K.M., De la tour C.B., Forterre P. Both DNA gyrase and reverse gyrase are present in the hyperthermophilic bacterium Thermotoga maritima . Proc. Natl. Acad. Sci. USA. 1997;94:10606–10611.. doi: 10.1073/pnas.94.20.10606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.Henne A., Bruggemann H., Raasch C., et al. The genome sequence of the extreme thermophile Thermus thermophilus . Nat. Biotechnol. 2004;22:547–553. doi: 10.1038/nbt956. [DOI] [PubMed] [Google Scholar]
- R18.Huber H., Hohn M.J., Rachel R., Fuchs T., Wimmer V.C., Stetter K.O. A new phylum of Archaea represented by a nanosized hyperthermophilic symbiont. Nature. 2002;417:63–67. doi: 10.1038/417063a. [DOI] [PubMed] [Google Scholar]
- R19.Kampmann M., Stock D. Reverse gyrase has heat-protective DNA chaperone activity independent of supercoiling. Nucleic Acids Res. 2004;32:3537–3545. doi: 10.1093/nar/gkh683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Kikuchi A., Asai K. Reverse gyrase—a topoisomerase which introduces positive superhelical turns into DNA. Nature. 1984;309:677–681. doi: 10.1038/309677a0. [DOI] [PubMed] [Google Scholar]
- R21.Koonin E.V., Makarova K.S., Aravind L. Horizontal gene transfer in prokaryotes: quantification and classification. Annu. Rev. Microbiol. 2001;55:709–742. doi: 10.1146/annurev.micro.55.1.709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R22.Koski L.B., Golding G.B. The closest BLAST hit is often not the nearest neighbor. J. Mol. Evol. 2001;52:540–542. doi: 10.1007/s002390010184. [DOI] [PubMed] [Google Scholar]
- R23.Lopez-Garcia P., Forterre P. DNA topology and the thermal stress response, a tale from mesophiles and hyperthermophiles. Bioessays. 2000;22:738–746. doi: 10.1002/1521-1878(200008)22:8<738::AID-BIES7>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- R24.Marguet E., Forterre P. DNA stability at temperatures typical for hyperthermophiles. Nucleic Acids Res. 1994;22:1681–1686. doi: 10.1093/nar/22.9.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R25.Napoli A., Valenti A., Salerno V., Nadal M., Garnier F., Rossi M., Ciaramella M. Reverse gyrase recruitment to DNA after UV light irradiation in Sulfolobus solfataricus . J. Biol. Chem. 2004;279:33192–33198. doi: 10.1074/jbc.M402619200. [DOI] [PubMed] [Google Scholar]
- R26.Nelson K.E., Clayton R.A., Gill S.R., et al. Evidence for lateral gene transfer between archaea and bacteria from genome sequence of Thermotoga maritima . Nature. 1999;399:323–329. doi: 10.1038/20601. [DOI] [PubMed] [Google Scholar]
- R27.Nisbet E.G., Fowler C.M.R. Some like it hot. Nature. 1996;382:404–406. [Google Scholar]
- R28.Philippe H. MUST, a computer package of Management Utilities for Sequences and Trees. Nucleic Acids Res. 1993;21:5264–5272. doi: 10.1093/nar/21.22.5264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R29.Ronquist F., Huelsenbeck J.P. MrBayes 3: Bayesian phylogenetic inference under mixed models. Bioinformatics. 2003;19:1572–1574. doi: 10.1093/bioinformatics/btg180. [DOI] [PubMed] [Google Scholar]
- R30.Stepkowski T., Brzezinski K., Legocki A.B., Jaskolski M., Bena G. Bayesian phylogenetic analysis reveals two-domain topology of S-adenosylhomocysteine hydrolase protein sequences. Mol. Phylogenet. Evol. 2005;34:15–28. doi: 10.1016/j.ympev.2004.09.008. [DOI] [PubMed] [Google Scholar]
- R31.Stetter K.O. Extremely thermophilic chemolithoautotrophic archaebacteria. In: Schlegel H.G., Bowien B., editors. Autotrophic Bacteria. Berlin: Science Tech. Publishers and Springer-Verlag; 1989. pp. 167–171. [Google Scholar]
- R32.Stetter K.O. Vol. 202. Chichester, U.K.: Ciba Found Symp. J. Wiley ; 1996. Hyperthermophiles in the history of life; pp. 1–10. [PubMed] [Google Scholar]
- R33.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.Waters E., Hohn M.J., Ahel I., et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA. 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R35.Woese C.R., Olsen G.J., Ibba M., Soll D. Aminoacyl-tRNA synthetases, the genetic code, and the evolutionary process. Microbiol. Mol. Biol. Rev. 2000;64:202–236. doi: 10.1128/mmbr.64.1.202-236.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]