Abstract
Previously, we reported from the Sulfolobus solfataricus open reading frame (ORF) SSO2517 the cloning, overexpression and characterization of an esterase belonging to the hormone-sensitive lipase (HSL) family and apparently having a deletion at the N-terminus, which we named SSoNΔ. Searching the recently reported Sulfolobus acidocaldarius genome by sequence alignment, using SSO2517 as a query, allowed identity of a putative esterase (ORF SAC1105) sharing high sequence similarity (82%) with SSO2517. This esterase displays an N-terminus and total length similar to other known esterases of the HSL family. Analysis of the upstream DNA sequence of SS02517 revealed the possibility of expressing a longer version of the protein with an extended N-terminus; however, no clear translation signal consistent with a longer protein version was detected. This new version of SSO2517 was cloned, over-expressed, purified and characterized. The resulting protein, named SSoNΔlong, was 15-fold more active with the substrate p-nitrophenyl hexanoate than SSoNΔ. Furthermore, SSoNΔlong and SSoNΔ displayed different substrate specificities for triacylglycerols. These results and the phylogenetic relationship between S. solfataricus and S. acidocaldarius suggest a common origin of SSO2517 and SAC1105 from an ancestral gene, followed by divergent evolution. Alternatively, a yet-to-be discovered mechanism of translation that directs the expression of SSoNΔlong under specific metabolic conditions could be hypothesized.
Keywords: archaea, HSL family, N-terminus, thermophilic carboxylesterase
Introduction
Most esterases/lipases belonging to the hormone-sensitive lipase (HSL) family, comprising either characterized enzymes or open reading frames (ORFs), display similar primary sequences of about 300 amino acids (see the ESTHER database at http://bioweb.ensam.inra.fr/esther). Upstream of the consensus signature HGGA/G (Sussman et al. 1991), several of the microbial versions of these proteins have an N-terminus of about 60 amino acids that is characterized by low similarity among these proteins and sometimes by different structural organization (De Simone et al. 2001). Recently, we cloned and characterized an esterase belonging to the HSL family from Sulfolobus solfataricus P2 (She et al. 2001), which we named SsoNΔ. SsoNΔ is characterized by a large deletion at the N-terminus (Mandrich et al. 2005). In parallel, we performed a comparative analysis with a truncated version of another protein of the HSL family, namely, Alicyclobacillus acidocaldarius esterase 2 (EST2; Manco et al. 1997, 1998), and demonstrated that the N-terminus of EST2 is involved in substrate specificity, catalytic efficiency, thermostability, thermophilicity and even regioselectivity (Mandrich et al. 2005). At about the same time, Kim and Lee (2004) published a paper describing the cloning and expression of three ORFs (SSO2493, SSO2517 and SSO2521) from S. solfataricus, coding for putative lipases/esterases. Kim and Lee (2004) reported on the characterization of an esterase, called Est3, resulting from the cloning of SSO2493, but they erroneously attributed the sequence of SSO2517 to Est3, corresponding to SsoNΔ protein, and the protein sequence of SSO2493 was named Est2. Kim and Lee (2004) successfully cloned SSO2517 from S. solfataricus P2 but with an extra 60 amino acids at the N-terminus compared with the SsoNΔ version that we previously characterized (Mandrich et al. 2005). Although Kim and Lee (2004) neither purified nor characterized their longer version, they showed that its N-terminus shares weak similarity to other HSL family proteins, and demonstrated by in situ colony assay that the protein has enzymatic activity. According to the GeneMark program output (Besemer and Borodovsky 1999), the nucleotide sequence of SSO2517, as annoted in the genome database, does not start with a typical AUG start codon but with UUG, coding for a leucine, and codes for a protein of 251 amino acids. Kim Lee (2004) reported the cloning of the longer version of SSO2517 starting with an upstream AUU codon coding for isoleucine, without giving reasons for this choice. Here, we report an analysis of the upstream sequence of SSO2517 and related genes to verify their organization in terms of start codons, upstream transcriptional signals and the presence or absence of an N-terminus related to other HSL family members. In addition, we cloned, expressed and characterized the long version of SSO2517 and compared its biochemical properties with the shorter version.
Materials and methods
Bacterial strains, media and growth conditions
Escherichia coli strains Top10 and BL21(DE3) were grown in Luria-Bertani (LB) medium (Sigma Chemical Co. St. Louis, MO) at 37 °C. Ampicillin (Sigma) was used at a concentration of 100 µg ml–1.
Comparative analysis in silico
The comparative analysis of SsoNΔ with the S. acidocaldarius genome database was performed using similarity searches (BlastP) and alignment programs available on the ExPASy Proteomic Server (http://www.expasy.org). Gene identification analysis was performed by means of the EasyGene program available at http://www.binf.ku.dk/services/easygene (Larsen and Krogh 2003).
Analysis of the DNA sequence upstream of SSO2517
Based on the genomic DNA sequence of S. solfataricus P2 (She et al. 2001), we designed an oligonucleotide in a region about 300 bp upstream of SSO2517, the ORF from which we previously cloned SsoNΔ (Mandrich et al. 2005). This oligonucleotide, SsoNΔ 5′upstream (5′-TCCTCAAATTCAGTTAATTAAG-3′), and a second one, SsoNΔ3′ (5′-CTTGGGGTCGACTTAACCCCTCATTAAGATGTCCTTT-3′), were used as forward and reverse primers, respectively, in a 30-cycle polymerase chain reaction (1 min at 94 °C, 1 min at 50 °C and 2 min at 72 °C), with genomic DNA as template. The PCR product was purified on a 1% agarose gel, eluted from the gel and sequenced.
Cloning and overexpression of SsoNΔlong in E. coli
Manipulations of E. coli plasmid DNA were carried out by standard methods (Sambrock et al. 1989). The SsoNΔlong gene was amplified by PCR directly from the genomic DNA of S. solfataricus P2. Oligonucleotides used for cloning were SsoNΔ5′long 5′-AAATTAGGTGGAAGAATTCCCCTAG-3′ and SsoNΔ3′ (see above) as forward and reverse primers, respectively. The PCR conditions were 1 min at 94 °C, 1 min at 50 °C and 2 min at 72 °C, for 30 cycles. The amplification primers were designed to include the enzyme restriction sites (underlined) EcoRI and SalI upstream of the initiation site and downstream of the stop signal, respectively. The PCR products were separated by electrophoresis on 1% agarose gel, purified, digested with EcoRI and SalI, and ligated into the EcoRI- SalI-linearized expression vector pT7-SCII (Invitrogen) to produce the construct called pT7-SCII-SsoNΔlong. The ligation mixture was transformed into E. coli Top10. The cloned fragment was completely sequenced.
Overexpression and protein purification
After overnight growth of E. coli BL21(DE3) cells transformed with pT7-SCII-SsoNΔlongin 5 liters of LB medium containing 100 µg ml–1 of ampicillin at 37 °C, 1 mM isopropyl-b-D-thiogalactopiranoside (IPTG) was added to induce protein expression. After a 3-h induction, cells were harvested by centrifugation. SsoNΔlong was purified as described by Manco et al. (1998).
Electrophoretic analysis
Electrophoresis on 15% (w/v) SDS-PAGE was performed with a Bio-Rad Mini-Protean III cell unit at room temperature, essentially as described by Laemmli (1970). The “Prestained Protein Marker, Broad Range” (Cell Signaling) was used as the molecular mass standard and contained MBP-β-galactosidase (175.0 kDa), MBP-paramyosin (83.0 kDa), glutamic dehydrogenase (62.0 kDa), aldolase (47.5 kDa), triosephosphate isomerase (32.5 kDa), β-lactoglobulin A (25.0 kDa), lysozyme (16.5 kDa) and aprotinin (6.5 kDa).
Esterase activity
The time course of the esterase-catalyzed hydrolysis of p-nitrophenyl-hexanoate (pNP-hexanoate; Sigma) was followed as described by Manco et al. (1998). The dependence of the initial velocity on pH was monitored at 348 nm (the pH-independent isosbestic point of p-nitrophenol and p-nitrophenoxide ion), as previously described (Manco et al. 1998), with pNP-hexanoate as substrate.
The dependence of enzymatic activity on temperature was studied over the range 60–90 °C, with pNP-hexanoate as substrate and phosphate buffer at pH 6.5 and pH 7.1 for SsoNΔ and SsoNΔlong, respectively.
Esterase activity on triacylglycerols
Enzyme activity toward triacylglycerols with acyl chain lengths from 4 to 18 carbon atoms (all triacylglycerol esters were purchased from Sigma) was measured by pH titration with a pH-stat controller apparatus (Radiometer Copenhagen, Brønshøj, Denmark). The reaction mixture was 20 mM Na2HPO4/NaH2PO4 buffer (pH 7.1 or 6.5), containing 4% (v/v) acetonitrile and 20 mM triacylglycerols. Stock solutions of triacylglycerols were prepared by dissolving substrates in pure acetonitrile. Assays were performed at 50 °C in duplicate or triplicate and results are the mean of two independent experiments. Esterase activity was defined as µeq of NaOH used to neutralize the free acid formed by the enzymatic reaction in one minute by 1 mg of enzyme (Mandrich et al. 2005).
Results and discussion
Analysis of the DNA sequence upstream of ORF SSO2517 and related genes
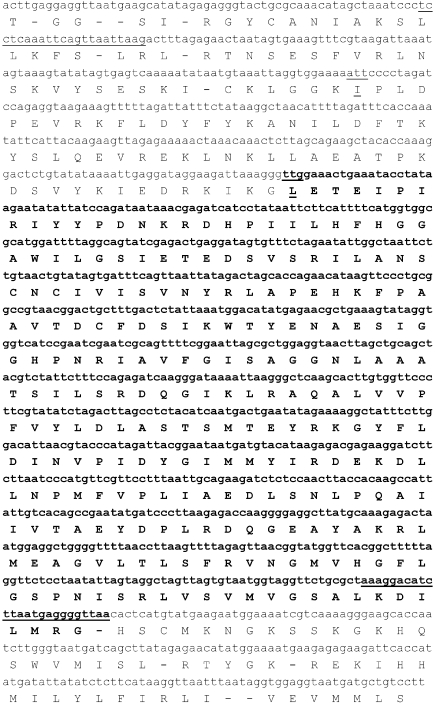
We designed two oligonucleotides for the amplification and re-sequencing of the entire region (nucleotides 2,285,942 to 2,287,200) containing SSO2517 to confirm the absence of sequencing errors in the S. solfataricus P2 genome database. The sequence obtained, shown in Figure 1, was identical to the sequence submitted to the database, and the translation of this region resulted in an ORF extending up to a cysteine residue and containing the sequence of enzyme SsoNΔ that we previously characterized (Mandrich et al. 2005). The coding sequence was assumed to start at a TTG codon (underlined in Figure 1) to yield a polypeptide of 251 amino acids; hereafter, we will refer to this protein as SsoNΔlong. From the same ORF, Kim and Lee (2004) reported the cloning of a protein starting with isoleucine (codon ATT, underlined in Figure 1). This hypothetical start codon is located 18 nucleotides downstream of the aforementioned cysteine residue.
Figure 1.
Nucleotide sequence of the S. solfataricus SsoNΔ gene (in bold), and its upstream sequence, reported from the S. solfataricus P2 genomic database. The predicted amino acid sequence is given below the nucleotide sequence in the standard one-letter code. The oligonucleotides used to amplify the gene from the genome for re-sequencing are underlined. The sequence between oligonucleotides was confirmed by re-sequencing. The start site for SsoNΔlong is underlined.
Because the complete genome sequence of S. acidocaldarius has recently been reported (Chen et al. 2005), and about 80% of its genes are related to those of S. solfataricus, we searched this genome for SsoNΔ-related ORFs by BlastP (http://www.expasy.org). High sequence identity was detected with SAC1116 (50% identity), coding for a putative acetyl esterase, and 72% identity (84% similarity) was found with SAC1105 (Figure 2), annoted as an esterase/lipase. Furthermore, SAC1105 displays an N-terminus similar to other members of the HSL family. In Figure 3, the first 60 amino acids of A. acidocaldarius EST2, belonging to the HSL family and previously used in comparative analyses with SsoNΔ (Mandrich et al. 2005), was aligned with the N-terminus of SsoNΔlong and the N-terminus of SAC1105. The alignment shows that the N termini are related, with SsoNΔlong and SAC1105 displaying around 30 and 28% identity with EST2, respectively.
Figure 2.
Sequence alignment of SsoNΔ and the putative esterase SAC1105 from S. acidocaldarius.
Figure 3.
Sequence alignment of the N termini of the putative SsoNΔlong, the esterase EST2 from A. acidocaldarius, and the putative S. acidocaldarius esterase from SAC1105 belonging to the HSL family. Asterisks and crosses indicate identical and similar residues, respectively, between EST2 and the two other sequences.
According to the program GeneMark, used for gene discovery in the S. solfataricus genome (She et al. 2001), the more likely candidate product of SSO2517 is protein SsoNΔ rather than SsoNΔlong. This gene version should start with the AUU codon (underlined in Figure 1), a canonical start codon for 10% of archaeal genes. To determine which SSO2517 version is expressed, we further analyzed the nucleotide sequence of SSO2517, encompassing 400 upstream nucleotides, using the gene finder program EasyGene (Larsen and Krogh 2003). For this analysis we used a default R-value cut-off of 2, and five other values ranging from 0.1 to 1000. The lower the R value, the more the false-positive hits are reduced. In all cases, the unique coding sequence detected in this region of the S. solfataricus genome was the short version, namely SsoNΔ. However, if a setting of 2 for start codon leniency was used, a less likely AUG start codon was detected, but this was located downstream from the UUG; no upstream start codon was detected. These automatic methods of gene detection are based on the statistical relevance of the critical constraints that define a gene, start codon and translational signals, and were originally developed for the analysis of bacterial genomes. A recent comparative analysis of the available archaeal genomes has confirmed the presence of specific translational signals (Torarinsson et al. 2005); in particular the BRE motif constituted by two to four A/Ts (Bell et al. 1999) at position –32, which follows an AT-rich peak between positions –29 and –23, called the Box A motif (Hain et al. 1992, Palmer and Daniels 1995, Danner and Soppa 1996). These signals are prevalent in leaderless transcripts, which mostly correspond to unique genes or to the first gene of an operon (Type 1 genes). In contrast, leadered transcripts usually display, in a conserved G-rich region between positions –15 and –4, a Shine-Dalgarno (SD) sequence that differs from those of bacteria, with the four common sequence motifs being GUGA, GGUG, AGGU and GAGG. The leadered transcripts are produced from genes inside operons (Type 2 genes) (Torarinsson et al. 2005). For both type of transcripts, the start codons GUG and UUG in addition to AUG are frequently found. The sequencing of a few proteins has indicated that GUG and UUG start codons in archaea both encode methionine via the non-formylated archaeal initiator tRNA (Krah et al. 1996, Helianti et al. 2001, Peng et al. 2003). The enzyme ATP-glucokinase of A. pernix, which has been reported to have a GTG start codon encoding a valine as first residue, represents one exception (Hansen et al. 2002). At present, data are too limited to allow a generalization.
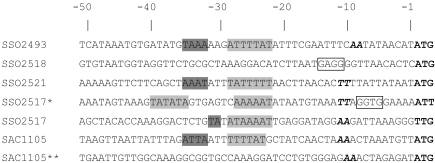
Figure 4 shows the results of a comparative analysis of start codons and translational signals, identified upstream of S. solfataricus ORFs encoding three esterases (SSO2493, SSO2518 and SSO2521), the short and long forms of SSO2517, the S. acidocaldarius SAC1105 encoding the putative esterase displaying high similarity to SsoNΔ, and a short version of this latter ORF, which was obtained by deleting the N-terminus up to methionine 53. All of the ORFs we analyzed have an AT-rich peak between positions –29 and –23, three out of five have the BRE motifs, and all have AUG as the start codon, except SSO2517, which has UUG. In contrast, the long version of SSO2517 presents an SD sequence and two AT-rich regions (one at position –23/–28 and one at position –36/–41), but lacks both the BRE motif and a canonical start codon, which should prevent translation of SSO2517 in its long version. However, some rare cases of genes starting with AUU have been reported in Eukarya (http://bioinfo.iitk.ac.in/bio- info/news.php), and in Bacteria (Gualerzi and Pon 1990), and we cannot exclude the possibility that a similar situation holds also for the Archaea. The ORF SAC1105**, which corresponds to the artificial short version of the esterase SacEst (Figure 2), has AUG as the start codon but it lacks all translational signals and cannot be expressed.
Figure 4.
Sequences comparison of 50 nucleotides upstream of the start codon (bold) for putative esterases/lipases of S. solfataricus (SSO2493, SSO2517, SSO2518, SSO2521), S. acidocaldarius SAC1105, for the putative long version of SSO2517 (indicated with an asterisk) and for a short version of SAC1105 (indicated with a double asterisk). The putative translational signals were highlighted as follows: BRE motif, shadowed dark gray; Box A motif, shadowed gray; Shine-Dalgarno sequence, boxed; A/T peak centered at –10, italicized and bold; start codon, bold. A single asterisk (*) denotes SsoNΔlong, and the double asterisk (**) denotes the short version of the putative esterase from SAC1105.
We next considered the classification of the SsoNΔ/ SsoNΔlong gene, because analysis of the relevant genomic region suggests that this should be the first gene of an operon, being the AUG start for the next gene (SSO2518), which is only 5 nt downstream from the stop codon of the SsoNΔ/ SsoNΔlong gene. In agreement with the general rules proposed for archaeal signals, SSO2518 should yield a leadered transcript and should contain an SD sequence, which it does (Figure 4). In contrast, SSO2517 should give a leaderless transcript, should lack the SD sequence and should possess canonical Box A and BRE sequences upstream of a canonical start codon, as we observed in the case of SsoNΔ.
Cloning, overexpression and characterization of SsoNΔlong
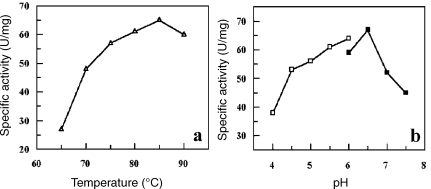
All of the above results indicate that translation of SsoNΔlong should be impossible. Given that esterase activity was reported for this protein (Kim and Lee 2004), we decided to compare its biochemical properties with those of the shorter version. The gene was cloned by PCR amplification into the expression vector pT7-SCII. SsoNΔlong was expressed and purified (data not shown) and Figure 5 shows the electrophoretic analysis of the pure protein. A band of about 34 kDa was obtained in agreement with the mass deduced from the sequence (34,567). The optimal pH for SsoNΔlong measured at 70 °C with pNP-hexanoate as substrate was 6.5 (Figure 6A). This pH value is quite similar to the pH of 7.1 reported for the short version (Mandrich et al. 2005). Substantial differences between the two enzymes included: (1) the optimal temperature for activity; (2) the kinetic parameters; and (3) the activity on triacylglycerols. As shown in Figure 6B, the temperature optimum was 85 °C, 15°C above the value of 70 °C reported for the short version. Deletion of 35 amino acids at the N-terminus of A. acidocaldarius EST2 caused a similar change in thermophilicity (Mandrich et al. 2005), confirming the role of the N-terminus in the activity–temperature relationship and likely also in thermal stability. The kinetic parameters for the two enzymes were measured at their respective optimal temperatures (Table 1). Values for SsoNΔ are in agreement with previously published data (Mandrich et al. 2005). SsoNΔlong displayed a specific activity 15-fold higher than SsoNΔ. The KM of SsoNΔlong was substantially lower than that of SsoNΔ, resulting in a 25-fold higher catalytic efficiency compared with SsoNΔ.
Figure 5.
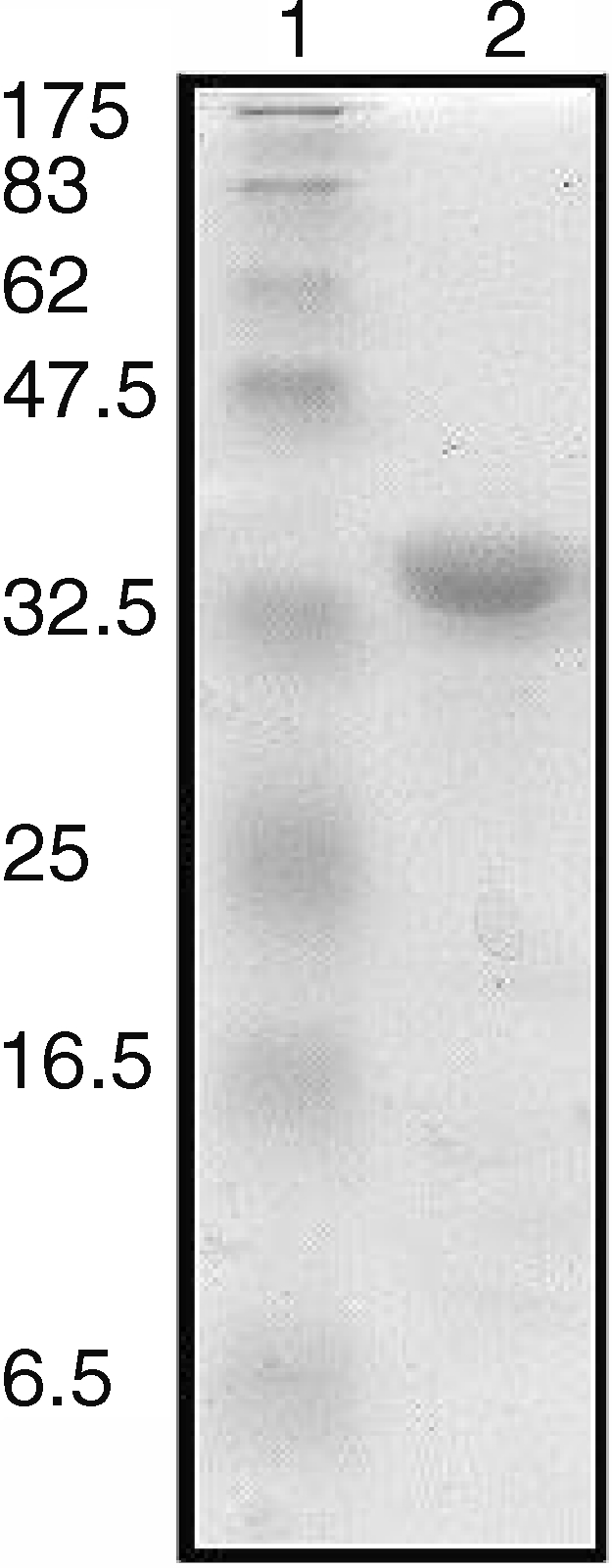
Analysis by SDS-PAGE of SsoNΔlong. Lane 1: molecular mass markers. Lane 2: SsoNΔlong (3 µg).
Figure 6.
Affinity of SsoNΔlong: temperature (a) and pH (b) responses.
Table 1.
Comparison of kinetic parameters of SsoNΔ and SsoNΔlong in their respective optimal conditions with the substrate pNP-hexanoate. Assays were done in duplicate or triplicate and the results shown are the means of two independent experiments.
| Enzyme | Specific activity (U mg–1) | kcat (s–1) | KM (µM) | kcat/KM (µM–1 s–1) |
| SsoNΔ | 4.5 ± 0.3 | 2.5 ± 0.1 | 54.0 ± 4.0 | 0.050 ± 0.001 |
| SsoNΔlong | 60.0 ± 3.0 | 34.5 ± 0.5 | 30.0 ± 5.0 | 1.15 ± 0.10 |
The next question we attempted to answer was whether the N-terminus in some way favors activity with the natural lipase substrates, namely triacylglycerols. We previously reported that A. acidocaldarius EST2 is devoid of a true lipase activity, e.g., activity with triglyceryloleate emulsions, and is devoid of interfacial activation (Manco et al. 1998). However, EST2 and its N-terminal deleted forms were able to degrade triacylglycerols dissolved in high concentrations of acetonitrile, and the same was observed for SsoNΔ (Mandrich et al. 2005). SsoNΔ displayed a specific activity comparable with the EST2 N-terminal deleted forms, and only marginally decreased activity on substrates with increasing acyl chain lengths ranging from 4 to 18 carbon atoms (Mandrich et al. 2005). The activities measured for SsoNΔlong and SsoNΔ on different triacylglycerols substrates are reported in Table 2. SsoNΔlong activity on triacylglycerols decreased with increasing acyl chain length in the range from 4 to 18 carbon atoms, with SsoNΔlong activity on glyceryl tributanoate being 6-fold higher than SsoNΔ activity. In contrast, SsoNΔlong activity on glyceryl trioleate was drastically reduced (10-fold) compared with SsoNΔ activity. The profile of SsoNΔlong activity was similar to that of the intact form of EST2, which showed maximum activity on glyceryl tributanoate and minimum activity on glyceryl trioleate, as previously reported (Mandrich et al. 2005). EST2 and SsoNΔlong had similar activity on glyceryl trioleate, confirming that the N-terminus has a role in modulating substrate specificity, probably by restricting access of the substrate to the active site. SsoNΔ showed 10-fold higher activity than SsoNΔlong with glyceryl trioleate as substrate. Therefore, if SsoNΔ is expressed in S. solfataricus in vivo, it could be regarded as a form evolving toward a lipase-like enzyme, in terms of length of acyl chain of substrate hydrolyzed.
Table 2.
Esterase activity toward pNP-esters and triacyglycerols was measured at 50 °C by pH titration with a radiometer apparatus. Each assay was done in duplicate or triplicate with the standard assay. Results are the means of two independent experiments.
| Specific activity (µeq mg–1 min–1) | |||||||
| pNP-hexanoate | pNP-dodecanoate | Glyceryl tributyrate | Glyceryl trihexanoate | Glyceryl trioctanoate | Glyceryl tridecanoate | Glyceryl trioleate | |
| SsoNΔ | 1.90 ± 0.05 | 0.34 ± 0.03 | 2.5 ± 0.1 | 2.4 ± 0.1 | 2.3 ± 0.1 | 2.1 ± 0.1 | 1.9 ± 0.1 |
| SsoNΔlong | 11.6 ± 0.9 | 1.50 ± 0.07 | 15.0 ± 0.7 | 9.5 ± 0.4 | 4.1 ± 0.2 | 0.45 ± 0.02 | 0.20 ± 0.01 |
In conclusion, which version of SSO2517 is expressed in vivo remains an open question. In the absence of in vivo data we can draw only the following conclusions. On the basis of known translation signals in the archaea, the SsoNΔ sequence contains all of the required translation signals. In contrast, SsoNΔlong has a canonical SD sequence but lacks a canonical start codon, which should prevent expression of SsoNΔlong. However, data from Kim and Lee (2004) and biochemical data reported here demonstrate that SsoNΔlong is a functional enzyme with properties similar to other thermophilic esterases of the HSL family that we have previously characterized. Furthermore, the N-terminus of SsoNΔlong seems to comply with the functional role we previously described (Mandrich et al. 2005), ruling out a genetic drift of this region toward a non-functional sequence. Furthermore, in S. acidocaldarius, only the long version of the homologous SAC1105 possesses signals for translation. Based on the phylogenetic relationship between S. solfataricus and S. acidocaldarius (Matte-Tailliez et al. 2002), we hypothesize two different scenarios. (1) The organization in S. solfataricus is an example of divergent evolution. The long version, inherited also by S. acidocaldarius, could have been lost in S. solfataricus in favor of the more specialized short version. (2) A different mechanism that does not require the use of a canonical start codon (AUG, UUG, GUG) may exist, as reported in Eukarya and Bacteria. If scenario 2 is correct, we should be able to detect both proteins in S. solfataricus, albeit perhaps under different metabolic conditions. This will be the subject of future analyses.
Acknowledgments
This work was supported by the MIUR grant “Piano Nazionale Ricerca per le Biotecnologie Avanzate II – Tema 6 “Biocatalisi” and “Studio dei meccanismi molecolari dell’adattamento della vita alle alte temperature: risvolti conoscitivi e applicativi.”
References
- R1.Bell S.D., Kosa P.L, Sigler P.B., Jackson S.P. Orientation of the transcription preinitiation complex in archaea. Proc. Natl. Acad. Sci. USA. 1999;96:13662–13667. doi: 10.1073/pnas.96.24.13662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Besemer J., Borodovsky M. Heuristic approach to deriving models for gene finding. Nucleic Acids Res. 1999;27:3911–3920. doi: 10.1093/nar/27.19.3911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Chen L., Brugger K., Skovgaard M., et al. The genome of Sulfolobus acidocaldarius, a model organism of crenarchaeota. J. Bacteriol. 2005;187:4992–4999. doi: 10.1128/JB.187.14.4992-4999.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Danner S., Soppa J. Characterization of the distal promoter element of halobacteria in vivo using saturation mutagenesis and selection. Mol. Microbiol. 1996;19:1265–1276. doi: 10.1111/j.1365-2958.1996.tb02471.x. [DOI] [PubMed] [Google Scholar]
- R5.De Simone G., Menchise V., Manco G., Mandrich L., Sorrentino N., Lang D., Rossi M., Pedone C. The crystal structure of a hyper-thermophilic carboxylesterase from the archaeon Archaeoglobus fulgidus . J. Mol. Biol. 2001;314:507–518. doi: 10.1006/jmbi.2001.5152. [DOI] [PubMed] [Google Scholar]
- R6.Gualerzi C.O., Pon C.L. Initiation of mRNA translation in prokaryotes. Biochemistry. 1990;29:5881–5889. doi: 10.1021/bi00477a001. [DOI] [PubMed] [Google Scholar]
- R7.Hain J., Reiter W.D., Hudepohl U., Zillig W. Elements of an archaeal promoter defined by mutational analysis. Nucleic Acids Res. 1992;20:5423–5428. doi: 10.1093/nar/20.20.5423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R8.Hansen T., Reichstein B., Schmid R., Schonheit P. The first archaeal ATP-dependent glucokinase, from the hyperthermophilic crenarchaeon Aeropyrum pernix, represents a monomeric, extremely thermophilic ROK glucokinase with broad hexose specificity. J. Bacteriol. 2002;184:5955–5965. doi: 10.1128/JB.184.21.5955-5965.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.Helianti I., Morita Y., Yamamura A., Murakami Y., Yokoyama K., Tamiya E. Characterization of native glutamate dehydrogenase from an aerobic hyperthermophilic archaeon Aeropyrum pernix K1. Appl. Microbiol. Biotechnol. 2001;56:388–394. doi: 10.1007/s002530100575. [DOI] [PubMed] [Google Scholar]
- R10.Kim S., Lee S.B. Thermostable esterase from a thermoacidophilic archaeon: purification and characterization for enzymatic resolution of a chiral compound. Biosci. Biotechnol. Biochem. 2004;68:2289–2298. doi: 10.1271/bbb.68.2289. [DOI] [PubMed] [Google Scholar]
- R11.Krah R., Kozyavkin S.A., Slesarev A.I. A two-subunit type I DNA topoisomerase (reverse gyrase) from an extreme hyperthermophile. Proc. Natl. Acad. Sci. USA. 1996;93:106–110. doi: 10.1073/pnas.93.1.106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R12.Laemmli U.K. Cleavage of structural protein during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- R13.Larsen T., Krogh A. EasyGene—a prokariotic gene finder that ranks ORFs by statistical significance. BMC Bioinformatics. 2003;4:21. doi: 10.1186/1471-2105-4-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R14.Manco G., Adinolfi E., Pisani F.M., Carratore V., Rossi M. Identification of an esterase from Bacillus acidocaldarius with sequence similarity to a hormone sensitive lipase subfamily. Protein Pept. Lett. 1997;4:375–382. [Google Scholar]
- R15.Manco G., Adinolfi E., Pisani F.M., Ottolina G., Carrea G., Rossi M. Overexpression and properties of a new thermophilic and thermostable esterase from Bacillus acidocaldarius with sequence similarity to hormone sensitive lipase subfamily. Biochem. J. 1998;332:203–212. doi: 10.1042/bj3320203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R16.Mandrich L., Merone L., Pezzullo M., Cipolla L., Nicotra F., Rossi M., Manco G. Role of the N-terminus in enzyme activity, stability and specificity in thermophilic esterases belonging to the HSL family. J. Mol. Biol. 2005;345:501–512. doi: 10.1016/j.jmb.2004.10.035. [DOI] [PubMed] [Google Scholar]
- R17.Matte-Tailliez O., Brochier C., Forterre P., Philippe H. Archaeal phylogeny based on ribosomal proteins. Mol. Biol. Evol. 2002;19:631–639. doi: 10.1093/oxfordjournals.molbev.a004122. [DOI] [PubMed] [Google Scholar]
- R18.Palmer J.R., Daniels C.J. In vivo definition of an archaeal promoter. J. Bacteriol. 1995;177:1844–1849. doi: 10.1128/jb.177.7.1844-1849.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R19.Peng X., Chen L., She Q. Genus-specific protein binding to the large clusters of DNA repeats (short regularly spaced repeats) present in Sulfolobus genomes. J. Bacteriol. 2003;185:2410–2417. doi: 10.1128/JB.185.8.2410-2417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Sambrock J., Fritsch E.F., Maniatis T. Cold Spring Harbor: 3rd Edn. Spring Harbor Laboratory Press; 1989. Molecular cloning: a laboratory manual; pp. 31–116. [Google Scholar]
- R21.She Q., Sing R.K., Confalonieri F., et al. The complete genome of the crenarchaeon Sulfolobus solfataricus P2. Proc. Natl. Acad. Sci. USA. 2001;98:7835–7840. doi: 10.1073/pnas.141222098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R22.Sussman J.L., Harel M., Frolow F., Oefner C., Goldman A., Toker L., Silman I. Atomic structure of acetylcholinesterase from Torpedo californica: a prototypic acetylcholine-binding protein. Science. 1991;253:872–879. doi: 10.1126/science.1678899. [DOI] [PubMed] [Google Scholar]
- R23.Torarinsson E., Klenk H.P., Garrett R.A. Divergent transcriptional and translational signals in archaea. Environ. Microbiol. 2005;7:47–54. doi: 10.1111/j.1462-2920.2004.00674.x. [DOI] [PubMed] [Google Scholar]