Abstract
The genomic associations of the archaeal ribosomal proteins, (r-proteins), were examined in detail. The archaeal versions of the universal r-protein genes are typically in clusters similar or identical and to those found in bacteria. Of the 35 nonuniversal archaeal r-protein genes examined, the gene encoding L18e was found to be associated with the conserved L13 cluster, whereas the genes for S4e, L32e and L19e were found in the archaeal version of the spc operon. Eleven nonuniversal protein genes were not associated with any common genomic context. Of the remaining 19 protein genes, 17 were convincingly assigned to one of 10 previously unrecognized gene clusters. Examination of the gene content of these clusters revealed multiple associations with genes involved in the initiation of protein synthesis, transcription or other cellular processes. The lack of such associations in the universal clusters suggests that initially the ribosome evolved largely independently of other processes. More recently it likely has evolved in concert with other cellular systems. It was also verified that a second copy of the gene encoding L7ae found in some bacteria is actually a homolog of the gene encoding L30e and should be annotated as such.
Keywords: operons, ribosome evolution, transcription
Introduction
The archaeal translation machinery has a long evolutionary history and it is of interest to know how it have evolved and how its interactions with other cellular processes have changed over time (Fox and Naik 2004). In general, genes that are translated together share similar origins, physical interactions, functions or regulatory mechanism (Dandekar et al. 1998). In the work reported here, we identified previously unrecognized genomic associations of ribosomal proteins (r-proteins) in archaea and discuss the implications of these associations for the early history of the translation machinery.
Among the three domains of life, there are approximately 102 recognized r-protein families. Of these, 17 large subunit and 19 small subunit r-proteins are universal These 36 r-proteins likely appeared before the last common ancestor, (LUCA), and thus are considered to be ancient in origin (Kyrpides et al. 1999, Lecompte et al. 2002). The Archaea and the Eucaryota share 11 large subunit (LSU) r-proteins and 20 small subunit (SSU) r-proteins but share no r-protein with the Bacteria other than the universal proteins (Lecompte et al. 2002). The large numbers of r-protein families shared by the Archaea and the Eucaryota suggest that the eukaryotic translation system originated from an archaeal version (Hartman et al. 2006).
In early studies on the regulation of the synthesis of ribosomal components in E. coli, it was found that many r-proteins are encoded together; there being 32 r-proteins from both subunits and two translation-related proteins grouped into seven well-studied clusters. These are the alpha, L10 (rif), L11, S10, S20, str and spc operons (Nomura et al. 1984). In each case, gene expression was found to be regulated by one of the r-protein components within the cluster, although the detailed mechanisms differ (Nomura et al. 1984, Zengel and Lindahl 1994) between clusters and even between species in the same cluster (Allen et al. 1999). As more complete genomes of other bacterial species became available, it was found that these seven r-protein clusters were widely conserved among bacterial species (Mushegian and Koonin 1996, Siefert et al. 1997). Among these, the S10 and the spc operon are the largest, each encoding about twelve r-proteins.
Multiple complete genomes from both archaeal and bacterial species are now available (http://www.ncbi.nlm.nih.gov/genomes/lproks.cgi). It has been confirmed that the S10, str, spc, and S13 clusters are also found in the Archaea (Tatusov et al. 2001). These clusters have been characterized experimentally in several archaeal species including Haloarcula marismortui, Sulfolobus acidocaldarius, and Desulfurococcus mobilis and their analogy to the bacterial operons confirmed, (Scholzen and Arndt 1992, Ramirez et al. 1994, Ceccarelli et al. 1995, Yang et al. 1999). The universality of these gene clusters suggests that the proteins for which they code not only have a long history of association, but may represent extremely early regulatory relationships (Kyrpides et al. 1999, Lecompte et al. 2002, Mushegian 2005). In particular, it has been suggested that the progenotic entities of this era used groups of gene clusters (i.e., mini-chromosomes) to carry genetic information instead of what would be considered a complete genome (Olsen and Woese 1997, Siefert et al. 1997). If so, these extant extremely conserved r-protein gene clusters with their RNA level regulation may be a remnant of this earlier time.
Although these clusters of universal r-proteins are of great interest, there are other less conserved r-proteins that may also be informative. In particular, the Archaea and Eucaryota share 11 large subunit r-proteins and 20 small subunit r-proteins which are not found in bacteria (Lecompte et al. 2002). In the work presented here, we investigated the genomic associations of each of these proteins and identified 10 previously unrecognized, but significantly conserved gene clusters.
Materials and methods
Genomes included in the study
Representative archaeal species whose complete genomes were available from the National Center for Biotechnology Information (NCBI) GenBank database (http://www.ncbi.nlm. nih.gov/genomes/lproks.cgi) were considered for analysis. When multiple species of a single genus had been sequenced, only one was selected for inclusion here. The 27 genomes chosen included seven from the Crenarchaeota (Aeropyrum pernix, Hyperthermus butylicus, Pyrobaculum aerophilum, Pyrobacterium islandicum, Staphylothermus marinus, Sulfolobus solfataricus, and Thermofilum pendens) and 20 from the Euryarchaeota. The latter were Archaeoglobus fulgidus, Haloarcula marismortui, Halobacterium sp. NRC-1, Haloquadratum walsbyi, Methanocaldococcus jannaschii, Methanococcoides burtonii, Methanococcus maripaludis, Methanoculleus marisnigri, Methanopyrus kandleri, Methanosaeta thermophila, Methanosarcina acetivorans, Methanospirillum hungatei, Methanosphaera stadtmanae, Methanothermobacter thermautotrophicus, Natronomonas pharaonis, Picrophilus torridus, Pyrococcus abyssi, Thermococcus kodakarensis and Thermoplasma volcanium.
The small genome of the parasitic Nanoarchaeum equitans, which is known to have been subject to extreme rearrangement (Waters et al. 2003, Makarova and Koonin 2005) likely relating to its life style, was excluded from the analysis. Many of the r-proteins are absent, and even highly conserved gene clusters, such as the S10 and spc operons, are disrupted such that their components are dispersed throughout the genome.
Determination of gene neighborhood for the ribosomal proteins
All 27 genomes were examined to identify the genomic neighborhood of each r-protein gene. In addition, sequence alignments were developed for each r-protein using the ClustalW algorithm (Thompson et al. 1994) as implemented in the multiple sequence alignment editor BioEdit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Proteins were initially assigned according to their Cluster of Orthologous Groups (COG) database number (Tatusov et al. 2003). This facilitated proper annotation of the proteins.
Identification of conserved r-protein gene clusters
Extended lists of genes upstream and downstream of each r-protein gene in each genome under consideration were prepared. Both decoding direction and spacer sequences were used to establish likely start and end positions of possible gene clusters. The positions of each r-protein gene in the 27 genomes were compared and clusters with similar gene organization that occur in multiple organisms were identified. In this manner, complete genetic maps of both r-protein genes and their downstream and upstream neighbors were generated. A cluster was considered to be significant if it occurred in at least half of the species considered including at least three representatives of both the Crenarchaeota and Euryarchaeota.
Identification of possible lateral transfer events
The intention here was to examine the extent of similarity in gene order in distantly related taxa. However, similar patterns of gene organization may be found in distantly related organisms as a result of horizontal gene transfer. To ascertain the extent to which lateral transfer events involving the r-proteins have occurred, the phylogenetic tree of each protein was compared with a standard tree (Daubin et al. 2003). If an organism was clearly misplaced in the protein tree, for example, a halophile among the Crenarchaeota, this was considered to be suggestive of a lateral transfer event.
The r-proteins are typically small and as a result individual protein trees differ considerably from one another in many details, but typically they are consistent in major long-range groupings, e.g., crenarchaeota and euryarchaeota are separated, whereas there are strong local relationships, e.g., all halophiles will be clustered. To obtain a tree of relationships, 16S rRNA sequences of the 27 archaeal species were retrieved from GenBank and aligned manually in accordance with known secondary structural features using the BioEdit Sequence Alignment Editor (http://www.mbio.ncsu.edu/BioEdit/bioedit.html). Phylogenetic and molecular evolutionary analyses were conducted using the Neighbor Joining algorithm in MEGA version 3.1 (Kumar et al. 2004). Individual trees were generated from each protein alignment in a similar manner and visually compared with the 16S rRNA tree to identify possible lateral transfer events involving the r-proteins.
Results
Nomenclatural clarifications
In assembling the data sets used here, the earlier comparative studies (Lecompte et al. 2002) were, in effect, repeated. Although the majority of the earlier conclusions were confirmed, two important discrepancies were encountered. The more straightforward case involved bacterial L16 and archaeal L10e which were originally regarded as distinct (Lecompte et al. 2002). In the COG database (Tatusov et al. 2001), L10e is placed in the same family as bacterial L16. With crystal structures available from both archaeal and bacterial ribosomes, it is apparent that these proteins share almost identical tertiary structures (Klein et al. 2004, Schuwirth et al. 2005). Our alignment (not shown) of sequences of L10e from 17 archaeal species and L16 from various bacterial species confirmed that L10e is orthologous with bacterial L16. Thus, the COG database placement of L10e in the same family as bacterial protein L16 is correct. In bacteria, L16 is usually encoded within the S10 operon, which typically includes 11 other universal r-protein genes. In most archaeal genomes, however, although the S10 operon is largely intact, the gene encoding L10e/L16 is typically found elsewhere.
The second discrepancy involved L7ae which occurs in all archaeal species and some bacteria (Lecompte et al. 2002). The gene encoding L7ae is found in most of the Firmicutes, as well as some species that belong to the Actinobacteria and Thermotogae families. Moreover, in complete genomes of some bacteria, two genes that share sequence similarity are both annotated as rpl7ae. When this occurs, the second copy is found upstream of the genes for S12 and S7 which mark the beginning of the str operon. As we will show, this second copy of rpl7ae has more similarity with rpl30e and should be annotated as such. Thus, rpl30e although not universal, also occurs in all three domains of life.
Gene clusters
The extent to which each archaeal r-protein gene was found in a characteristic gene cluster is summarized in Table 1. Twelve proteins, L10e (=L1), L13e (COG4352), L35ae (COG2451), L38e, L40e (COG1552), L41e, S3ae (COG1890), S8e (COG2007), S17e (COG1383), S25e (COG4901), S26e (COG4830) and S30e (COG4919), were not routinely associated with the same genes and thus were not assigned to a cluster. For the clusters that were observed, Table 1 indicates how many members of the Crenarchaeota<> (Max = 7) and the Euryarchaeota (Max = 19) contain the indicated cluster. As observed previously (Nomura et al. 1984), genes for four of the non-universal archaeal r-proteins were located in one of the classic r-protein clusters that encode universal r-proteins in bacteria. In particular, rpl18e was found to be associated with the conserved L13 cluster and rps4e, rpl32e and rpl19e were found in the archaeal version of the spc operon. Of the genes encoding the remaining 19 archaeal-specific proteins, 17 were convincingly assigned to one of 10 previously unrecognized clusters that are conserved over varying phylogenetic distances. The remaining two genes, rps6e and rpl15e, showed some conservation in genomic neighborhood, but it was uncertain if they should be regarded as conserved clusters.
Table 1.
List of common clusters containing genes for archaea-unique ribosomal proteins. The nonuniversal archaeal ribosome proteins (in bold) are shown in their most common genomic contexts. The numbers of crenarchaeota (Max = 7) and euryarchaeota (19) containing the particular arrangement are shown along with the total % of organisms containing that version. If the cluster did not completely meet our criteria (L14e, L7ae, and S6e), it is listed as uncertain. In two cases (L14e and L34e; S24e and S27ae), alternative but similar arrangements are shown. Gene/Protein names inside parentheses are not always present in the cluster and hence do not form the core of the cluster.
| Gene cluster | Number of | Number of | % that | Typical cluster arrangement |
| crenarchaeota | euryarchaeota | agree | ||
| L14e, L34e | 3 | 6 | 34.6 | Uncertain: L34e-cytidylate kinase-L14e |
| L14e | 4 | 0 | 15.3 | Alternative: L14e-truB in some Crenarchaeota |
| L15e | 5 | 10 | 57.6 | L15e-COG1325-COG1603 |
| L18e | 7 | 19 | 100 | L18e-L13-S9-rpoN |
| L19e, L32e, S4e | 6 | 18 | 92.3 | L14-L24-S4e-L5-S14-S8-L6-L32e-L19e-L18-S5-L30-L15-secY |
| L21e | 6 | 17 | 88.4 | (COG1258)-L21e-rpoF-COG1491 |
| L24, L7ae, S28e | 3 | 18 | 80.7 | ( L7ae)-S28e-L24e-(ndK)-(eIF2) |
| L30e | 7 | 13 | 76.9 | rpoH-rpoB2-rpoB1-rpoA1-rpoA2-L30e-NusA |
| L31e, L39, LXae | 4 | 11 | 57.6 | L39e-L31e-LXa-eIF6 |
| L37ae | 5 | 8 | 50 | L37ae-(rpoP)-COG2136 |
| L37e | 4 | 15 | 73 | snRNP-L37e |
| L44e, S27e | 6 | 15 | 80.7 | L44e-S27e-eIF2_α |
| S6e | 4 | 8 | 46.1 | Uncertain: S6e-eIF2_γ |
| S19e | 7 | 16 | 88.4 | S19e-COG2118, usually before L39e-L31e-LXa-eIF6 |
| S24e, S27ae | 2 | 18 | 76.9 | rpoE1-rpoE2-HP-S24e-S27ae-(COG5330) |
| S24e, S27ae | 3 | 1 | 15.3 | Alternative: S24e-S27ae-COG5330 |
For each of these 12 clusters (10 plus 2), the distribution of each variant was mapped onto a representative phylogenetic tree derived from 16S rRNA. Because of the small size of many of the r-proteins, considerable variation in the trees obtained from individual proteins was expected. Nevertheless, the major groupings typically remain together thereby allowing the identification of likely horizontal gene transfer events between organisms in different major clusters. Overall, the number of such likely horizontal gene transfer events seen was modest and did not significantly affect the results. The most obvious examples of likely horizontal gene transfer involved rpl18e in Methanobacter thermoautotrophicum, rpl31e in Archaeoglobus fulgidus and rpl37e in Methanosphaera stadtmanae. Finally, questions about the nomenclature of rpl7ae and rpl30e were clarified.
Archaeal r-protein genes associated with universal r-protein clusters
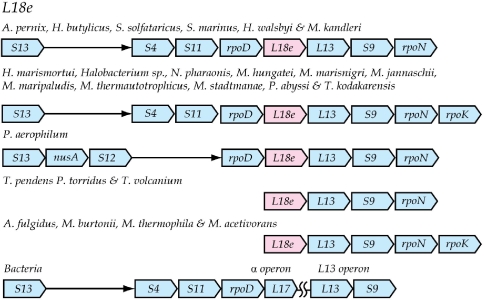
L18e-L13-S9-rpoN
The archaeal r-protein L18e (COG1727) gene is located immediately upstream of the previously known bacterial L13 operon, which consists of universal proteins L13 and S9 as a single transcription unit (Isono et al. 1985). In most archaeal species, this cluster is followed by rpoN, which codes for the omega subunit of DNA-directed RNA polymerase. In some species, the universal alpha operon is upstream of the L18e cluster (Figure 1). The alpha operon itself is structured as in bacterial genomes except for the absence of an ortholog for r-protein L17. Thus, in archaea, the core gene cluster for the alpha operon is rps13-rps4-rps11-rpoD, and is frequently followed by the grouping rpl18e-rpl13-rps9-rpoN. This huge cluster has been shown to form a single transcription unit in H. marismortui and includes the gene for phosphopyruvate hydratase (eno), thus revealing coupled transcription of a glycolytic enzyme and an r-protein (Kromer and Arndt 1991). The position of L18e in this extended cluster suggests this protein may be a homolog of bacterial r-protein L17 which is missing in the Archaea. However, sequence comparisons did not support this hypothesis. Instead, L18e is homologous to universal r-protein L15 (Sano et al. 1999) and is a partial duplication of it. From a structural perspective it is L31e, not L18e, that incorporates into the ribosome in the same vicinity as L17. Thus, the gene for L18e is likely a late addition to the L13 operon.
Figure 1.
Aligned versions of the L18e gene cluster found in various archaeal genomes. Organisms containing a particular arrangement are indicated. In this figure and subsequent figures, genes are shown as boxes with a pointed end that indicates relative transcription direction but not gene length. Each box is labeled with its gene name except the r-protein genes, which are labeled to indicate their gene product. The non-universal archaeal r-protein genes are colored in pink and all other genes are shown in cyan. The thin arrows connect immediately adjoining genes and serve as alignment gaps. Blank spaces indicate the absence of a gene, e.g., rpoK in this figure, from the cluster but not necessarily the genome. These conventions are followed in all the subsequent figures.
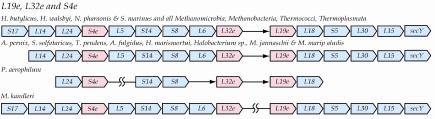
L19e, L32e and S4e
This group of proteins is highly conserved among the Archaea and their genes are inserted into the spc operon, which otherwise maintains its bacterial gene order (Coenye and Vandamme 2005). The gene for S4e (COG1471) is usually located between the genes for L24 and L5, whereas the genes for L32e and L19e are usually located consecutively between the genes for L6 and L18 (Figure 2). Methanopyrus kandleri is an exception in that the cluster is broken after the gene encoding L32e. The second portion of the cluster beginning with the gene for L19e is found on the opposite strand approximately 1.2 MB upstream. This appears to be a special case of a recently disrupted spc operon. Pyrobaculum aerophilum is also an exception in that the genes for L24 and S4e remain together, but are separated from the rest of the operon components. In bacteria, the r-proteins transcribed from the spc operon are regulated by S8 (Zengel and Lindahl 1994). S8 typically binds to the rpl5 gene and represses translation of L5 and the other ribosomal proteins either by retroregulation or translational coupling (Cerretti et al. 1983, Mattheakis and Nomura 1988, Mattheakis et al. 1989). Given the conservation of the position of these proteins in the archaeal version of the cluster, it would not be surprising if a similar regulatory pathway were in use despite the addition of the genes for L19e, L32e and S4e, but this remains to be investigated.
Figure 2.
Alternative versions of the spc gene cluster in the Archaea. Labeling conventions are as in Figure 1. In addition, a double broken line indicates a major gap such that the two groups of genes separated by the gap are not in the same vicinity of the genome. It should be noted that the relative transcription directions indicated pertain only to members of each group not to the relative orientations between the groups. Thus, the two large subclusters indicated in M. kanderleri are in fact separated in the genome where they occur on opposite strands with transcription in opposite directions. This cluster always contains the archaeal-specific r-protein genes for L19e, L32e and S4e.
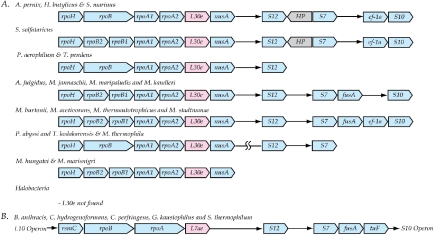
L30e and the str operon
Although the gene encoding L30e is found only in some archaeal species (Figure 3), it is associated with a set of upstream genes encoding DNA-directed RNA polymerase components including rpoH, rpoB and rpoA. The latter two are split into two genes in many species. rpl30e is followed by transcription elongation factor gene nusA and the genes for S12 and S7. In bacteria, rpoB and rpoA are at the downstream end of the L10 operon (Klenk et al. 1999). rps12, rps7, fusA (translation elongation factor G), and tufB (translation elongation factor Tu) then form the conserved str operon whose expression is regulated by S7. Previous comparisons of the RNA polymerase subunits among archaeal showed that the rpoB and rpoA are sometimes split genes that together encode the DNA-directed RNA polymerase beta (COG0085K) and beta prime subunits (COG0086K). Together these genes form the core of the multisubunit DNA-dependent RNA polymerase (Archambault and Friesen 1993, Cramer et al. 2001, Schuwirth et al. 2005). In bacteria, the split rpoB genes and rpoA genes are replaced by the single genes, rpoB and rpoC, (equivalent of archaeal rpoA), thereby maintaining the equivalent gene order upstream of rps12 and rps7. The archaeal version of the cluster frequently includes translational elongation factor 2 and the alpha subunit of translational elongation factor 1 after the rps7 gene. These are functional equivalents of the bacterial genes. In some species (P. aerophilum, T. pendens), these three genes are scattered elsewhere in the genome. Methanospirillum hungatei and M. marisnigri have a distinct cluster that includes rps12-rps7-EF2. This might be a case of late separation of the str operon from the older and larger archaeal cluster. The major distinction of the two systems is the presence in archaea of the gene for L30e and the transcription elongation factor nusA gene upstream of the r-protein S12 gene. Based on this comparison, it appears that a gene cluster analogous to the str operon is found in the Archaea too.
Figure 3.
Alternative versions of the gene cluster containing the gene encoding L30e in the Archaea. Labels are as in Figures 1 and 2. Group A shows the archaeal arrangements and Group B is the arrangement typically found in the Firmicutes. The gene which is homologous to the gene for L30e in the Firmicutes is typically incorrectly annotated as a second copy of the gene encoding L7ae as discussed in the text.
In most bacteria, there is no gene between rpoA and nusA. However, an examination of bacterial genomes revealed that in a few cases (Firmicutes, actinobacterial and thermotogal genomes) there is a gene, typically annotated as a second copy of rpl7ae, in the position occupied by the gene coding for L30e in the Archaea. To explore this further, multiple sequence alignments were constructed (not shown). The second copy of rpl7ae in various bacteria was aligned with archaeal versions of rpl30e and rpl7ae. In addition the archaeal rpl7ae and rpl30e sequences were aligned. Clear evidence of similarity was found in all three cases suggesting that rpl7ae and rpl30e are related. However, the version of the protein found in the bacterial str operon is far more similar to archaeal L30e than to archaeal L7ae and should therefore be annotated as L30e.
Archaea-unique clusters
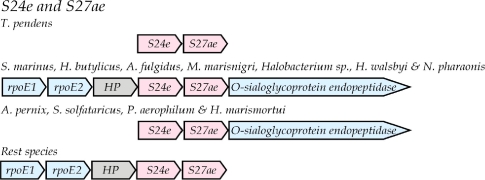
S24e-S27ae
S24e (COG2004) and S27ae (COG1998) are encoded consecutively in all 27 archaeal species (Figure 4). The surrounding genes are such that three alternative arrangements are seen. In the Euryarchaeota, two DNA-directed RNA polymerase subunits E1 and E2 and an archaeal-conserved hypothetical protein of unknown function are always located upstream. The unidentified protein is usually 150 to 195 amino acids long. In the case of Halobacterium sp. NRC-1, rpoE1 and rps24e are separated by 594 bases which is large enough to encode a protein of 198 amino acids. However, the largest open reading frame would only encode a 32-residue hypothetical protein.
Figure 4.
Alternative versions of the S24e-S27ae gene cluster in Archaea. Notation is as in Figure 1. The hypothetical protein is indicated as HP.
A multiple sequence alignment of thirteen examples of this hypothetical protein’s sequence revealed several conserved segments as well as highly variable region. The three crenarchaeal species and H. marismortui lack this upstream group of proteins but instead have a shared gene encoding O-sialoglycoprotein endopeptidase immediately downstream. Finally, several archaeal species have both the upstream and downstream neighbors discussed above.
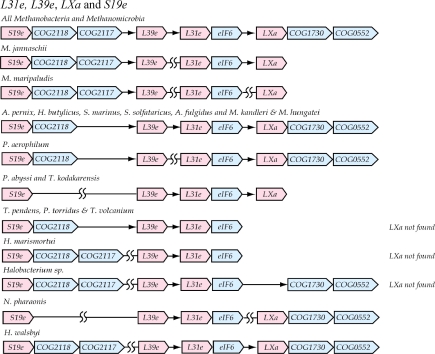
L39e, L31e, S19e and LXa
The genes for these four unique archaeal proteins (Figure 5) are always found in close proximity in a conserved cluster in conjunction with genes encoding several other proteins. There are, however, disruptions in several cases. The core of the cluster is rps19e-COG2118-COG2117-rpl39e. The COG2118 gene encodes a hypothetical DNA-binding protein and the COG2117 gene encodes a subunit of a tRNA methyltransferase. rpl31e (COG2097) typically follows rpl39e (COG2167). However, in P. aerophilum, M. jannaschii and M. maripaludis, these genes are separated from each other with the cluster in effect being broken into two pieces. In several other cases, the cluster is again broken into two fragments with the rpl39e-rpl31e proximity kept intact. In all species, the gene for translation initiation factor 6 (eIF6) is immediately downstream of the gene for L31eand is frequently followed by rpl20A, which encodes ther-protein LXa (COG2157). Translation initiation factor 6 occurs in both the Archaea and Eucaryota and prevents premature association between the 60S and 40S ribosomal subunits. The gene for LXa is missing in the genomic sequences of some species belonging to the Halobacteria or Thermoplasmata families (Lecompte et al. 2002). The genes for COG1730 and COG0552 are frequently associated with the cluster too. They encode a molecular chaperone and a signal recognition protein, respectively.
Figure 5.
Structure of the L31e-L39e-LXa-S19e gene cluster in the Archaea. Notation is as in Figure 1 and 2.
L7ae-S28e-L24e-ndK-infB
The genes for S28e (COG2053) and L24e appear next to each other in all the archaeal genomes with the exception of H. walsbyi, where the gene for S28e was not found, and T. pendens, where the two genes are separated. Additionally, in three crenarchaeal species, A. pernix, S. solfataricus, and S. marinus, the genes for S28e and L24e, although adjacent, are coded by different strands. In twenty of the twenty-six archaeal genomes, the gene for r-protein L7ae, (COG1358), is found immediately upstream of the gene for S28e as shown in Figure 6. The two crenarchaeal species A. pernix and S. solfataricus, along with a euryarchaeon, M. kandleri, deviate from this pattern with the rpl7ae gene found elsewhere in these genomes. Two more genes, ndk (nucleoside-diphosphate kinase) and infB (translation initiation factor 2) are frequently found immediately downstream of rps28e and rpl24e. Thus, a clear picture emerges of a core conserved cluster involving the genes rpl7ae, rps28e, rpl24e, ndk and infB.
Figure 6.
Alternative arrangement of ribosomal protein genes in the S28e-L24e gene cluster. Notation is as in Figure 1.
Characterization of other smaller archeal r-protein gene clusters
The genes encoding the remaining nonuniversal proteins found in the Archaea are in small clusters containing only two or three genes. These clusters are summarized in Figure 7 and are briefly discussed below.
Figure 7.
Additional clusters of archaeal unique r-protein genes. Notation is as in Figures 1 and 2.
L21e (COG2139) is almost always encoded between a gene for a pseudouridylate-synthase-like protein (COG1258J) and rpoF, a DNA-directed RNA polymerase component. Typically a gene for an RNA-binding protein, COG1491J, is immediately downstream of rpoF. However, in M. stadtmanae and P. abyssi, genes encoding different proteins are found.
L34e and L14e are the gene products of an uncertain cluster. It is only found in some archaeal species, but when both genes are present they are always in proximity to one another. When present, the gene for L34e(COG2174) is always upstream of cmk, which encodes a cytidylate kinase. The gene for L14e, (COG2163), either immediately follows cmk or is downstream from it. Neither protein was found in species from the families Archaeoglobi, Halobacteria, Methanomicrobia or Thermoplasmata.
L44e and S27e are always encoded together. rpsUI2, which codes for the alpha subunit of initiation factor 2 (COG1093J) is typically found downstream of the genes for L44e(COG1631) and S27e(COG1998). S27e has a novel C4 zinc finger motif (Herve du Penhoat et al. 2004). L44e has a zinc finger motif with a similar structure, except that the first C2 motif is replaced by a CH motif with several amino acids in between (Ban et al. 2000, Laity et al. 2001). Although they lack obvious primary sequence similarity, the structural similarity between these proteins and their co-occurrence in the genome suggests they may have arisen in the same time frame of the archaeal lineage.
L37e (COG2126) is found in many archaea, but it is absent in P. aerophilum, P. torridus and H. marismortui. Since several closely related species have L37e, these absences may reflect poor annotation or sequencing problems. Excluding these species, the gene for L37e is typically located downstream of snRNP, which encodes the protein component of a small nuclear ribonucleoprotein (Collins et al. 2001). The only exception is M. jannaschii.
The gene for L37ae (COG1997) is found in conjunction with either or both of two other genes, one for a DNA-directed RNA polymerase subunit P (COG1996) and the other for RNA processing protein (COG2136). These genes are encoded after rpl37ae, and when both are present, rpoP occurs first.
L15e (COG1632) is found in all archaea and although it meets the cluster criteria, the position of its gene is not strongly conserved. Most frequently the gene is located upstream of genes, encoding an exosome subunit and the protein part of a ribonuclease P subunit.
S6e (COG2125) does not completely meet our cluster criteria and it is uncertain whether it represents a conserved cluster. The coding region of rps6e is frequently located immediately upstream of eif2G, which is the gene for the gamma subunit of the elongation initiation factor 2. However, in the genomes of all members of the families Halobacteria and Methanomicrobia as well as several other archaeal species these genes are separated. Moreover, in species from the families Methanobacteria, Methanomicrobia and Methanopyri, rps6e-eif2G are typically located downstream of the rpl7ae-rps28e-rpl24e-ndK-infB cluster. This latter arrangement is consistent with the observation that L30e, L7ae/S6e and the eukaryotic unique S12e may share a conserved RNA-binding motif (Mushegian and Koonin 1996).
Discussion and conclusions
It must be considered whether the specific clusters reported here represent chance occurrences or transcriptional relationships between the clustered genes. In the absence of experimental data showing that clusters are operons, one must rely largely on statistical arguments. Although the significance of clustering has been considered in the literature (Durand and Sankoff 2003), no one recognized test has been established for clusters of orthologous genes. The main considerations influencing cluster conservation are the extent of proximity and order of the genes, genome size, and evolutionary distance between the genomes being considered. The more rigorous the proximity requirements and the more distantly related the genomes, the less likely that gene order will be conserved by chance. For example, when just five diverse bacterial genomes were compared (Siefert et al. 1997), only 16 universal clusters in total were found, and essentially all of them reflect genes sharing a transcriptional relationship in those organisms (mainly E. coli) where experimental data are available. In our study, the manner in which the clusters were identified required that they have nearly perfect proximity as well as the same gene order within the cluster. The extent to which the clusters are conserved in the two archaeal orders is shown in Table 1. Twelve of the clusters occurred in more than half of the genomes considered with multiple representatives in both the Crenarchaeota and Euryarchaeota. Six of the clusters occurred in more than 80% of the species.
In bacteria and archaea, genes that have the same origins or share similar functions tend to form conserved gene clusters that likely coordinate expression of the gene products. Such conserved gene order may be an indicator of historical association. In the case of the universal r-proteins, these associations have remained largely unchanged since LUCA and almost exclusively involve universal r-proteins. The major exceptions are the core RNA polymerase genes, which are found immediately upstream of the str operon in many or most bacteria.
To explore such associations further, a comparative analysis of the gene context of the known archaeal r-proteins in seventeen complete genomes was conducted. As expected from prior studies, the universal r-protein genes were found in the same major clusters that occur in bacteria (Siefert et al. 1997, Coenye and Vandamme 2005). Of the nonuniversal archaeal r-protein genes, four were found in these universal clusters. L18e was found in the L13 operon, whereas the genes for L19e, L32e and S4e were in the spc operon. In addition to the conserved operons, at least ten previously unrecognized conserved clusters, each containing at least one non-universal archaeal r-protein gene, were identified. These clusters were typically smaller compared with the universal r-protein operons and they frequently included more than one nonuniversal r-protein gene. The remaining 11 nonuniversal archaeal r-proteins were not consistently associated with any particular gene cluster. Two proteins, L7ae and L16e, are universal in the Archaea and also found in a modest number of mostly high G-C content Gram-positive bacteria.
The nonuniversal r-proteins would likely be later additions, if one excludes the possibility of universal loss in the Bacteria. In support of this, the nonuniversal r-proteins are primarily associated with regions of the rRNA thought to be newer (Hury et al. 2006, Bokov and Steinberg 2009). Among the nonuniversal protein genes, those that belong to conserved clusters, are likely older than those not associated with any cluster. In contrast to the universal r-proteins, the clusters that contain nonuniversal r-protein genes frequently contain non-r-protein genes. Since the nonuniversal r-proteins are likely later additions to the ribosomal machinery, we speculate that the proteins that they are associated with are later additions, likely coming into existence in the same time frame. Thus, it may be possible to gain insight in to the relative time of emergence of various cellular processes.
Among the genes associated with the nonuniversal r-proteins are a number coding for proteins involved in the initiation of translation. These are infB, eIF2α, eIF2-GTPase, eIF2γ and eIF6. eIF6 is an anti-association protein that binds to the large subunit and prevents the formation of the functional ribosome complex in both the Archaea and Eucaryota (Ceci et al. 2003). eIF2α and eIF2γ are components of eIF2, a translation initiation factor that is composed of several heterogeneous subunits in the Archaea. It is responsible for initiator tRNA binding and translation start site recognition. The sequence of eIF2γ shows homology to a protein belonging to universal elongation factor family that includes EF-Tu in the Bacteria and eEF1A in the Archaea (Marintchev and Wagner 2004). Because the two archaeal initiation factors can function only as a complex, it is likely that they appeared at approximately the same time during ribosome evolution. It is also possible that the proteins, S6e, S27e and L44e, which belong to the same clusters as the two initiation factors, also appeared in the same time frame. L44e is known to be a protein component of the E site of the eukaryotic and archaeal ribosome (Schroeder et al. 2007) and might be one of the more ancient non-universal r-proteins. S6e has a unique RNA-binding domain that is shared between translation termination suppressors, ribosomal protein S12e and rRNA modifying enzyme rimK in E. coli (Mushegian and Koonin 1996).
In bacteria, the initiation factors IF2 and IF3 play an identical role in protein synthesis as their archaeal counterparts, but they do not share any sequence similarity to the archaeal proteins in question. Initiation of bacterial protein synthesis utilizes a Shine-Dalgarno sequence that is typically found upstream of the start codon, whereas in eukaryotes the Kozak scanning mechanism is typically employed. Thus, it is clear that the mechanisms of translation initiation underwent further development after the Bacteria and Archaea diverged.
As with the translation initiation factors discussed above, several genes associated with transcription, including various DNA-dependent RNA polymerase (RNAP) subunits, are found in archaeal r-protein clusters. The L30e cluster in archaea is in effect a homolog of the Beta cluster and str operon, which are found together in many bacteria. The genes rpoH, rpoA and rpoB are typically upstream of rpl30e which is then followed, in analogy with the str operon by the genes for S12 and S7. This similarity is extended by our observation that what has been reported as a second copy of rpl7ae in some bacteria is not only rpl31e, but is also in the same cluster position as archaeal rpl31e. In the archaea, rpoA and rpoB are split genes encoding the β′ and β subunits of the DNA-dependent RNA polymerase. These core components of the RNA polymerase (RNAP) are conserved in all three domains (Cramer 2002, Cramer et al. 2001, Darst 2001). In addition, in the archaeal cluster, rpoH encodes RNA polymerase subunit H, which is a counterpart of rpb5, a subunit shared by the RNA polymerases I, II and III in the Eucaryota. No corresponding gene was found in bacteria (Cramer 2002).
When the nonuniversal r-protein gene clusters are considered, the linkage between transcription and translation is expanded further. Thus, the L21e and S24e/S27ae clusters also contain components of the archaeal RNAP. The gene rpoF is usually found downstream of the gene for L21e, whereas genes rpoE1 and rpoE2 are usually found upstream of the genes for S24e and S27ae. Their products, the archaeal RNAP E1 and E2 subunits, are homologous to rpb4 and rpb7, subunits of eukaryotic RNA polymerase II. They are reported to form a heterodimer in their own polymerase complex (Todone et al. 2001, Armache et al. 2003, Bushnell and Kornberg 2003).
In addition to the genes involved in transcription and the initiation of translation, the nonuniversal archaeal r-protein gene clusters contain a variety of other genes many of which code for products of uncertain function. Genes coding for proteins of known function include those for cytidylate kinase and nucleoside diphosphate kinase, both of which are involved in nucleotide synthesis. Likewise, truB, snRNP and COG1258 are involved in post-transcriptional RNA maturation.
In summary, unlike those of the universal genes, the nonuniversal r-protein gene clusters contain many non-r-protein genes. If these gene clusters are regulatory groupings, then the expression of the newer r-protein genes is likely to be more strongly coordinated with other cellular processes. In particular, there is a significant increase in the amount of coordination with transcription. In addition, there is likely to be significant coordination with the initiation of protein synthesis, and to a lesser extent RNA processing and nucleotide synthesis. The overall picture suggests an evolutionary history in which the core ribosome and transcription machinery initially arose before LUCA. Subsequently, after the archaeal and bacterial domains separated, the transcription process was refined and new mechanisms for the initiation of protein synthesis introduced in the same time frame. These were likely coordinated with enhancements to the protein synthesis machinery represented by the various nonuniversal r-proteins rather than with the older core machinery.
Acknowledgments
This research was supported in part by grants to GEF from the NASA Exobiology program (NNG05GN75G), the Robert A. Welch Foundation (E-1451), the Texas Advanced Research Program, and the Institute of Space Systems Operations.
References
- R1.Allen T., Shen P., Samsel L., Liu R., Lindahl L., Zengel J.M. Phylogenetic analysis of L4-mediated autogenous control of the S10 ribosomal protein operon. J. Bacteriol. . 1999;181:6124–6132. doi: 10.1128/jb.181.19.6124-6132.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R2.Archambault J., Friesen J.D. Genetics of eukaryotic RNA polymerases I, II, and III. Microbiol. Rev. . 1993;57:703–724. doi: 10.1128/mr.57.3.703-724.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Armache K.J., Kettenberger H., Cramer P. Architecture of initiation-competent 12-subunit RNA polymerase II. Proc. Natl. Acad. Sci. USA . 2003;100:6964–6968. doi: 10.1073/pnas.1030608100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Ban N., Nissen P., Hansen J., Moore P.B., Steitz T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science. 2000;289:905–920. doi: 10.1126/science.289.5481.905. [DOI] [PubMed] [Google Scholar]
- R5.Bokov K., Steinberg S.V. A hierarchical model for evolution of 23S ribosomal RNA. Nature. 2009;457:977–980. doi: 10.1038/nature07749. [DOI] [PubMed] [Google Scholar]
- R6.Bushnell D.A., Kornberg R.D. Complete, 12-subunit RNA polymerase II at 4.1-A resolution: implications for the initiation of transcription. Proc. Natl. Acad. Sci. USA. 2003;100:6969–6973. doi: 10.1073/pnas.1130601100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R7.Ceccarelli E., Bocchetta M., Creti R., Sanangelantoni A.M., Tiboni O., Cammarano P. Chromosomal organization and nucleotide sequence of the genes for elongation factors EF-1 alpha and EF-2 and ribosomal proteins S7 and S10 of the hyperthermophilic archaeum Desulfurococcus mobilis . Mol. Gen. Genet. . 1995;246:687–696. doi: 10.1007/BF00290714. [DOI] [PubMed] [Google Scholar]
- R8.Ceci M., Gaviraghi C., Gorrini C., Sala L.A., Offenhauser N., Marchisio P.C., Biffo S. Release of eIF6 (p27BBP) from the 60S subunit allows 80S ribosome assembly. Nature . 2003;426:579–584. doi: 10.1038/nature02160. [DOI] [PubMed] [Google Scholar]
- R9.Cerretti D.P., Dean D., Davis G.R., Bedwell D.M., Nomura M. The spc ribosomal protein operon of Escherichia coli: sequence and cotranscription of the ribosomal protein genes and a protein export gene. Nucleic Acids Res. 1983;11:2599–2616. doi: 10.1093/nar/11.9.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.Coenye T., Vandamme P. Organisation of the S10, spc and alpha ribosomal protein gene clusters in prokaryotic genomes. FEMS Microbiol. Lett. . 2005;242:117–126. doi: 10.1016/j.femsle.2004.10.050. [DOI] [PubMed] [Google Scholar]
- R11.Collins B.M., Harrop S.J., Kornfeld G.D., Dawes I.W., Curmi P.M., Mabbutt B.C. Crystal structure of a heptameric Sm-like protein complex from archaea: implications for the structure and evolution of snRNPs. J. Mol. Biol. . 2001;309:915–923. doi: 10.1006/jmbi.2001.4693. [DOI] [PubMed] [Google Scholar]
- R12.Cramer P. Multisubunit RNA polymerases. Curr. Opin. Struct. Biol. . 2002;12:89–97. doi: 10.1016/s0959-440x(02)00294-4. [DOI] [PubMed] [Google Scholar]
- R13.Cramer P., Bushnell D.A., Kornberg R.D. Structural basis of transcription: RNA polymerase II at 2.8 angstrom resolution. Science. 2001;292:1863–1876. doi: 10.1126/science.1059493. [DOI] [PubMed] [Google Scholar]
- R14.Dandekar T., Snel B., Huynen M., Bork P. Conservation of gene order: a fingerprint of proteins that physically interact. Trends Biochem. Sci. . 1998;23:324–328. doi: 10.1016/s0968-0004(98)01274-2. [DOI] [PubMed] [Google Scholar]
- R15.Darst S.A. Bacterial RNA polymerase. Curr. Opin. Struct. Biol. . 2001;11:155–162. doi: 10.1016/s0959-440x(00)00185-8. [DOI] [PubMed] [Google Scholar]
- R16.Daubin V., Moran N.A., Ochman H. Phylogenetics and the cohesion of bacterial genomes. Science . 2003;301:829–832. doi: 10.1126/science.1086568. [DOI] [PubMed] [Google Scholar]
- R17.Durand D., Sankoff D. Tests for gene clustering. J. Comput. Biol. . 2003;10:453–482. doi: 10.1089/10665270360688129. [DOI] [PubMed] [Google Scholar]
- R18.Fox G.E., Naik A.K. New York: Kluwer Academic/Plenum; 2004. The evolutionary history of the translation machinery; pp. 92–105. [Google Scholar]
- R19.Hartman H., Favaretto P., Smith T.F. The archaeal origins of the eukaryotic translational system. Archaea . 2006;2:1–9. doi: 10.1155/2006/431618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R20.Herve du Penhoat C., Atreya H.S., Shen Y., Liu G., Acton T.B., Xiao R., Li Z., Murray D., Montelione G.T., Szyperski T. The NMR solution structure of the 30S ribosomal protein S27e encoded in gene RS27_ARCFU of Archaeoglobus fulgidis reveals a novel protein fold. Protein Sci. 2004;13:1407–1416. doi: 10.1110/ps.03589204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R21.Hury J., Nagaswamy U., Larios-Sanz M., Fox G.E. Ribosome origins: the relative age of 23S rRNA domains. Orig. Life Evol. Biosph. . 2006;36:421–429. doi: 10.1007/s11084-006-9011-z. [DOI] [PubMed] [Google Scholar]
- R22.Isono S., Thamm S., Kitakawa M., Isono K. Cloning and nucleotide sequencing of the genes for ribosomal proteins S9 (rpsI) and L13 (rplM) of Escherichia coli . Mol. Gen. Genet. . 1985;198:279–282. doi: 10.1007/BF00383007. [DOI] [PubMed] [Google Scholar]
- R23.Klein D.J., Moore P.B., Steitz T.A. The roles of ribosomal proteins in the structure assembly, and evolution of the large ribosomal subunit. J. Mol. Biol. . 2004;340:141–177. doi: 10.1016/j.jmb.2004.03.076. [DOI] [PubMed] [Google Scholar]
- R24.Klenk H.P., Meier T.D., Durovic P., Schwass V., Lottspeich F., Dennis P.P., Zillig W. RNA polymerase of Aquifex pyrophilus: implications for the evolution of the bacterial rpoBC operon and extremely thermophilic bacteria. J. Mol. Evol. . 1999;48:528–541. doi: 10.1007/pl00006496. [DOI] [PubMed] [Google Scholar]
- R25.Kromer W.J., Arndt E. Halobacterial S9 operon. Three ribosomal protein genes are cotranscribed with genes encoding a tRNA(Leu), the enolase, and a putative membrane protein in the archaebacterium Haloarcula (Halobacterium) marismortui. J. Biol. Chem. 1991;266:24573–24579. [PubMed] [Google Scholar]
- R26.Kumar S., Tamura K., Nei M. MEGA3: Integrated software for molecular evolutionary genetics analysis and sequence alignment. Brief. Bioinform. . 2004;5:150–163. doi: 10.1093/bib/5.2.150. [DOI] [PubMed] [Google Scholar]
- R27.Kyrpides N., Overbeek R., Ouzounis C. Universal protein families and the functional content of the last universal common ancestor. J. Mol. Evol. . 1999;49:413–423. doi: 10.1007/pl00006564. [DOI] [PubMed] [Google Scholar]
- R28.Laity J.H., Lee B.M., Wright P.E. Zinc finger proteins: new insights into structural and functional diversity. Curr. Opin. Struct. Biol. 2001;11:39–46. doi: 10.1016/s0959-440x(00)00167-6. [DOI] [PubMed] [Google Scholar]
- R29.Lecompte O., Ripp R., Thierry J.C., Moras D., Poch O. Comparative analysis of ribosomal proteins in complete genomes: an example of reductive evolution at the domain scale. Nucleic Acids Res. . 2002;30:5382–5390. doi: 10.1093/nar/gkf693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R30.Makarova K.S., Koonin E.V. Evolutionary and functional genomics of the Archaea. Curr. Opin. Microbiol. . 2005;8:586–594. doi: 10.1016/j.mib.2005.08.003. [DOI] [PubMed] [Google Scholar]
- R31.Marintchev A., Wagner G. Translation initiation: structures, mechanisms and evolution. Q. Rev. Biophys. . 2004;37:197–284. doi: 10.1017/S0033583505004026. [DOI] [PubMed] [Google Scholar]
- R32.Mattheakis L.C., Nomura M. Feedback regulation of the spc operon in Escherichia coli: translational coupling and mRNA processing. J. Bacteriol. . 1988;170:4484–4492. doi: 10.1128/jb.170.10.4484-4492.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R33.Mattheakis L., Vu L., Sor F., Nomura M. Retroregulation of the synthesis of ribosomal proteins L14 and L24 by feedback repressor S8 in Escherichia coli . Proc. Natl. Acad. Sci. USA . 1989;86:448–452. doi: 10.1073/pnas.86.2.448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R34.Mushegian A. Protein content of minimal and ancestral ribosome. Rna . 2005;11:1400–1406. doi: 10.1261/rna.2180205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R35.Mushegian A.R., Koonin E.V. A minimal gene set for cellular life derived by comparison of complete bacterial genomes. Proc. Natl. Acad. Sci. USA . 1996;93:10268–10273. doi: 10.1073/pnas.93.19.10268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R36.Nomura M., Gourse R., Baughman G. Regulation of the synthesis of ribosomes and ribosomal components. Annu. Rev. Biochem. . 1984;53:75–117. doi: 10.1146/annurev.bi.53.070184.000451. [DOI] [PubMed] [Google Scholar]
- R37.Olsen G.J., Woese C.R. Archaeal genomics: an overview. Cell . 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- R38.Ramirez C., Shimmin L.C., Leggatt P., Matheson A.T. Structure and transcription of the L11-L1-L10-L12 ribosomal protein gene operon from the extreme thermophilic archaeon Sulfolobus acidocaldarius . J. Mol. Biol. . 1994;244:242–249. doi: 10.1006/jmbi.1994.1723. [DOI] [PubMed] [Google Scholar]
- R39.Sano K., Taguchi A., Furumoto H., Uda T., Itoh T. Cloning, sequencing, and characterization of ribosomal protein and RNA polymerase genes from the region analogous to the alpha-operon of Escherichia coli in halophilic archaea, Halobacterium halobium . Biochem. Biophys. Res. Commun. 1999;264:24–28. doi: 10.1006/bbrc.1999.1480. [DOI] [PubMed] [Google Scholar]
- R40.Scholzen T., Arndt E. The alpha-operon equivalent genome region in the extreme halophilic archaebacterium Haloarcula (Halobacterium) marismortui . J. Biol. Chem. . 1992;267:12123–12130. [PubMed] [Google Scholar]
- R41.Schroeder S.J., Blaha G., Tirado-Rives J., Steitz T.A., Moore P.B. The structures of antibiotics bound to the E site region of the 50 S ribosomal subunit of Haloarcula marismortui: 13-deoxytedanolide and girodazole. J. Mol. Biol. . 2007;367:1471–1479. doi: 10.1016/j.jmb.2007.01.081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R42.Schuwirth B.S., Borovinskaya M.A., Hau C.W., Zhang W., Vila-Sanjurjo A., Holton J.M., Cate J.H. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- R43.Siefert J.L., Martin K.A., Abdi F., Widger W.R., Fox G.E. Conserved gene clusters in bacterial genomes provide further support for the primacy of RNA. J. Mol. Evol. 1997;45:467–472. doi: 10.1007/pl00006251. [DOI] [PubMed] [Google Scholar]
- R44.Tatusov R.L., Natale D.A., Garkavtsev I.V., Tatusova T.A., Shankavaram U.T., Rao B.S., Kiryutin B., Galperin M.Y., Fedorova N.D., Koonin E.V. The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. . 2001;29:22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R45.Tatusov R.L., Fedorova N.D., Jackson J.D., et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R46.Thompson J.D., Higgins D.G., Gibson T.J. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. . 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R47.Todone F., Brick P., Werner F., Weinzierl R.O., Onesti S. Structure of an archaeal homolog of the eukaryotic RNA polymerase II RPB4/RPB7 complex. Mol. Cell. . 2001;8:1137–1143. doi: 10.1016/s1097-2765(01)00379-3. [DOI] [PubMed] [Google Scholar]
- R48.Waters E., Hohn M.J., Ahelet Al I., et al. The genome of Nanoarchaeum equitans: insights into early archaeal evolution and derived parasitism. Proc. Natl. Acad. Sci. USA . 2003;100:12984–12988. doi: 10.1073/pnas.1735403100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R49.Yang D., Kusser I., Kopke A.K., Koop B.F., Matheson A.T. The structure and evolution of the ribosomal proteins encoded in the spc operon of the archaeon (Crenarchaeota) Sulfolobus acidocaldarius . Mol. Phylogenet. Evol. . 1999;12:177–185. doi: 10.1006/mpev.1998.0607. [DOI] [PubMed] [Google Scholar]
- R50.Zengel J.M., Lindahl L. Diverse mechanisms for regulating ribosomal protein synthesis in Escherichia coli . Prog. Nucleic Acid Res. Mol. Biol. . 1994;47:331–370. doi: 10.1016/s0079-6603(08)60256-1. [DOI] [PubMed] [Google Scholar]