Abstract
Adenosine monophosphate (AMP)-forming acetyl-CoA synthetase (ACS; acetate:CoA ligase (AMP-forming), EC 6.2.1.1) is a key enzyme for conversion of acetate to acetyl-CoA, an essential intermediate at the junction of anabolic and catabolic pathways. Phylogenetic analysis of putative short and medium chain acyl-CoA synthetase sequences indicates that the ACSs form a distinct clade from other acyl-CoA synthetases. Within this clade, the archaeal ACSs are not monophyletic and fall into three groups composed of both bacterial and archaeal sequences. Kinetic analysis of two archaeal enzymes, an ACS from Methanothermobacter thermautotrophicus (designated as MT-ACS1) and an ACS from Archaeoglobus fulgidus (designated as AF-ACS2), revealed that these enzymes have very different properties. MT-ACS1 has nearly 11-fold higher affinity and 14-fold higher catalytic efficiency with acetate than with propionate, a property shared by most ACSs. However, AF-ACS2 has only 2.3-fold higher affinity and catalytic efficiency with acetate than with propionate. This enzyme has an affinity for propionate that is almost identical to that of MT-ACS1 for acetate and nearly tenfold higher than the affinity of MT-ACS1 for propionate. Furthermore, MT-ACS1 is limited to acetate and propionate as acyl substrates, whereas AF-ACS2 can also utilize longer straight and branched chain acyl substrates. Phylogenetic analysis, sequence alignment and structural modeling suggest a molecular basis for the altered substrate preference and expanded substrate range of AF-ACS2 versus MT-ACS1.
Keywords: acetate, Archaeoglobus fulgidus, Methanothermobacter thermautotrophicus
Introduction
Acetyl-CoA plays a central role in carbon metabolism in the Bacteria, Archaea and Eukarya as an essential intermediate at the junction of various anabolic and catabolic pathways. Adenosine monophosphate (AMP)-forming acetyl-CoA synthetase (ACS; acetate:CoA ligase (AMP-forming), EC 6.2.1.1) is widespread in all three domains of life and is the predominant enzyme for activation of acetate to acetyl-CoA (Equation 1).
Based on isotopic exchange, labeling experiments and detection of an enzyme-bound acetyl-AMP, a mechanism (Equations 2a and 2b) in which the reaction proceeds through an acetyl-AMP intermediate has been proposed (Berg 1956a, Berg 1956b, Webster 1963, Anke and Spector 1975):
The first step of the reaction, which requires acetate and ATP, but not CoA, involves formation of the acetyl-AMP intermediate and release of pyrophosphate (PPi). In the second step, the acetyl group is transferred to the sulfhydryl group of CoA and AMP is released. An inorganic pyrophosphatase draws the reaction in this forward direction by removing PPi, a potent inhibitor of ACS.
The ACS is a member of the acyl-adenylate forming enzyme superfamily in which all members undergo a similar two-step reaction mechanism with an enzyme-bound acyl-adenylate intermediate formed in the first step of the reaction. Although members of this superfamily all catalyze mechanistically similar reactions, they share little identity and similarity in amino acid sequence with the exception of a few signature motifs and conserved core sequence motifs (Babbitt et al. 1992, Kleinkauf and Von Dohren 1996, Chang et al. 1997, Marahiel et al. 1997). Structures for several members of this family have been determined (Conti et al. 1996, Conti et al. 1997, May et al. 2002), but provide little information regarding the active site and catalytic mechanism of ACS, because they catalyze unrelated reactions in which the intermediates serve different functions and share too little homology to allow structural modeling of ACS.
The structures of the Salmonella enterica ACS and Saccharomyces cerevisiae ACS1 now provide direct insight into the catalytic mechanism of ACS. The S. cerevisiae enzyme was crystallized in the presence of ATP (Jogl and Tong 2004) and the S. enterica enzyme (Gulick et al. 2003) was crystallized in the presence of CoA and adenosine-5′-propylphosphate, which mimics the acyl-adenylate intermediate (Grayson and Westkaemper 1988, Horswill and Escalante-Semerena 2002). These structures demonstrate the enzyme in two different conformations. The structure of the yeast enzyme is thought to represent the conformation of the enzyme in the first step of the reaction, which involves acetate and ATP but not CoA. In this structure, the smaller C-terminal domain is in an open position away from the active site. The structure of the bacterial enzyme is thought to represent the conformation for the second step of the reaction, in which the acetyl-adenylate intermediate reacts with CoA. In this structure, the C-terminal domain has rotated 140° toward the N-terminal domain, thus rearranging the active site upon CoA binding for catalysis of the second step of the reaction.
Most ACSs have a limited substrate range, showing a strong preference for acetate as the acyl substrate, although propionate can serve as a less efficient substrate. However, the Pyrobaculum aerophilum ACS (PA-ACS) has been shown to utilize butyrate and isobutyrate in addition to acetate and propionate (Brasen et al. 2005). Furthermore, PA-ACS is octameric, unlike other ACSs which have been shown to be monomeric, dimeric or trimeric (Brasen et al. 2005). These findings call into question whether ACSs are more diverse than previously expected.
We report here the biochemical and kinetic characterization of two ACSs from the archaea Methanothermobacter thermautotrophicus (MT-ACS1) and Archaeoglobus fulgidus (AF-ACS2). MT-ACS1 is a typical ACS in that acetate is the strongly preferred substrate over propionate and it cannot utilize larger substrates such as butyrate. Through modeling of MT-ACS1 on the S. enterica and yeast ACS structures, Ingram-Smith et al. (2006) identified four residues that comprise at least part of the acetate binding pocket of MT-ACS1 and have shown that alterations of these residues can greatly influence acyl substrate range and preference.
As observed for the P. aerophilum enzyme, AF-ACS2 is unusual in that it shows only a weak preference for acetate versus propionate and can also utilize butyrate and isobutyrate. The presence of the four acetate pocket residues in both AF-ACS2 and PA-ACS2 suggests that additional residues play an important role in determining substrate range and preference. The possible molecular basis for the broad substrate specificity of these two enzymes relative to MT-ACS1 and other characterized ACSs is discussed.
Materials and methods
Sequence and phylogenetic analysis
Putative ACS amino acid sequences were identified in BLASTP and TBLASTN searches (Altschul et al. 1990, 1997) of the finished genome sequences at the National Center for Biotechnology Information (NCBI) using the M. thermautotrophicus Z245 MT-ACS1 deduced amino acid sequence as the query. Acetyl-CoA synthetase sequences were aligned by Clustal X (Thompson et al. 1997) using a Gonnet PAM 250 weight matrix with a gap opening penalty of 10.0 and a gap extension penalty of 0.05. Aligned sequences were analyzed with the MEGA program (Kumar et al. 1994) using a neighbor joining algorithm with a gamma distance estimation (g = 2). The phylogeny was constructed based on pairwise distance estimates of the expected number of amino acid replacements per site (0.2 in the scale bar). One thousand bootstrap replicates were performed and values of 80% or higher are shown. Sequences from only one strain or species of closely related bacteria were included in the analysis for brevity and readability.
Sequence similarity and identity were determined using the BLAST2 pairwise alignment program (http://www.ncbi.nlm.nih.gov/blast/bl2seq/wblast2.cgi). ConSurf (Armon et al. 2001, Glaser et al. 2003, Landau et al. 2005) (http://consurf.tau.ac.il) was used to determine evolutionary conserved residues that are likely to be important for protein structure and function.
Cloning and sequencing the acs genes
The genes encoding MT-ACS1 and MT-ACS2 from M. thermautotrophicus strains DH, Z245, and FTF, and AF-ACS2 from A. fulgidus were PCR amplified from genomic DNA using the following primer pairs: MT-ACS1, 5′-ATGTCAAAGGATACCTCAGTTCTCC-3′ and 5′-CATCAAATATGAAGGGAGGGTATGG-3′; MT-ACS2, 5′-ATGAGAGGACAGCTTGATGCTCTG-3′ and 5′-CTGATTCTCCCATCGGCAAATGG-3′; AF-ACS2, 5′-ATGGCAGACCCGATGGAAGCTATG-3′ and 5′-CCACTTTGAAGCCATACTACCACC-3′. The PCR products were purified from agarose gel using the SpinPrep Gel DNA Kit (Novagen, Madison, WI). The PCR products were cloned into the pETBlue-1 expression vector (Novagen). The sequences of the cloned acs genes were confirmed by Li-Cor bidirectional sequencing at the Nucleic Acid Facility at Clemson University.
Heterologous enzyme production in Escherichia coli
The enzymes MT-ACS1, MT-ACS2, and AF-ACS2 were heterologously produced in E. coli Rosetta Blue(DE3). Cultures were grown at 37 °C in LB medium containing 50 µg ml–1 ampicillin and 34 µg ml–1 chloramphenicol to A600 = 0.6. Heterologous protein production was induced by the addition of 0.5 mM IPTG. Cells were grown overnight at 22–25 °C and harvested.
Enzyme purification
A similar purification scheme was used for both MT-ACS1 and AF-ACS2. Cells suspended in ice-cold buffer A (25 mM Tris (pH 7.5)) were disrupted by two passages through a French pressure cell at 138 MPa and the cell lysate was clarified by ultracentrifugation. The supernatant was applied to a Q-sepharose fast-flow anion exchange column (GE Healthcare, Piscataway, NJ) that was developed with a linear gradient from 0 to 1 M KCl in buffer A. Fractions containing active enzyme were pooled and diluted with 0.5 volumes of buffer B (25 mM Tris (pH 7.0)) containing 2 M ammonium sulfate and applied to a phenyl sepharose fast-flow hydrophobic interaction column (GE Healthcare) that was developed with a gradient from 0.7 to 0 M ammonium sulfate in buffer B. The purified enzymes were dialyzed against buffer B and concentrated to > 1 mg ml–1. The enzymes were purified to apparent homogeneity as judged by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) (Laemmli 1970). Aliquots of the purified protein were stored at –20 °C. Protein concentrations were determined by the Bradford method (Bradford 1976) with bovine serum albumin as the standard.
Molecular mass determination
The native molecular mass of each enzyme was determined by gel filtration chromatography on a Superose 12 column (GE Healthcare) calibrated with chymotrypsinogen (25 kDa), ovalbumin (43 kDa), albumin (67 kDa), aldolase (158 kDa), catalase (232 kDa), ferritin (440 kDa) and blue dextran (2000 kDa). Protein samples (0.2 ml) were loaded onto the column pre-equilibrated with 50 mM Tris (pH 7.5) containing 150 mM KCl and the column was developed at a flow rate of 0.5 mlmin–1. The subunit molecular mass of each enzyme was determined by SDS-PAGE and was in agreement with the predicted size based on the deduced amino acid sequence.
Enzymatic assays for ACS activity
Enzymatic activity was determined by monitoring acetyl-CoA formation from acetate, ATP and CoA by the hydroxamate reaction (Lipmann and Tuttle 1945, Rose et al. 1954), in which activated acyl groups are converted to an acyl-hydroxamate and subsequently to a ferric hydroxamate complex that can be detected spectrophotometrically at 540 nm. Reaction mixtures contained 100 mM Tris (pH 7.5), 600 mM hydroxylamine-HCl (pH 7.0) and 2 mM glutathione (reduced form), with varied concentrations of acyl substrate, HSCoA, and MgCl2-ATP. A standard reaction temperature of 65 °C was used, as this was determined to be the optimal temperature for both enzymes. Reactions were terminated by the addition of two volumes of stop solution (1 N HCl, 5% trichloroacetic acid, 1.25% FeCl2). Acetyl-CoA formation was quantified by comparison with a standard curve prepared using known concentrations of acetyl-CoA in the reaction mixture. Reaction times for each enzyme were empirically determined such that the rate of the reaction remained linear and was within the acetyl-CoA standard curve.
For determination of apparent kinetic parameters, the concentration of one substrate (acyl substrate, HSCoA or equimolar MgATP) was varied and the other two substrates were held constant at saturating concentrations as follows: MT-ACS1, 40 mM acetate, 20 mM MgATP, 0.5 mM CoA; AF-ACS2, 50 mM acetate, 20 mM MgATP, 1 mM CoA. Concentrations for the varied substrate generally ranged from 0.2 to 5–10 times the Km value. For determination of apparent kinetic parameters for metals, the ATP concentration was held at 20 mM for both enzymes and the metal concentrations were varied from 0.1 to 10 mM.
The apparent steady-state kinetic parameters kcat and kcat/Kmand their standard errors were determined by nonlinear regression to fit the data to the Michaelis-Menten equation using Kaleidagraph (Synergy Software). The enzymes followed Michaelis-Menten kinetics for all substrates with the exception that inhibition was observed above 0.5 mM HSCoA for MT-ACS1, in which case the kinetic parameters for acetate and ATP were performed in the presence of 0.5 mM HSCoA in the reaction mixture.
Modeling the MT-ACS1 and AF-ACS2 structures
The M. thermautotrophicus MT-ACS1 and the A. fulgidus AF-ACS2 structures were modeled on the S. enterica ACS structure (PDB ID: 1PG4) using DS Modeler (Accelrys Inc., San Diego, CA) and the default parameters. The structures were visualized with DS Visualizer (Accelrys) and DS Viewer Pro 5.0 (Accelrys). The models were visually compared with the S. enterica ACS structure to ensure there were no major structural anomalies.
Genbank accession numbers
Methanothermobacter thermautotrophicus DH ACS1 (MTH217-MTH216), NP_275360 and NP_275359; and M. thermautotrophicus DH ACS2 (MTH1603-MTH1604), NP_276715 and NP_276716. M. thermautotrophicus Z245 ACS1, DQ274062; and M. thermautotrophicus Z245 ACS2, DQ355203. M. thermautotrophicus FTF ACS1, DQ355204; and M. thermautotrophicus FTF ACS2, DQ355205; and A. fulgidus ACS2, NP_069202.
Results
Presence of two ACS open reading frames (ORFs) in M. thermautotrophicus
Analysis of the genome sequence of M. thermautotrophicus DH revealed the presence of two putative ACSs, designated here as MTDH-ACS1 and MTDH-ACS2. The M. thermautotrophicus DH genome sequence annotation indicates the gene encoding MTDH-ACS1 is interrupted by a stop codon and a frame shift, resulting in two adjacent ORFs (MTH217-MTH216, gi:2621263 and 2621262) that together have homology to full length ACS. The DNA region from the start ATG of MTH217 to the stop codon of MTH216 was amplified and cloned into the pETBlue-1 expression vector. A soluble truncated protein of about 63 kDa was heterologously produced in E. coli but did not exhibit ACS activity (data not shown). This size is consistent with the position of the stop codon indicated in the published genome sequence. The sequence of the cloned M. thermautotrophicus DH ACS1 gene (determined concurrently with the overexpression studies) confirmed the stop codon for MTH217 is authentic.
The genes encoding the ACS1 homologs from M. thermautotrophicus strains Z245 and FTF were also cloned for heterologous expression in E. coli. The three MT-ACS1 homologs share 98.5% amino acid sequence identity, with only nine positions that are not identical among all three (data not shown). Sequence analysis of the genes encoding the MT-ACS1 homologs from strains Z245 and FTF revealed the presence of a Glu codon at the equivalent position to the stop codon in the M. thermautotrophicus DH MT-ACS1. Alteration of the stop codon in the gene encoding M. thermautotrophicus DH MT-ACS1 to a Glu codon resulted in production of a full length, but insoluble, protein that was not characterized (data not shown).
The M. thermautotrophicus DH genome sequence annotation for the gene encoding MT-ACS2 indicates that this gene is also interrupted by a stop codon, as for MT-ACS1, resulting in two adjacent ORFs (MTH1603-MTH1604) that together have homology to full length ACS. The DNA region from the start ATG of MTH1603 to the stop codon of MTH1604 was amplified and cloned into the pETBlue-1 expression vector, as were the genes encoding the MT-ACS2 homologs from M. thermautotrophicus strains Z245 and FTF. The three MT-ACS2 homologs share 97% identity in deduced amino acid sequence, with only 12–14 amino acid differences between any two (data not shown). Sequence analysis of the cloned M. thermautotrophicus DH MT-ACS2 gene indicated an error in the M. thermautotrophicus DH genome sequence. An additional nucleotide is present that shifts the reading frame such that a full length protein of 73 kDa is encoded.
Heterologous expression of the M. thermautotrophicus Z245 and FTF MT-ACS1 genes in E. coli resulted in soluble, active proteins of the expected size for a full length ACS. The M. thermautotrophicus Z245 MT-ACS1 (henceforth referred to as MT-ACS1) was purified to electrophoretic homogeneity and subjected to biochemical and kinetic characterization. Expression of each of the three MT-ACS2 genes in E. coli resulted in production of a 73 kDa protein (data not shown), in agreement with the size predicted for the full length ACS. However, all three MT-ACS2 proteins were insoluble and were not characterized.
Phylogenetic analysis of ACS
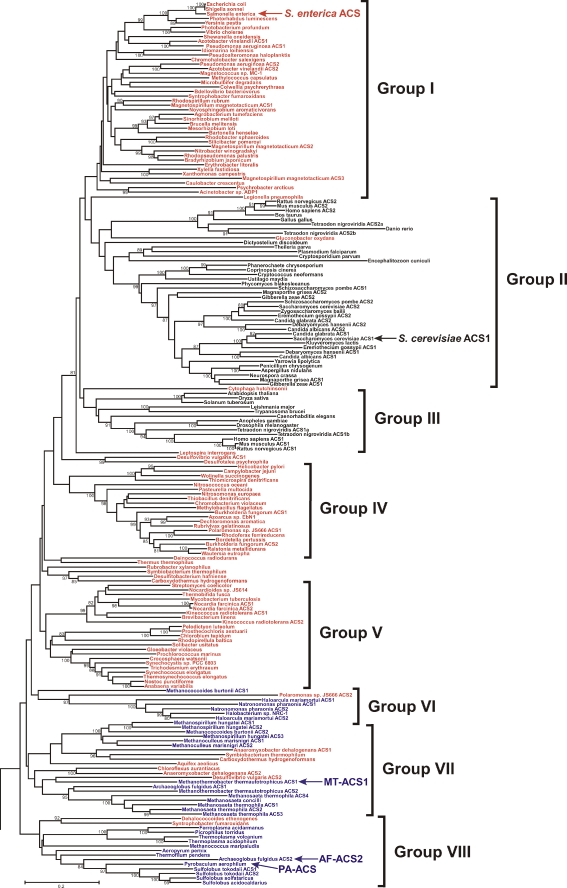
Phylogenetic analysis of putative short and medium chain acyl-CoA synthetase sequences revealed several distinct clades, one of which contains all of the proven ACSs. Propionyl-CoA synthetase (Horswill and Escalante-Semerena 2002), Sa (Fujino et al. 2001b) and MACS1 (Fujino et al. 2001b) acyl-CoA synthetases that show a preference for propionate, isobutyrate and octanoate, respectively, reside in other clades outside of the ACS clade. Within the ACS clade, shown in Figure 1, the sequences form eight major groups. Most ACS sequences group according to domain. Groups II and III are composed solely of eukaryotic sequences, with the exception of a single bacterial sequence in each. There are three large groups (I, IV and V) composed exclusively of bacterial sequences and several small bacterial clusters. The archaeal sequences fall into three groups (VI, VII and VIII), each of which also contains one or more bacterial sequences (Figure 1).
Figure 1.
Phylogeny of ACS sequences. A phylogeny of putative short and medium chain acyl-CoA synthetases from the finished genome sequences available at NCBI was constructed using the neighbor joining algorithm of MEGA (Kumar et al. 1994). Only the major clade containing the proven ACSs is shown here. For most genera, sequences from only one species were used in constructing the phylogeny for brevity and readability. Eukaryotic sequences are indicated in black, bacterial sequences in red and archaeal sequences in blue.
The S. enterica ACS and S. cerevisiae ACS1 sequences, representing the only two ACSs whose structures have been solved (Gulick et al. 2003, Jogl and Tong 2004), reside in Groups I and II. Among the putative archaeal ACSs, the sequences of the Haloarcula marismortui ACS1 and P. aerophilum ACS (PA-ACS), for which enzymatic activities have been proven (Brasen and Schonheit 2005, Brasen et al. 2005), reside in Groups VII and VIII, respectively (Figure 1). The Methanosaeta concilii ACS sequence in Group VII is identical to that of the Methanothrix soehngenii ACS, which has been purified and characterized (Jetten et al. 1989, Eggen et al. 1991).
To obtain a more thorough representation of the characteristics of archaeal enzymes across the ACS phylogeny, the M. thermautotrophicus MT-ACS1 and A. fulgidus AF-ACS2, whose sequences reside in groups VII and VIII of the phylogeny (Figure 1), were biochemically and kinetically characterized.
Biochemical analysis of MT-ACS1 and AF-ACS2
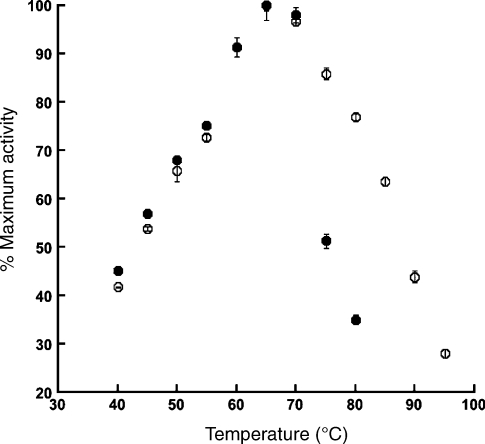
The enzymes MT-ACS1 and AF-ACS2 were heterologously produced in E. coli as soluble, active proteins and purified. The calculated masses for the MT-ACS1 and AF-ACS2 monomers are 71,556 Da and 77,587 Da, respectively. The molecular masses of the native enzymes were determined by gel filtration chromatography to be 144.8 kDa and 221.2 kDa, respectively, suggesting MT-ACS1 is a dimer and AF-ACS2 is a trimer. The temperature optimum was determined to be about 65–70 °C for each enzyme (Figure 2). Less than 50% activity was observed below 45 °C for both enzymes. At 80 °C, MT-ACS1 retained only 35% activity, whereas AF-ACS2 retained 44% activity at 90 °C and still had 20% activity at 100 °C.
Figure 2.
Temperature optima for MT1-ACS and AF-ACS2. Enzyme reactions were performed at the indicated temperatures in triplicate. Activities are reported as a percentage of the maximum activity determined for each enzyme. Symbols: filled circles = MT1-ACS; and open circles = AF-ACS2.
Kinetic analysis of MT-ACS1 and AF-ACS2
The acyl substrate range for MT-ACS1 was limited to acetate and propionate; the enzyme was unable to utilize butyrate. However, AF-ACS2 was able to utilize acetate, propionate and butyrate. This enzyme also had strong, but unsaturable, activity with isobutyrate and weak, but unsaturable, activity with valerate, but could not utilize larger acyl substrates or other branched chain acyl substrates. The kinetic parameters determined for each enzyme are shown in Table 1.
Table 1.
Kinetic parameters for MT-ACS1 and AF-ACS2.
| Substrate | Enzyme | Km | kcat | kcat/Km |
| (mM) | (s–1) | (s–1 mM–1) | ||
| Acetate | MT-ACS11 | 3.5 ± 0.1 | 65.4 ± 0.3 | 18.6 ± 0.5 |
| AF-ACS2 | 1.7 ± 0.06 | 35.9 ± 0.58 | 21.2 ± 0.4 | |
| Propionate | MT-ACS11 | 36.5 ± 1.9 | 46.3 ± 0.7 | 1.3 ± 0.04 |
| AF-ACS2 | 3.9 ± 0.02 | 35.7 ± 0.17 | 9.1 ± 0.06 | |
| Butyrate | MT-ACS11 | — | — | — |
| AF-ACS2 | 133.0 ± 14.6 | 1.68 ± 0.07 | 0.013 ± 0.0009 | |
| Valerate | MT-ACS1 | — | — | — |
| AF-ACS2 | Unsaturable | |||
| Isobutyrate | MT-ACS1 | — | — | — |
| AF-ACS2 | Unsaturable | |||
| ATP | MT-ACS11 | 3.3 ± 0.2 | 66.6 ± 0.9 | 20.2 ± 0.9 |
| AF-ACS2 | 2.9 ± 0.01 | 38.1 ± 0.12 | 13.2 ± 0.09 | |
| CoA | MT-ACS1 | 0.19 ± 0.003 | 81.6 ± 0.7 | 423.7 ± 4.7 |
| AF-ACS2 | 0.30 ± 0.003 | 44.3 ± 0.2 | 144.9 ± 1.2 | |
1 Kinetic parameters for MT-ACS1 were determined in the presence of 0.5 mM CoA.
There are a number of noteworthy points to be made from the results in Table 1. The two enzymes have similar affinity (Km) for acetate but very different affinities for propionate. The affinity of AF-ACS2 for propionate is almost identical to that of MT-ACS1 for acetate and nearly tenfold higher than the affinity of MT-ACS1 for propionate. Whereas the difference in affinity between the two substrates is over tenfold for MT-ACS1, AF-ACS2 has only a 2.3-fold higher affinity for acetate than propionate. MT-ACS1 has a strong preference for acetate as the substrate as shown by the 14-fold higher catalytic efficiency (kcat/Km) with acetate versus propionate. However, AF-ACS2 has only a 2.3-fold higher preference for acetate over propionate. Finally, AF-ACS2 was able to utilize butyrate as substrate, although the Km was 78-fold higher than that for acetate and 34-fold higher than that for propionate, and the turnover rate was 21-fold reduced with butyrate compared with acetate or propionate.
The Km values for CoA for both enzymes showed less than twofold difference (Table 1), and the Km values for ATP were similar for both enzymes. Both enzymes demonstrated a strong preference for ATP versus CTP, GTP, TTP, UTP, ITP or ADP, for which less than 5% activity was observed (data not shown).
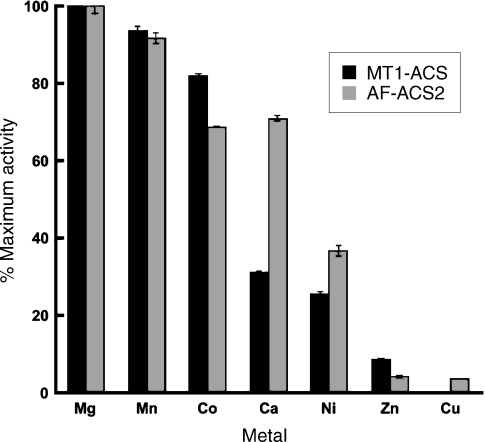
The metal specificity was tested for each enzyme using a standard metal concentration of 20 mM and 20 mM ATP. Both MT-ACS1 and AF-ACS2 showed strong preference for Mg2+ and Mn2+ as the divalent metal (Figure 3) and Co2+ also gave high activity. Strong activity was observed with Ca2+ for AF-ACS2 but not for MT-ACS1. Moderate activity was observed for both enzymes with Ni2+, whereas Cu2+ and Zn2+ worked poorly for both enzymes. The kinetic parameters determined for those metals that gave the highest activity are shown in Table 2. For MT-ACS1, the highest affinity and turnover rate were observed with Mg2+. Although the highest turnover rate was observed with Mg2+, Mn2+ and Ca2+ gave the highest catalytic efficiencies for AF-ACS2.
Figure 3.
Divalent metal specificity for MT1-ACS and AF-ACS2. Enzyme reactions were performed in triplicate at 65 °C in the presence of 20 mM metal (as the chloride salt) +20 mM ATP. Activities are reported as a percentage of the maximum activity determined for each enzyme with Mg2+ as the metal substrate.
Table 2.
Kinetic parameters for divalent metals for MTI-ACS and AF-ACS2.
| Enzyme | Metal | Km (mM) | kcat (s–1) | kcat/Km (s–1 mM–1) |
| MT1-ACS1 | Mg2+ | 0.38 ± 0.02 | 56.5 ± 1.0 | 147.5 ± 4.5 |
| Mn2+ | 0.61 ± 0.02 | 44.6 ± 0.5 | 72.8 ± 2.3 | |
| Co2+ | 0.99 ± 0.03 | 39.4 ± 0.3 | 40.0 ± 1.1 | |
| AF-ACS2 | Mg2+ | 1.08 ± 0.05 | 23.4 ± 0.3 | 21.8 ± 0.7 |
| Mn2+ | 0.70 ± 0.02 | 20.6 ± 0.23 | 29.7 ± 0.76 | |
| Ca2+ | 0.64 ± 0.006 | 18.2 ± 0.05 | 28.3 ± 0.27 | |
| Co2+ | 1.50 ± 0.02 | 19.0 ± 0.13 | 12.2 ± 0.09 | |
1 Kinetic parameters for MT-ACS1 were determined in the presence of 0.5 mM CoA.
Discussion
Although our kinetic and biochemical characterization of MT-ACS1 and AF-ACS2 expands our knowledge of the properties of ACSs, there is still a paucity of information on this important class of enzymes. With the advent of whole genome sequencing, gene functions are usually assigned based on homology with other sequences in the sequence databases. In many cases, a particular enzymatic function may be assigned based on homology to just a single sequence whose function has been proven. Acetyl-CoA synthetase is widespread in all three domains, and most putative ACSs have been assigned this function through homology. Of the 193 ACS sequences shown in the phylogeny in Figure 1, only a handful have been biochemically characterized. The S. enterica ACS and S. cerevisiae ACS1, whose structures have been solved (Gulick et al. 2003, Jogl and Tong 2004), are quite distant from the archaeal ACSs, for which there is no structure.
Although genes predicted to encode for ACS are widespread in the Archaea, only a few archaeal ACSs have been biochemically characterized. Acetyl-CoA synthetase activity was first detected in archaea in Methanothermobacter marburgensis (formerly Methanobacterium thermoautotrophicum Marburg) (Oberlies et al. 1980), a thermophilic chemolithoautotrophic methanoarchaeon that can utilize H2/CO2 as the sole carbon and energy source (Zeikus and Wolfe 1972). When M. marburgensis (closely related to M. thermautotrophicus) was grown on H2/CO2 in the presence of acetate, 10% of cellular carbon was derived from acetate with the remainder derived from CO2 (Fuchs et al. 1978). Oberlies et al. (1980) subsequently demonstrated ACS activity in M. marburgensis cells grown with limiting H2/CO2 and proposed that ACS allows assimilation of acetate as a cellular carbon source in order to spare limited supplies of CO2.
The first archaeal ACSs purified and characterized were those from M. soehngenii and Methanothrix thermophila CALS-1 (now Methanosaeta concilii and Methanosaeta thermophila CALS-1) (Jetten et al. 1989, Teh and Zinder 1992). Acetyl-CoA synthetase is the first enzyme in the activation of acetate to acetyl-CoA for methanogenesis in the obligately acetoclastic Methanosaeta (Jetten et al. 1989, Teh and Zinder 1992, Allen and Zinder 1996). In fact, the recently completed genome sequences of Methanosaeta species reveal that M. thermophila PT has four genes that encode ACS and M. concilii has five (K.S. Smith and C. Ingram-Smith, unpublished data). All of the methanoarchaea with ACS have at least two acs genes, with the exception of Methanococcus maripaludis (Figure 1).
A number of halophilic archaea are able to utilize acetate as a carbon and energy source. Brasen and Schonheit (Brasen and Schonheit 2001, Brasen and Schonheit 2004) demonstrated that Halococcus saccharolyticus, Haloferax volcanii, Halorubrum saccharovorum and H. marismortui grown on acetate as a carbon and energy source exhibited ACS activity. During growth on glucose, these halophiles excreted acetate into the media and exhibited ADP-forming acetyl-CoA synthetase activity (ADP-ACS; acetyl-CoA + ADP + Pi« acetate + ATP + CoA) but not ACS activity. Upon entry into stationary phase, ACS activity was induced and the excreted acetate was consumed. Thus, ADP-ACS was determined to be responsible for acetate and ATP production from excess acetyl-CoA during growth on glucose but ACS was responsible for activation of acetate for use as a carbon and energy source.
Although the H. marismortui, M. soehngenii, and M. thermophila CALS-1 ACSs and the M. thermautotrophicus MT-ACS1 all show a strong preference for acetate, the characterized ACSs from A. fulgidus and P. aerophilum have an expanded substrate range. What physiological purpose could this broad substrate range serve? One possibility is that A. fulgidus and P. aerophilum can utilize a more diverse array of carbon or energy sources than other archaea. Both P. aerophilum and A. fulgidus can utilize complex organics such as yeast extract, meat extract, tryptone, and peptone as growth substrates (Stetter 1988, Volkl et al. 1993), whereas the others cannot. The presence of an ACS with an expanded substrate range may provide a means for utilization of other short chain fatty acids either scavenged from the environment or from the breakdown of complex organics without the need for additional enzymes.
The findings of the phylogenetic analysis presented here (Figure 1) contrast with those of Brasen and Schonheit (2005) with respect to the archaeal ACSs. In their analysis, the archaeal sequences formed one distinct clade, leading to the conclusion that the archaeal sequences form a separate branch within the prokaryotic sequences and have a monophyletic origin (Brasen and Schonheit 2005). In our analysis, the archaeal sequences form three groups, each of which also contains bacterial sequences. This may be a result of the larger number of sequences from each domain used in this analysis (193 sequences composed of 107 bacterial, 33 archaeal, and 53 eukaryotic sequences versus 51 total sequences by Brasen and Schonheit (2005)) and differences in methodology. However, a separate phylogenetic analysis using the minimum evolution algorithm of the MEGA package (Kumar et al. 1994) showed a similar result (data not shown).
Overall, the ACSs show strong sequence conservation regardless of domain, as indicated by pairwise amino acid sequence comparisons that show MT-ACS1 and AF-ACS2 share 38–49% identity and 57–67% similarity with S. cerevisiae ACS1 and S. enterica ACS. However, the subunit composition of the ACSs is quite diverse, with monomeric (S. enterica and H. marismortui ACSs (Brasen and Schonheit 2005, Gulick et al. 2003), dimeric (MT-ACS1, M. concilii, and Bradyrhizobium japonicum ACSs (Jetten et al. 1989, Preston et al. 1990, Lee et al. 2001)), trimeric (AF-ACS2 and S. cerevisiae ACS1 (Jogl and Tong 2004)), and octameric enzymes (PA-ACS (Brasen et al. 2005)) thus far characterized.
Characterized enzymes from groups I, II, III, VI, and VII, which include the bacterial ACSs from S. enterica (Gulick et al. 2003) and B. japonicum (Preston et al. 1990, Lee et al. 2001), the eukaryotic ACS1 and ACS2 enzymes from human (Luong et al. 2000, Fujino et al. 2001a) and S. cerevisiae (van den Berg et al. 1996), and archaeal ACSs from M. concilii (M. soehngenii) (Jetten et al. 1989, Eggen et al. 1991), H. marismortui (Brasen and Schonheit 2005), and M. thermautotrophicum (MT-ACS1), all show a strong preference for acetate as the acyl substrate, with propionate being the only alternative acyl substrate. AF-ACS2 and the P. aerophilum PA-ACS (Brasen et al. 2005), both from group VIII, have an expanded substrate range that includes butyrate and the branched chain isobutyrate as well as acetate and propionate. AF-ACS2 shows only a weak (twofold) preference for acetate over propionate, unlike most ACSs that generally have a 10- to 20-fold preference for acetate, as determined by the higher catalytic efficiency (kcat/Km).
Although a full kinetic characterization of PA-ACS was not reported, the Km value for acetate was determined to be 3 µM, over 500-fold lower than that observed for AF-ACS2. Although this may indicate that these enzymes have very different kinetic properties, it may also be due to differences in the enzyme assay used in the two studies. The Km value for acetate observed for PA-ACS is at least 20- to several hundredfold lower than that observed for any other ACS. The Km value for acetate for AF-ACS2 is well in line with the values determined using the hydroxamate assay with other archaeal ACSs including MT-ACS1 (Jetten et al. 1989, Preston et al. 1990, Teh and Zinder 1992). It is not known whether these differences are meaningful with regard to whether these enzymes may represent a subgroup of ACSs that show only weak preference for acetate and expanded substrate range.
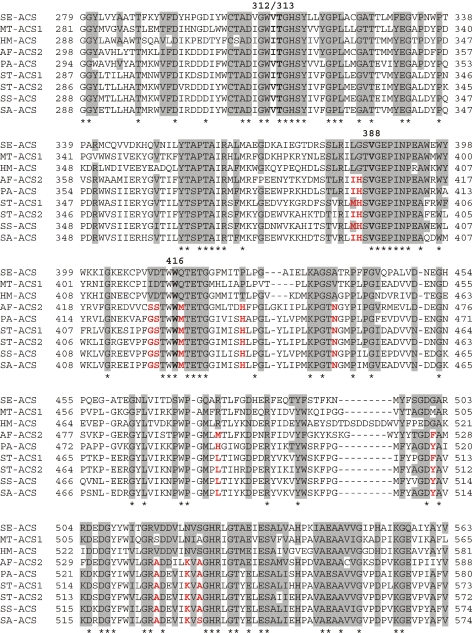
These findings lead one to question whether AF-ACS2 and PA-ACS are anomalies within the ACSs or whether they represent a subset of enzymes with different properties from the “traditional” ACSs that have a strong preference for acetate and a narrow substrate range. Four residues (Ile312, Thr313, Val388, and Trp416) have been shown to form the acetate binding pocket of MT-ACS1 and have been shown to be important in acyl substrate selection (Ingram-Smith et al. 2006). Alteration of any of these four residues influences substrate affinity or substrate range, or both, as well as catalysis (Ingram-Smith et al. 2006). For example, alteration of Trp416 to Gly, the residue found in short and medium chain acyl-CoA synthetases other than acetyl- and propionyl-CoA synthetases, expands the substrate range of MT-ACS1 such that the enzyme can utilize substrates ranging from acetate to octanoate (including some branched chain acyl substrates) and changes the substrate preference from acetate to valerate. In propionyl-CoA synthetase, Val388 is replaced by Ala. A Val388Ala MT-ACS1 variant has higher affinity for propionate than acetate and a slightly greater preference for propionate as well. Among the ACS sequences in Figure 1, including both AF-ACS2 and PA-ACS, Thr313, Val388, and Trp416 are completely conserved and Ile312 is highly conserved, with Val as the only other amino acid observed at the equivalent position.
Within group VIII in the ACS phylogeny in Figure 1, a subclade consisting of AF-ACS2 and PA-ACS as well as four Sulfolobus sequences is strongly supported by bootstrapping (84%). These sequences were aligned with the S. enterica ACS, MT-ACS1, and H. marismortui ACS sequences and a partial alignment is shown in Figure 4. ConSurf analysis (Armon et al. 2001, Glaser et al. 2003, Landau et al. 2005) of the alignment was performed to help delineate amino acid residues that might contribute to the broad substrate range of AF-ACS2 and PA-ACS and suggest whether members of this subclade within group VIII represents a subclass of ACSs with expanded substrate range. ConSurf calculates evolutionary conservation scores for each residue within a protein sequence based on protein structure, multiple sequence alignment, evolutionary distance between sequences, and evolutionary tree topology. The evolutionary conservation scores are then mapped onto the protein structure to define probable regions of structural and functional importance.
Figure 4.
Alignment and ConSurf analysis of ACS sequences. The M. thermautotrophicus MT-ACS1 and A. fulgidus AF-ACS2 amino acid sequences were aligned with the S. enterica ACS sequence (SE-ACS) and the ACS sequences from P. aerophilum (PA-ACS), Sulfolobus tokodaii (ST-ACS1 and ACS2), Sulfolobus solfataricus (SS-ACS), and Sulfolobus acidocaldarius (SA-ACS) using Clustal X (Thompson et al. 1997). A partial alignment is shown here. Residues of the S. enterica ACS found to have high evolutionary conservation scores by ConSurf analysis (Armon et al. 2001, Glaser et al. 2003, Landau et al. 2005) (http://consurf.tau.ac.il) are shaded, as are residues in the other ACS sequences that are identical or among the alternative residues listed for each of these highly conserved positions. Asterisks indicate those positions that are identical in all nine sequences. The acetate binding pocket residues are boldfaced and numbered above the aligned sequences according to their position within MT-ACS1. Those residues at positions with high ConSurf scores that differ in AF-ACS2, PA-ACS, and the Sulfolobus sequences from the three ACS sequences representing enzymes with “traditional” characteristics are indicated in red.
Using the S. enterica ACS structure as the query for ConSurf analysis, the evolutionary conservation score for each position along with the alternative residues from the fifty most closely related sequences were determined. Salmonella enterica ACS residues determined to be the most highly conserved by ConSurf analysis are shaded in the partial alignment (Figure 4), along with residues in the other sequences that are identical or among the alternative residues for that position. In the complete alignment, eleven residues with high ConSurf scores (all of which are shown in the partial alignment in Figure 4) are conserved in the three ACS sequences representing enzymes with “traditional” characteristics, but differ in AF-ACS2 and PA-ACS. In addition, the four Sulfolobus sequences that reside in the same subclade in group VIII as AF-ACS2 and PA-ACS of the phylogeny also differ at these same positions.
MT-ACS1 and AF-ACS2 have been modeled on the S. enterica ACS structure to determine whether the acetate binding pockets show any major differences that could be attributed to the differences in substrate preference and substrate range of these two enzymes. The models depicted in Figure 5 show residues within 10 Å of the propyl group of the adenosine-5′-propylphosphate ligand. In the S. enterica structure, the adenosine-5′-propylphosphate mimics the acetyl-adenylate intermediate and the propyl group approximates the position of acetate in the active site (Gulick et al. 2003). Overall, the acetate binding sites of the two modeled enzymes are very similar (Figure 5). However, there are key differences in the positioning of certain residues that are conserved in both enzymes and in residues that were identified by the ConSurf analysis (Figure 4) to be conserved in the traditional ACSs but not the AF-ACS2/PA-ACS subclade of group VIII (Figure 1).
Figure 5.
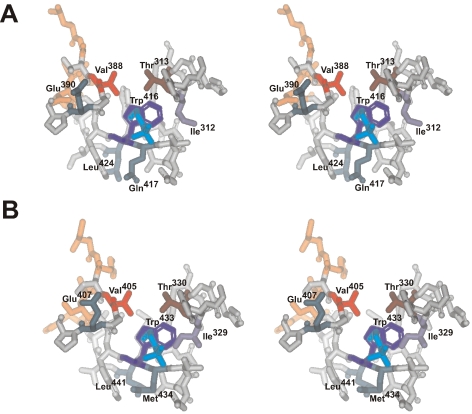
The acetate binding pocket of MT-ACS1 and AF-ACS2. (A) MT-ACS1 and (B) AF-ACS2 were modeled on the S. enterica ACS structure (PDB: 1PG4) using Accelrys DS Modeler 1.1 and the stereo image of the putative active site acetate binding pocket was created using DS ViewerPro 5.0. Residues within a 10 Å sphere of the propyl moiety of the propylphosphate group of the adenosine-5’-propylphosphate mimic of the acetyl-adenylate intermediate are shown. The acetate pocket residues (Ile312, Thr313, Val388, and Trp416 of MT-ACS1 and Ile329, Thr330, Val405, and Trp433 of AF-ACS2) are colored and labeled. Those residues with high ConSurf scores that differ between MT-ACS1 and AF-ACS2 are shown in orange. Residues discussed in the text are labeled and shown in dark green. The propylphosphate group is shown in aqua.
The propyl group is in a similar position in both the MT-ACS1 and AF-ACS2 models, and three of the acetate pocket residues occupy similar positions as well. However, whereas Ile312 of MT-ACS points away from the propylphosphate group (Figure 5A), Ile329 of AF-ACS2 points inwards (Figure 5B). This may increase the hydrophobicity of the acetate pocket and could account in part for the higher affinity of AF-ACS2 for acyl substrates than observed for MT-ACS1. Among residues identical to both enzymes, the other major difference observed in the acetate pocket region is that Leu424 of MT-ACS1 is positioned quite differently from the equivalent residue Leu441 of AF-ACS2. The overall increased hydrophobicity of the acyl substrate binding pocket of AF-ACS2 may positively influence substrate affinity, but it is likely that the combined effects of multiple residues are required to determine whether the enzyme has a narrow or broad substrate range.
Of the eleven residues identified by ConSurf analysis to be highly conserved in traditional ACSs but not the AF-ACS2/PA-ACS subclade (Figure 4), six of these positions are in close proximity to Val388 and Trp416 of MT-ACS1 (Figure 5A), the two acetate pocket residues that were shown to have the greatest influence on substrate range and preference (Ingram-Smith et al. 2006). Five of these residues (Leu385, Gly386, Ile412, Asp413, and Pro427 of MT-ACS1 and Ile402, His403, Ser429, Ser430, and His444 of AF-ACS2) are clustered to one side of the pocket for both enzymes (Figure 5) and may influence the positioning of Glu390, which points slightly inward toward the pocket in MT-ACS1, and the equivalent residue Glu407 of AF-ACS2 that points slightly away from the acetate pocket. Withdrawal of the negative charge of this residue from the acetate pocket in AF-ACS2 may positively influence substrate binding.
The sixth residue, represented by Gln417 of MT-ACS1 and Met434 of AF-ACS2, is more intimately positioned near the acetate pocket (Figure 5). Glutamine417 points away from the acetate pocket and the propylphosphate group, whereas Met434 bends in toward the propylphosphate. Met434 may play a role in expanding the substrate range of AF-ACS2 by extending the hydrophobic pocket to better accommodate propionate and butyrate.
The results of our kinetic characterization of AF-ACS2 and the analysis of PA-ACS by Brasen et al. (2005), combined with phylogenetic analysis, sequence alignment, and structural modeling lead us to speculate that AF-ACS2 and PA-ACS, along with the Sulfolobus ACSs in group VIII in the phylogeny (Figure 4), represent a subclass of ACSs. We term these ACSs as “transitional,” meaning that these enzymes have kinetic characteristics intermediate between traditional ACSs and propionyl-CoA synthetase and other short and medium chain acyl-CoA synthetases that show a preference for substrates other than acetate and have an expanded substrate range (Fujino et al. 2001a, Horswill and Escalante-Semerena 2002). The transitional ACSs would be expected to have only a slight preference for acetate over other substrates, but would also be expected to have a broader substrate range than traditional ACSs that utilize only acetate and propionate. Clearly, more evidence is necessary before the concept of transitional ACSs can be accepted. However, the accumulated data suggest a direction for further studies to prove or disprove this idea.
Acknowledgments
This work was supported by NIH (Award GM69374-01A1 to K.S. Smith), the South Carolina Experiment Station (Project SC-1700198 to K.S. Smith) and Clemson University. We thank Jean Pierre Touzel for providing genomic DNA from M. thermautotrophicus Z245 and Imke Schroeder for providing genomic DNA from A. fulgidus. We also thank Kelcie Brunson for her technical assistance.
References
- R1.Allen G.W.J., Zinder S.H. Methanogenesis from acetate by cell-free extracts of the thermophilic acetotrophic methanogen Methanothrixthermophila CALS-1. Arch. Microbiol. 1996;166:275–281. doi: 10.1007/s002030050384. [DOI] [PubMed] [Google Scholar]
- R2.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- R3.Altschul S.F., Madden T.L., Schaffer A.A., Zhang J., Zhang Z., Miller W., Lipman D.J. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Anke H., Spector L.B. Evidence for an acetyl-enzyme intermediate in the action of acetyl-CoA synthetase. Biochem. Biophys. Res. Commun. 1975;67:767–773. doi: 10.1016/0006-291x(75)90879-7. [DOI] [PubMed] [Google Scholar]
- R5.Armon A., Graur D., Ben-tal N. ConSurf: an algorithmic tool for the identification of functional regions in proteins by surface mapping of phylogenetic information. J. Mol. Biol. 2001;307:447–463. doi: 10.1006/jmbi.2000.4474. [DOI] [PubMed] [Google Scholar]
- R6.Babbitt P.C., Kenyon G.L., Martin B.M., Charest H., Slyvestre M., Scholten J.D., Chang K.H., Liang P.H., Dunaway-mariano D. Ancestry of the 4-chlorobenzoate dehalogenase: analysis of amino acid sequence identities among families of acyl:adenyl ligases, enoyl-CoA hydratases/isomerases, and acyl-CoA thioesterases. Biochemistry. 1992;31:5594–5604. doi: 10.1021/bi00139a024. [DOI] [PubMed] [Google Scholar]
- R7.Berg P. Acyl adenylates: an enzymatic mechanism of acetate activation. J. Biol. Chem. 1956;222:991–1013. [PubMed] [Google Scholar]
- R8.Berg P. Adenylates: the synthesis and properties of adenyl acetate. J. Biol. Chem. 1956;222:1015–1034. [PubMed] [Google Scholar]
- R9.Bradford M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- R10.Brasen C., Schonheit P. Mechanisms of acetate formation and acetate activation in halophilic archaea. Arch. Microbiol. 2001;175:360–368. doi: 10.1007/s002030100273. [DOI] [PubMed] [Google Scholar]
- R11.Brasen C., Schonheit P. Regulation of acetate and acetyl-CoA converting enzymes during growth on acetate and/or glucose in the halophilic archaeon Haloarculamarismortui . FEMS Microbiol. Lett. 2004;241:21–26. doi: 10.1016/j.femsle.2004.09.033. [DOI] [PubMed] [Google Scholar]
- R12.Brasen C., Schonheit P. AMP-forming acetyl-CoA synthetase from the extremely halophilic archaeon Haloarculamarismortui: purification, identification and expression of the encoding gene, and phylogenetic affiliation. Extremophiles. 2005;9:355–365. doi: 10.1007/s00792-005-0449-0. [DOI] [PubMed] [Google Scholar]
- R13.Brasen C., Urbanke C., Schonheit P. A novel octameric AMP-forming acetyl-CoA synthetase from the hyperthermophilic crenarchaeon Pyrobaculumaerophilum . FEBS Lett. 2005;579:477–482. doi: 10.1016/j.febslet.2004.12.016. [DOI] [PubMed] [Google Scholar]
- R14.Chang K.H., Xiang H., Dunaway-mariano D. Acyl-adenylate motif of the acyl-adenylate/thioester-forming enzyme superfamily: a site-directed mutagenesis study with the Pseudomonassp. strain CBS3 4-chlorobenzoate:coenzyme A ligase. Biochemistry. 1997;36:15650–15659. doi: 10.1021/bi971262p. [DOI] [PubMed] [Google Scholar]
- R15.Conti E., Franks N.P., Brick P. Crystal structure of firefly luciferase throws light on a superfamily of adenylate-forming enzymes. Structure. 1996;4:287–298. doi: 10.1016/s0969-2126(96)00033-0. [DOI] [PubMed] [Google Scholar]
- R16.Conti E., Stachelhaus T., Marahiel M.A., Brick P. Structural basis for the activation of phenylalanine in the non-ribosomal biosynthesis of gramicidin S. EMBO J. 1997;16:4174–4183. doi: 10.1093/emboj/16.14.4174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.Eggen R.I., Geerling A.C., Boshoven A.B., De vos W.M. Cloning, sequence analysis, and functional expression of the acetyl coenzyme A synthetase gene from Methanothrixsoehngenii in Escherichiacoli . J. Bacteriol. 1991;173:6383–6389. doi: 10.1128/jb.173.20.6383-6389.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.Fuchs G., Stupperich E., Thauer R.K. Acetate assimilation and the synthesis of alanine, aspartate and glutamate in Methanobacteriumthermoautotrophicum . Arch. Microbiol. 1978;117:61–66. doi: 10.1007/BF00689352. [DOI] [PubMed] [Google Scholar]
- R19.Fujino T., Kondo J., Ishikawa M., Morikawa K., Yamamoto T.T. Acetyl-CoA synthetase 2, a mitochondrial matrix enzyme involved in the oxidation of acetate. J. Biol. Chem. 2001;276:11420–11426. doi: 10.1074/jbc.M008782200. [DOI] [PubMed] [Google Scholar]
- R20.Fujino T., Takei Y.A., Sone H., Ioka R.X., Kamataki A., Magoori K., Takahashi S., Sakai J., Yamamoto T.T. Molecular identification and characterization of two medium-chain acyl-CoA synthetases, MACS1 and the Sa gene product. J. Biol. Chem. 2001;276:35961–35966. doi: 10.1074/jbc.M106651200. [DOI] [PubMed] [Google Scholar]
- R21.Glaser F., Pupko T., Paz I., Bell R.E., Bechor-shental D., Martz E., Ben-tal N. ConSurf: identification of functional regions in proteins by surface-mapping of phylogenetic information. Bioinformatics. 2003;19:163–164. doi: 10.1093/bioinformatics/19.1.163. [DOI] [PubMed] [Google Scholar]
- R22.Grayson N.A., Westkaemper R.B. Stable analogs of acyl adenylates. Inhibition of acetyl- and acyl-CoA synthetase by adenosine 5′-alkylphosphates. Life Sci. 1988;43:437–444. doi: 10.1016/0024-3205(88)90523-1. [DOI] [PubMed] [Google Scholar]
- R23.Gulick A.M., Starai V.J., Horswill A.R., Homick K.M. The 1.75 Å crystal structure of acetyl-CoA synthetase bound to adenosine-5′-propylphosphate and coenzyme A. Biochemistry. 2003;42:2866–2873. doi: 10.1021/bi0271603. [DOI] [PubMed] [Google Scholar]
- R24.Horswill A.R., Escalante-Semerena J.C. Characterization of the propionyl-CoA synthetase (PrpE) enzyme of Salmonellaenterica: residue Lys592 is required for propionyl-AMP synthesis. Biochemistry. 2002;41:2379–2387. doi: 10.1021/bi015647q. [DOI] [PubMed] [Google Scholar]
- R25.Ingram-Smith C., Woods B.I., Smith K.S. Characterization of the acyl substrate binding pocket of acetyl-CoA synthetase. Biochemistry. 2006 doi: 10.1021/bi061023e. In press. [DOI] [PubMed] [Google Scholar]
- R26.Jetten M.S., Stams A.J., Zehnder A.J. Isolation and characterization of acetyl-coenzyme A synthetase from Methanothrixsoehngenii . J. Bacteriol. 1989;171:5430–5435. doi: 10.1128/jb.171.10.5430-5435.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R27.Jogl G., Tong L. Crystal structure of yeast acetyl-coenzyme A synthetase in complex with AMP. Biochemistry. 2004;43:1425–1431. doi: 10.1021/bi035911a. [DOI] [PubMed] [Google Scholar]
- R28.Kleinkauf H., Von Dohren H. A nonribosomal system of peptide biosynthesis. Eur. J. Biochem. 1996;236:335–351. doi: 10.1111/j.1432-1033.1996.00335.x. [DOI] [PubMed] [Google Scholar]
- R29.Kumar S., Tamura K., Nei M. MEGA: molecular evolutionary genetic analysis. Comput. Appl. Biosci. 1994;10:189–191. doi: 10.1093/bioinformatics/10.2.189. [DOI] [PubMed] [Google Scholar]
- R30.Laemmli U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature. 1970;227:680–685. doi: 10.1038/227680a0. [DOI] [PubMed] [Google Scholar]
- R31.Landau M., Mayrose I., Rosenberg Y., Glaser F., Martz E., Pupko T., Ben-tal N. ConSurf 2005: the projection of evolutionary conservation scores of residues on protein structures. Nucleic Acids Res. 2005;33:W299–302. doi: 10.1093/nar/gki370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R32.Lee H.Y., Na K.B., Koo H.M., Kim Y.S. Identification of active site residues in Bradyrhizobiumjaponicum acetyl-CoA synthetase. J. Biochem. (Tokyo) 2001;130:807–813. doi: 10.1093/oxfordjournals.jbchem.a003052. [DOI] [PubMed] [Google Scholar]
- R33.Lipmann F., Tuttle L.C. A specific micromethod for determination of acyl phosphates. J. Biol. Chem. 1945;159:21–28. [Google Scholar]
- R34.Luong A., Hannah V.C., Brown M.S., Goldstein J.L. Molecular characterization of human acetyl-CoA synthetase, an enzyme regulated by sterol regulatory element-binding proteins. J. Biol. Chem. 2000;275:26458–26466. doi: 10.1074/jbc.M004160200. [DOI] [PubMed] [Google Scholar]
- R35.Marahiel M.A., Stachelhaus T., Mootz H.D. Modular peptide synthetases involved in nonribosomal peptide synthesis. Chem. Rev. 1997;97:2651–2674. doi: 10.1021/cr960029e. [DOI] [PubMed] [Google Scholar]
- R36.May J.J., Kessler N., Marahiel M.A., Stubbs M.T. Crystal structure of DhbE, an archetype for aryl acid activating domains of modular nonribosomal peptide synthetases. Proc. Natl. Acad. Sci. USA. 2002;99:12120–12125. doi: 10.1073/pnas.182156699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R37.Oberlies G., Fuchs G., Thauer R.K. Acetate thiokinase and the assimilation of acetate in Methanobacteriumthermoautotrophicum . Arch. Microbiol. 1980;128:248–252. doi: 10.1007/BF00406167. [DOI] [PubMed] [Google Scholar]
- R38.Preston G.G., Wall J.D., Emerich D.W. Purification and properties of acetyl-CoA synthetase from Bradyrhizobiumjaponicum bacteroids. Biochem. J. 1990;267:179–183. doi: 10.1042/bj2670179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R39.Rose I.A., Grunberg-manago M., Korey S.F., Ochoa S. Enzymatic phosphorylation of acetate. J. Biol. Chem. 1954;211:737–756. [PubMed] [Google Scholar]
- R40.Stetter K.O. Archaeoglobusfulgidus gen. nov., sp. nov.: a new taxon of extremely thermophilic archaebacteria. Syst. Appl. Microbiol. 1988;10:172–173. [Google Scholar]
- R41.Teh Y.L., Zinder S.H. Acetyl-coenzyme A synthetase in the thermophilic, acetate-utilizing methanogen Methanothrixsp. strain CALS-1. FEMS Microbiol. Lett. 1992;98:1–8. [Google Scholar]
- R42.Thompson J.D., Gibson T.J., Plewniak F., Jeanmougin F., Higgins D.J. The Clustal X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 1997;25:4876–4882. doi: 10.1093/nar/25.24.4876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R43.van den Derg M.A., De jong-gubbels P., Kortland C.J., Van dijken J.P., Pronk J.T., Steensma H.Y. The two acetyl-coenzyme A synthetases of Saccharomycescerevisiae differ with respect to kinetic properties and transcriptional regulation. J. Biol. Chem. 1996;271:28953–28959. doi: 10.1074/jbc.271.46.28953. [DOI] [PubMed] [Google Scholar]
- R44.Volkl P., Huber R., Drobner E., Rachel R., Burggraf S., Trincone A., Stetter K.O. Pyrobaculumaerophilum sp. nov., a novel nitrate-reducing hyperthermophilic archaeum. Appl. Environ. Microbiol. 1993;59:2918–2926. doi: 10.1128/aem.59.9.2918-2926.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R45.Webster L.T. Studies of the acetyl coenzyme A synthetase reaction I. Isolation and characterization of enzyme-bound acetyl adenylate. J. Biol. Chem. 1963;238:4010–4015. [PubMed] [Google Scholar]
- R46.Zeikus J.G., Wolfe R.S. Methanobacteriumthermoautotrophicus sp. n., an anaerobic, autotrophic, extreme thermophile. J. Bacteriol. 1972;109:707–715. doi: 10.1128/jb.109.2.707-713.1972. [DOI] [PMC free article] [PubMed] [Google Scholar]