Abstract
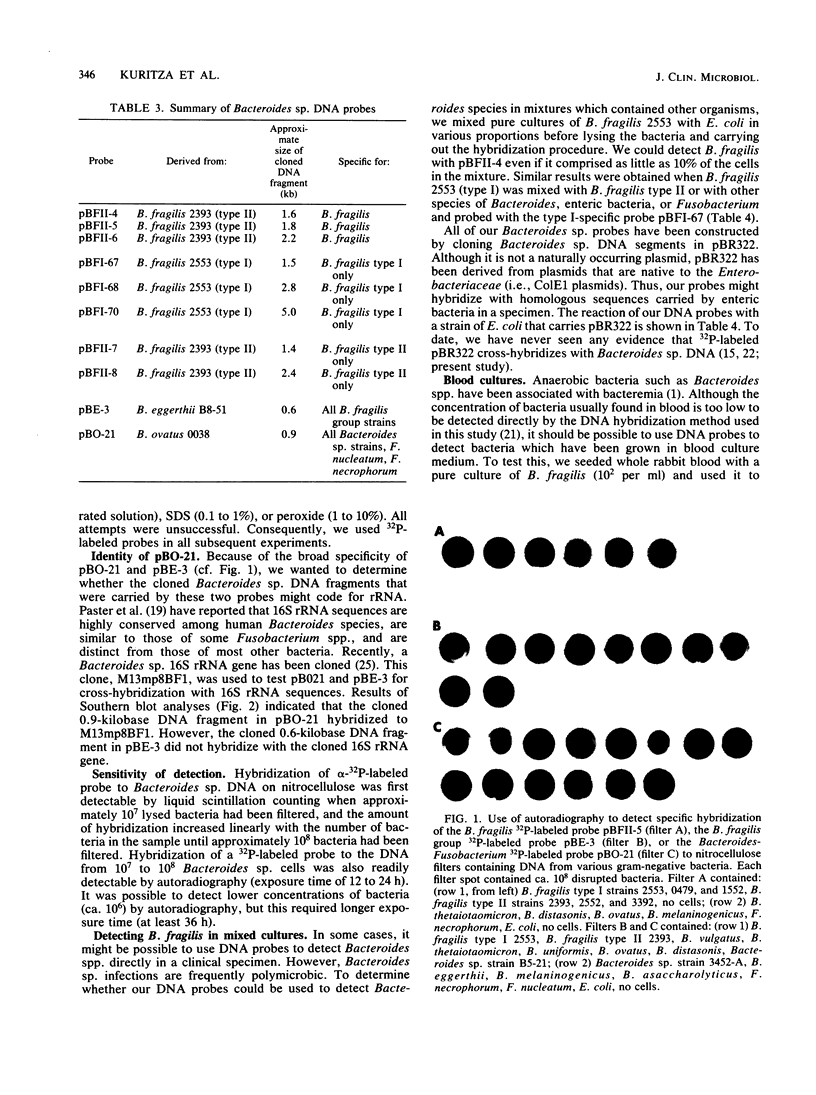
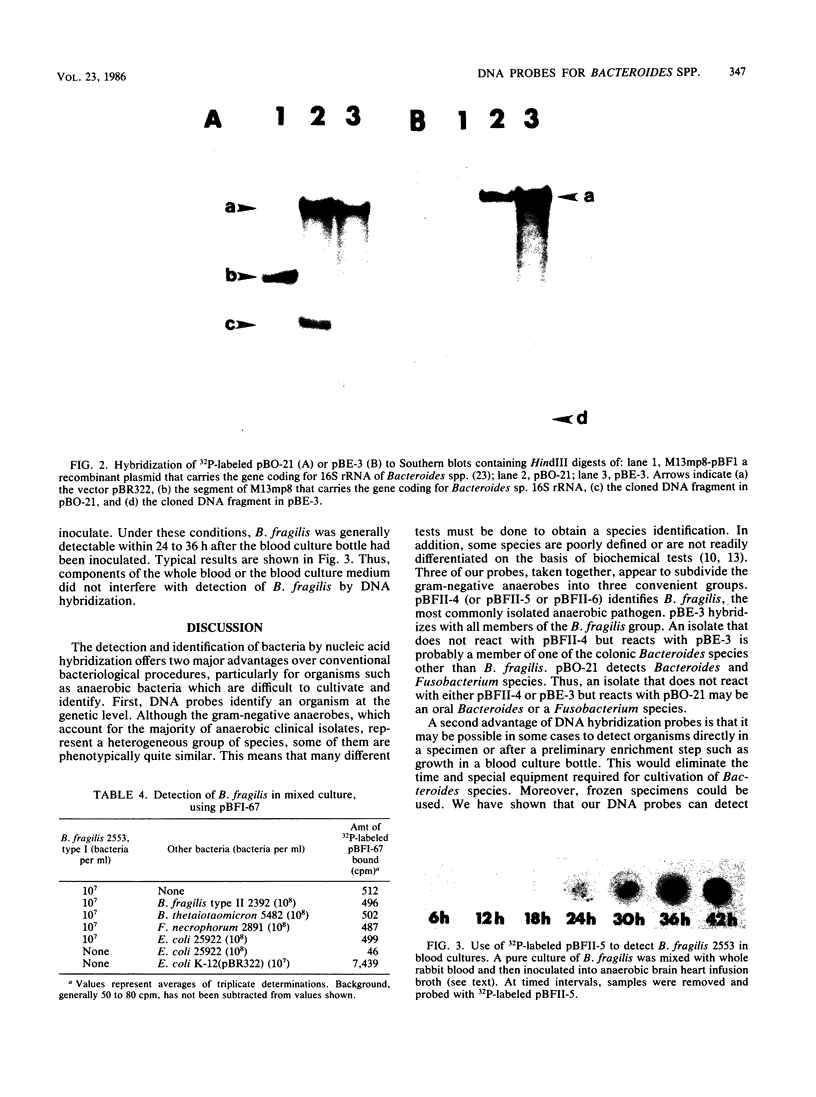
Conventional procedures for identifying gram-negative anaerobes such as Bacteroides spp. are cumbersome and time-consuming. A simpler approach would be to use DNA probes to identify these organisms. Since many different species of gram-negative anaerobes are isolated from clinical specimens, the most useful DNA probes would be probes that identify a few clinically significant groups of anaerobes. To obtain such probes, we cloned HindIII-digested chromosomal DNA from various Bacteroides species and then screened these probes, labeled with 32P by nick translation, for hybridization to DNA from various species of Bacteroides, Fusobacterium, and other gram-negative bacteria. We identified three DNA probes (pBFII-4, pBFII-5, and pBFII-6) that hybridized specifically to DNA from Bacteroides fragilis, the species that accounts for about half of all isolates from clinical specimens containing anaerobes. We identified one DNA probe (pBE-3) that hybridized to all members of the B. fragilis group, a group of species that resemble B. fragilis with respect to fermentation pattern and antibiotic resistances. We also identified one probe (pBO-21) that hybridized to DNA from all Bacteroides sp. strains as well as to DNA from strains of Fusobacterium necrophorum and Fusobacterium nucleatum. pBO-21, but not pBE-3, cross-hybridized with a cloned Bacteroides sp. 16S rRNA gene. The limit of detection for these probes was 10(6) bacteria. The probes could detect B. fragilis in blood culture medium and in mixed cultures with other gram-negative bacteria. Attempts to use biotin-labeled DNA probes instead of 32P-labeled probes were not successful because the Bacteroides sp. extracts contained material that bound the streptoavidin-peroxidase detection reagent.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Appelbaum P. C., Kaufmann C. S., Keifer J. C., Venbrux H. J. Comparison of three methods for anaerobe identification. J Clin Microbiol. 1983 Sep;18(3):614–621. doi: 10.1128/jcm.18.3.614-621.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buesching W. J., Svirbely J. R., Ayers L. W. Evaluation of the Anaerobe-Tek system for identification of anaerobic bacteria. J Clin Microbiol. 1983 May;17(5):824–829. doi: 10.1128/jcm.17.5.824-829.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fitts R., Diamond M., Hamilton C., Neri M. DNA-DNA hybridization assay for detection of Salmonella spp. in foods. Appl Environ Microbiol. 1983 Nov;46(5):1146–1151. doi: 10.1128/aem.46.5.1146-1151.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grimont P. A., Grimont F., Desplaces N., Tchen P. DNA probe specific for Legionella pneumophila. J Clin Microbiol. 1985 Mar;21(3):431–437. doi: 10.1128/jcm.21.3.431-437.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill W. E., Payne W. L., Aulisio C. C. Detection and enumeration of virulent Yersinia enterocolitica in food by DNA colony hybridization. Appl Environ Microbiol. 1983 Sep;46(3):636–641. doi: 10.1128/aem.46.3.636-641.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyypiä T., Stålhandske P., Vainionpä R., Pettersson U. Detection of enteroviruses by spot hybridization. J Clin Microbiol. 1984 Mar;19(3):436–438. doi: 10.1128/jcm.19.3.436-438.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kotarski S. F., Salyers A. A. Isolation and characterization of outer membranes of Bacteroides thetaiotaomicron grown on different carbohydrates. J Bacteriol. 1984 Apr;158(1):102–109. doi: 10.1128/jb.158.1.102-109.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuritza A. P., Salyers A. A. Use of a species-specific DNA hybridization probe for enumerating Bacteroides vulgatus in human feces. Appl Environ Microbiol. 1985 Oct;50(4):958–964. doi: 10.1128/aem.50.4.958-964.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseley S. L., Huq I., Alim A. R., So M., Samadpour-Motalebi M., Falkow S. Detection of enterotoxigenic Escherichia coli by DNA colony hybridization. J Infect Dis. 1980 Dec;142(6):892–898. doi: 10.1093/infdis/142.6.892. [DOI] [PubMed] [Google Scholar]
- Palva A. M. ompA gene in the detection of Escherichia coli and other Enterobacteriaceae by nucleic acid sandwich hybridization. J Clin Microbiol. 1983 Jul;18(1):92–100. doi: 10.1128/jcm.18.1.92-100.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patamaroj U., Seriwatana J., Echeverria P. Identification of enterotoxigenic Escherichia coli isolated from swine with diarrhea in Thailand by colony hybridization, using three enterotoxin gene probes. J Clin Microbiol. 1983 Dec;18(6):1429–1431. doi: 10.1128/jcm.18.6.1429-1431.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salyers A. A., Lynn S. P., Gardner J. F. Use of randomly cloned DNA fragments for identification of Bacteroides thetaiotaomicron. J Bacteriol. 1983 Apr;154(1):287–293. doi: 10.1128/jb.154.1.287-293.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Totten P. A., Holmes K. K., Handsfield H. H., Knapp J. S., Perine P. L., Falkow S. DNA hybridization technique for the detection of Neisseria gonorrhoeae in men with urethritis. J Infect Dis. 1983 Sep;148(3):462–471. doi: 10.1093/infdis/148.3.462. [DOI] [PubMed] [Google Scholar]
- Weisburg W. G., Oyaizu Y., Oyaizu H., Woese C. R. Natural relationship between bacteroides and flavobacteria. J Bacteriol. 1985 Oct;164(1):230–236. doi: 10.1128/jb.164.1.230-236.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wirth D. F., Pratt D. M. Rapid identification of Leishmania species by specific hybridization of kinetoplast DNA in cutaneous lesions. Proc Natl Acad Sci U S A. 1982 Nov;79(22):6999–7003. doi: 10.1073/pnas.79.22.6999. [DOI] [PMC free article] [PubMed] [Google Scholar]





