Abstract
Recombinant amidase from Sulfolobus solfataricus occurred as a dimer of 110 kDa comprising identical subunits. Only dimers were present at pHs above 7.0, but with decreasing pH, dimers associated into octamers, with complete oligomerization occurring at pH 3.0. Oligomerization showed reversible temperature-dependence, with octamer formation increasing with temperature from 36 °C to between 70 and 80° C. Increasing salt concentrations, favored dissociation of the octamers. Among the three investigated factors affecting the dimer–octamer equilibrium, the most important was pH. Among four mutants obtained by site-specific mutagenesis and selection for pH and temperature sensitivity, the T319I and D487N mutant amidases, like that of the native Sulfolobus solfataricus, responded to changes in pH and temperature with a conformational change affecting the dimer–octamer equilibrium. The Y41C and L34P mutant amidases were unaffected by pH and temperature, remaining always in the dimeric state. The differences among mutants in protein conformation must be related to the position of the introduced mutation. Although the L34P and Y41C mutations are located in the helical region 33–48 (LLKLQLESYERLDSLP), which is close to the amino-terminal segment of the protein, the T319I mutation is located in a strand on the surface of the protein, which is far from, and opposite to, the amino-terminal segment. The D487N mutation is located in the center of the protein, far distant from the 33–48 segment. These observations suggest that the segment of the protein closest to the amino-terminus plays a key role in the association of dimers into octamers.
Keywords: amidase, archaea, hyperthermophile, oligomerization, signature amidase
Introduction
Amidase of the hyperthermophile Sulfolobus solfataricus (SSAM) was obtained as a recombinant protein expressed in E. coli (Scotto d’Abusco et al. 2005). The protein belongs to the class of amidases characterized by the amidase signature sequence GGSS(S/G)GS that is included in a conserved stretch of about 130 amino acids and located in the center of the protein (Mayaux et al. 1991, Chebrou et al. 1996 ). The functions of the amidase signature family enzymes include formation of Gln-tRNAGln through the transamidation of misacylated Glu-tRNAGln by amidolysis of glutamine in many bacteria (Curnow et al. 1997, Schon et al. 1998), formation of indole-3-acetic acid in pathogenic plant bacteria (Gaffney et al. 1990), the metabolic turnover of foreign and endogenous amides in prokaryotes and eukaryotes (Mayaux et al. 1991, Gomi et al. 1991, Gopalakrishna et al. 2004, Cai et al. 2005) and the catabolism of neuromodulatory fatty acid amides in mammals (Koutek et al. 1994, Cravatt et al. 1996, Patricelli and Cravatt 2000, McKinney and Cravatt 2005, Wei et al. 2006).
The SSAM recombinant enzyme is a homodimer of 110 kDa which reversibly associates into octamers in a pH dependent reaction as established by gel permeation chromatography, analytical ultracentrifugation and dynamic light scattering (Scotto d’Abusco et al. 2005). At pH 7.0, all these techniques show the presence of two components in similar amounts, compatible with a dimeric and an octameric species. Low pH induces a reversible association of dimers into octamers. Above pH 8.0 only dimers are present, whereas below pH 3.0 only octamers. Association of dimers to octamers increases with temperature and is inhibited by nonpolar solvents. In contrast to the Sulfolobus solfataricus nativeamidase (WT SSAM), that of the Y41C mutant does not undergo oligomerization. From these results it was inferred that conformational changes in the protein induced by pH expose hydrophobic residues that stabilize the octamer, and that a specific tyrosine residue (Y41) may trigger the oligomerizion process (Scotto d’Abusco et al. 2005).
Here we report a detailed study of the effects of temperature, pH and ionic strength on the association of the WT protein and the amidase proteins of site-specific mutants selected on the basis of pH and temperature sensitivity. We aimed to: (1) confirm the importance of the region containing the Y41 residue as a trigger of oligomerization; (2) determine if other protein regions have a role in oligomerization; and (3) determine whether conformational changes induced by mutation influence the catalytic and physico-chemical properties of the enzyme. Proteins were characterized by analytical ultracentrifugation and according to their thermal stability, thermoactivity and thermal unfolding.
Materials and methods
Chemicals
Diethylphosphoramidate was purchased from Aldrich, phenylphosphorodiamidate from Alfa Aesar (Karlsruhe, Germany); α, β and γ cyclodextrins from Fluka (Switzerland); all other chemicalswere from Sigma-Aldrich (St Louis, MO, USA).
Protein expression and purification
The expression and the purification procedure of the WT protein was as described by Scotto d’Abusco et al. (2005).
Site-directed mutagenesis
Purified mutagenesis primers were designed following the QuickChange™ site-directed mutagenesis kit guidelines (Stratagene, La Jolla, CA). The sequences of the forward primers were (mutagenic positions underlined): T319I, 5′-GATTATGGAATAAAAGTAGAGGATATCTCCATACCTCTTCATAGG-3′; D487N, 5′-GGTCTGATGATAATAGGAAGGCATTTTGAAGAAAACAAGGTTCTAAAATTAGC-3′; L34P, 5′-GAGTTAAAATCCTTTTTGCAATTACCAAAACTGCAGTTGGAATCC-3′; Y41C, 5′-GCAATTACTAAAACTGCAGTTGGAATCCTGTGAGAGGTTAGACTCCTTGCC-3′. Reverse primers were complementary to forward primers. The mutagenesis reaction was done following the QuickChange™ site-directed mutagenesis kit instructions (Stratagene). Expression and purification of the mutants were performed following the same procedure used for the WT protein.
Protein determination
Protein concentration was determined from absorbance at 280 nm based on an extinction coefficient of 54,290 M–1 cm–1, calculated according to Gill and von Hippel (1989).
Gel permeation chromatography
Protein samples were incubated overnight at room temperature in the appropriate buffer at 20 mM and subjected to gel permeation chromatography (GPC) by injection onto a Superdex 200HR 10/30 column (Amersham Biosciences Europe) equilibrated with 20 mM sodium phosphate buffer pH 7.0, containing 0.15 M NaCl. The protein was eluted with the same buffer at 0.4 ml min–1 at room temperature. The absorbance of the eluate was monitored at 226 nm. The column was calibrated with the following standards (Amersham Biosciences Europe): thyroglobulin (669 kDa), ferritin (440 kDa), catalase (232 kDa), aldolase (158 kDa), albumin (67 kDa), ovoalbumin (43 kDa), and ribonuclease (13.7 kDa). The molecular mass of amidase was estimated from elution volume (Ve) based on a linear regression included in the Unicorn 4.10 package for control and data evaluation of the Äkta Chromatograph (Amersham Biosciences).
Analytical ultracentrifugation
Analytical ultracentrifugation (AUC) was conducted at 117,000<> g over the temperature range 4–36 °C in a Beckman Optima XL-I analytical ultracentrifuge, equipped with absorbance optics and an An60-Ti rotor. Protein was applied at a concentration of 0.5 mg ml–1. The buffer was 20 mM sodium phosphate pH 6.0, 7.0 or 8.0. Although equilibrium is attained in 120 min (Scotto d’Abusco et al. 2005), the samples were left in the ultracentrifuge to equilibrate overnight. Data were collected in continuous scan mode every 4 min at 278 nm at 0.005-cm intervals. Data were analyzed with the program Sedfit (Schuck and Rossmanith 2000) provided by Dr. Peter Schuck (NIH, Bethesda, MD).
Thermal stability
Measurements of enzymatic thermal stability were conducted at 70, 80 and 90 °C in Eppendorf tubes (0.2 ml) containing 70 µl of protein solution (0.2 mg ml–1) in 20 mM citrate buffer at pH 5.0 or 20 mM phosphate buffer at pH 8.0. Tubes were sealed and incubated in a Mastercycler personal (Eppendorf, Hamburg, Germany). After incubation, 50 µl aliquots were assayed for standard activity which was followed for at least 10 h after reaching 50% of initial activity.
Effect of sodium dodecyl sulfate
Samples containing 30 µg of WT SSAM were incubated overnight at 20°C in 100 µl of 20 mM sodium phosphate buffer pH 8.0 to induce the dimerization of the protein, as confirmed by GPC on a Superdex 200 column. Samples were also analyzed after heating at 80 °C for 2 h in the presence of 0, 0.0125 or 0.025% sodium dodecyl sulfate (SDS).
Differential scanning calorimetry
Heat capacity versus temperature profiles were obtained with a VP-DSC differential scanning calorimeter (MicroCal. Inc., Northampton, MA) at a heating rate of 60 °C h–1. Protein samples (0.1–0.3 mg ml–1) in the appropriate buffer were degassed at 25 °C before use. The reference cell of the calorimeter was filled with protein-free buffer. Measurement cells were kept under an excess pressure of 0.2 kPa to prevent bubbling during the scan. At the end of the run, solutions were subjected to a second heating cycle under the same conditions to determine the reversibility of the transitions. Thermograms were corrected by subtracting the instrument baseline, obtained with both cells filled with solvent, and normalized for protein concentration. The Tm value, i.e., the temperature at which excess heat capacity reaches a maximum, was determined with ORIGIN software provided by MicroCal, after subtraction of a cubic baseline connecting the pre- and post-transition traces.
Thermoactivity
The temperature dependence of the amidase reaction of the WT protein and its mutants was studied under standard assay conditions over the range 40–100 °C and the results evaluated with an Arrhenius plot.
Interaction with cyclodextrins
The WT protein was treated overnight with 50 mM phosphate buffer pH 8.0 to induce formation of the dimeric species. Cyclodextrins, α, β or γ (Fluka, Buchs SG, Swiss), were dissolved in water and added to the solution in a 1:20 or 1:40 molar excess over protein (on a dimer basis) before inducing oligomerization by addition of 0.5 M citrate buffer pH 5.0. After two hours at 20 °C the mixture was injected onto the polymer-based column TSK gel G3000 PWXL (7.8 × 300 mm, Tosoh Biosep, Kanagawa-Ken, Japan). Elution was carried out at room temperature with 20 mM phosphate buffer pH 7.0, containing 150 mM NaCl at 0.5 ml min–1 and monitored at 226 and 280 nm. Results represent areas beneath the UV absorbance curve.
Spectroscopic analyses
Spectroscopic analyses were performed as reported by Scotto d’Abusco et al. (2005).
Enzyme assay
The standard enzyme assay was performed at 70 °C for 5 min with 7.5 mM benzamide in 50 mM citrate buffer pH 5.0, under which conditions the WT protein and all the mutants amidases have maximal activity, whatever their state of association. At the end of the incubation, the reaction was stopped by adding 200 µl of 20% acetic acid; thereafter 50 µl of this solution was injected onto a Platinum EPS C18 column (53 × 7 mm, Altech, Deerfield, IL) and the amount of benzoic acid produced measured as described by Scotto d’Abusco et al. (2001).
Biochemical characterizations
The kinetic constants with aliphatic and aromatic amides were determined at 70 °C in 50 mM citrate buffer, pH 5.0. The free acid produced was measured by high performance liquid chromatography (HPLC) (Scotto d’Abusco et al. 2001). Kinetic data were calculated by nonlinear regression analysis using ENZFITTER (Leatherbarrows 1987), with at least seven velocity substrate data pairs per determination.
Inhibition studies were performed with the protease inhibitors chimostatine, diisopropylfluorophosphate (DFP) and phenylmethylsulfonyl fluoride (PMSF): 5 or 10 µg of enzyme in 50 mM citrate buffer pH 5.0 was treated for 10 min with 1–100 molar excess of the inhibitor (dissolved in DMSO) before the addition of 7.5 mM benzamide for the standard assay. Diethylphosphoramidate (DEPA) and phenylphosphorodiamidate (PPDA) dissolved in DMSO were used as competitive inhibitors with the benzamide substrate.
Construction of the model
Three signature amidases with different metabolic roles are presently known at atomic resolution: malonamidase E2 (MAE2) from Bradyrhizobium japonicum (Shin et al. 2003), the fatty acid amide hydrolase from Homo sapiens (FAAH) (Bracey et al. 2002), and the peptide amidase (PAM) from Stenotrophomonas maltophila (Labahn et al. 2002). Building the SSAM 3D structure was performed using PAM as a template. This choice was suggested both by a better sequence alignment of the target and template (complete coverage, identity level = 30%) compared with the other two sequences and the similarity in the substrates of SSAM and PAM. The 3D structure was computed after template/target alignment with Modeller v 6.2 (Sali and Blundell 1993). After prediction of the interaction patches with ISPRED (Fariselli et al. 2002), the dimer form was built with the ZDOCK tool (Chen and Weng 2002).
Results
Oligomerization, thermal stability and thermal unfolding
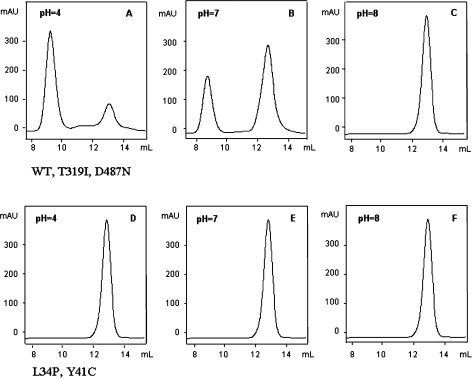
A set of GPC experiments was carried out to ascertain the association–dissociation behavior of the native protein (WT SSAM) and its mutants (T319I, D487N, L34P, Y41C) at 20 °C over the pH range 4.0–8.0. The elution patterns of WT SSAM, T319I and D487N proteins injected onto the Superdex 200 column after overnight incubation at pH 4.0 and 20 °C, displayed two peaks, one of apparent molecular mass (Mr) 400 kDa, corresponding to an octamer, and one of apparent Mr 100 kDa, corresponding to a dimer (Figure 1A).The dimer concentration increased when the incubation was performed at pH 7.0 (Figure 1B) and was the only peak when the incubation was performed at pH 8.0 (Figure 1C).
Figure 1.
Effect of pH on the oligomeric state of Sulfolobus solfataricus recombinant WT amidase and site-specific mutants assessed by gel permeation chromatography. WT protein, T319I and D487N mutants (panels A–C), L34P and Y41C mutants (panels D–F). Each protein (about 30 µg) was incubated overnight in 20 mM citrate buffer at pH 4.0 or in 20 mM phosphate buffer at pH 7.0 or 8.0 and was injected onto a Superdex 200 column eluted at room temperature at 0.4 ml min–1 with 20 mM phosphate buffer containing 150 mM NaCl. The eluate was monitored at 226 nm. Elution volumes: octamer = 9.5 ml; dimer = 12.8 ml.
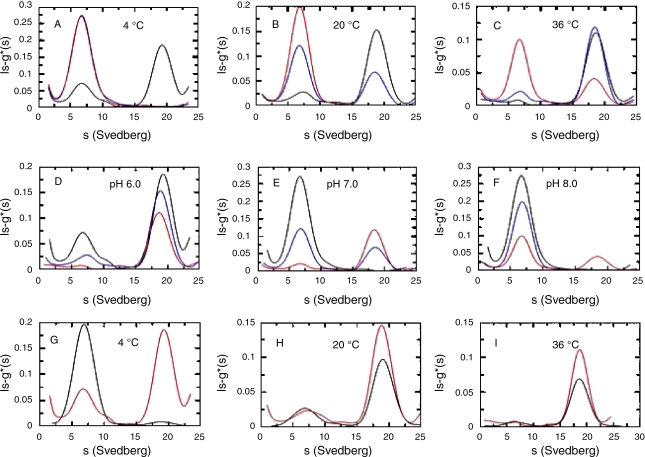
In contrast, at the same pH and temperature, the L34P and Y41C proteins eluted from the Superdex 200 column as a single peak of apparent Mr 100 kDa, corresponding to a dimer (Figures 1D–F). The combined effect of pH and temperature on the association–dissociation behavior of the WT protein was studied in greater detail by AUC between pH 6.0 and 8.0 at temperatures between 4 and 36 °C (Figure 2). Protein solutions were incubated at 4, 20 and 36 °C in 20 mM sodium phosphate buffers of pH 6.0, 7.0 and 8.0. The sedimentation velocity patterns of the samples incubated at 20 °C (Figure 2B) confirmed the GPC data and showed that a decrease in pH favored oligomerization of dimers (s20,w = 6 S) into octamers (s20,w = 18 S). An increase in temperature from 4 to 36 °C induced octamer formation at pH 6.0 but not at pH 7.0 and 8.0 (Figures 2D–F).
Figure 2.
The pH, temperature and salt dependence of the Sulfolobus solfataricus recombinant WT amidase oligomerization process assessed by analytical centrifugation. Experiments were conducted at 117,000 g after overnight incubation at about 0.5 mg ml–1 at the indicated pH and temperature. Each trace refers to a different experiment. Experiments conducted at 4 (A), 20 (B) and 36 (C) °C in 20 mM sodium phosphate pH 6.0 (black), 7.0 (blue) and 8.0 (red). Experiments conducted in 20 mM sodium phosphate pH 6.0 (D), 7.0 (E) and 8.0 (F) at 4 (black), 20 (blue), and 36°C (red). Experiments conducted at 4 (G), 20 (H), and 36°C (I) in 20 mM sodium phosphate pH 6.0 in the presence (black) or in the absence (red) of 1 M NaCl.
The effects of salt, pH and temperature on WT protein oligomerization were studied by AUC. At 4 °C and pH 6.0, the dimeric species amounted to about 30% in the absence of salt but about 95% in the presence of 1 M NaCl (black line) (Figure 2G). At 20 °C (Figure 2H) and 36 °C (Figure 2I), salt had no effect on protein oligomerization, and dimers were about 20 and 5%, respectively in either the presence or the absence of salt.
To further investigate the dependence of oligomerization on salt concentration, WT protein was dialyzed against 20 mM sodium phosphate buffer pH 7.0 at 20 °C, under which conditions both dimers and octamers are present. The protein was then incubated at 20 °C for 2 h with 0–300 mM NaCl followed by GPC on a Superdex 200 column equilibrated and eluted with buffer containing the same salt concentration as the incubation buffer. About 95% of the WT was disocciated at 20 mM or higher NaCl concentrations (Table 1). The behavior of the T319I and D487N mutants was similar to that of WT SSAM (results not shown), whereas salt did not affect the state of association of the L34P and Y41C mutant proteins.
Table 1.
Effect of salt concentration on the oligomer–dimer equilibrium of the Sulfolobus solfataricus WT protein. Percentages of octamer and dimer were determined in the elution pattern from a Superdex 200 column. After exhaustive dialysis against 20 mM sodium phosphate buffer pH 7.0, WT protein was incubated for 2 h in the same buffer containing the indicated salt concentration and immediately injected onto the Superdex 200 column eluted with 20 mMphosphate buffer pH 7.0 with the same salt concentration.
| NaCl (mM) | Octamer % | Dimer % |
| 0 | 60.6 | 39.4 |
| 5 | 53.8 | 46.6 |
| 10 | 35.3 | 64.7 |
| 20 | 6.5 | 93.5 |
| 30 | 5.5 | 94.5 |
| 100 | 5.5 | 94.5 |
| 150 | 5.0 | 95.0 |
| 300 | 5.0 | 95.0 |
| 1000 | 5.0 | 95.0 |
The effect of temperatures on protein oligomerization was investigated by GPC. The WT protein was incubated for 120 min in 20 mM sodium phosphate buffers at pHs of 6.0, 7.0 and 8.0 at temperatures of 20, 36, 70 and 80 °C, followed immediately by injection onto the Superdex 200 column. Relative amounts of dimeric and octameric WT SSAM as calculated from the AUC experiments and the GPC elution patterns were similar (Table 2), and in all cases the effects of pH and temperature were reversible.
Table 2.
Effects of pH and temperature on the percentage of dimeric (D) and octameric (O) species of the Sulfolobus solfataricus WT protein as indicated by AUC and GPC (in bold). Experiments were performed in 20 mM sodium phosphate buffers. Samples for AUC were incubated in the rotor for 12 h at 4, 20, and 36 °C, those for GPC were incubated for 2 h at 70 and 80 °C and immediately injected onto the Superdex 200 column.
| Temp. (° C) | pH 6.0 | pH 7.0 | pH 8.0 | |||||||||
| D | O | D | O | D | O | |||||||
| 4 | 25 | – | 75 | – | 100 | – | 0 | – | 100 | – | 0 | – |
| 20 | 20 | 40 | 80 | 60 | 66 | 57 | 34 | 43 | 100 | 100 | 0 | 0 |
| 36 | 10 | 15 | 90 | 85 | 25 | 30 | 75 | 70 | 70 | 75 | 30 | 25 |
| 70 | – | 0 | – | 100 | – | 28 | – | 72 | – | 65 | – | 35 |
| 80 | – | 0 | – | 100 | – | 25 | – | 75 | – | 60 | <>– | 40 |
The close agreement between the GPC and AUC methods indicates that the WT protein did not interact with the matrix of the Superdex column. The effects of pH and temperature on the SSAM mutants were, therefore, studied by GPC. The T319I and D487N proteins responded to pH and temperature similarly to the WT protein, whereas the L34P and Y41C mutants were insensitive to pH, remaining dimeric at all pHs and temperatures evaluated (results not shown).
The effect of SDS on temperature-induced oligomerization was tested with 30-µg samples of WT SSAM incubated overnight in 100 µl of 20 mM sodium phosphate buffer pH 8.0, which caused protein dimerization, as confirmed by GPC. Subsequently, samples were heated to 80 °C for 2 h, which transformed about 30% of the protein to the octameric form. Under the same conditions, only 15% was transformed to the octameric form in the presence of 0.0125% SDS (Table 3), whereas an SDS concentration of 0.025% prevented oligomerization entirely.
Table 3.
Effects of temperature and SDS on the octamer–dimer equilibrium. Samples containing 30 µg of WT SSAM were incubated overnight at 20 °C in 100 µl of 20 mM sodium phosphate buffer pH 8.0 before analysis by GPC on a Superdex 200 column. One sample was heated at 80 °C for 2 h and injected onto the column. Two samples were heated at 80 °C for 2 h in the presence of 0.0125 or 0.0250 % SDS and injected onto the column.
| pH | SDS (%) | Temp. (°C) | Time (h) | Dimer (%) | Octamer (%) |
| 8 | – | 20 | 15 | 100 | 0 |
| 8 | – | 80 | 2 | 70 | 30 |
| 8 | 0.0125 | 80 | 2 | 85 | 15 |
| 8 | 0.0250 | 80 | 2 | 100 | 0 |
The half-life of enzymatic activity in 50 mM citrate buffer pH 5.0, with 7.5 mM benzamide as substrate was determined at 70, 80 and 90 °C. Under these conditions, the WT protein and all mutant forms had a half-life in excess of 45 h at temperatures of 70 and 80 °C, and more than 20 h at 90 °C. At pH 8.0, which favors dissociation of the octamer, the half-life at 80 and 90 °C was less than at pH 5.0 (Table 4).
Table 4.
Thermal stability (half-life, h) of the WT Sulfolobus solfataricus recombinant amidase and its mutants. Incubations were conducted at the reported temperatures in Eppendorf tubes (0.2 ml) containing 0.2 mg ml–1 protein in 70 µl of 50 mM citrate buffer pH 5.0 or sodium phosphate buffer pH 8.0 . After incubation, a 50 µl sample was assayed for standard activity at pH 5.0 and 70 °C. Abbreviation: n.d.= not determined.
| 70 °C | 80 °C | 90 °C | ||||
| pH 5.0 | pH 8.0 | pH 5.0 | pH 8.0 | pH 5.0 | pH 8.0 | |
| WT | > 70 | > 70 | 103 | 25 | 60 | 0.5 |
| T319I | > 70 | n.d. | 101 | n.d. | 50 | 0.5 |
| D487N | > 70 | > 70 | 102 | n.d. | 42 | n.d. |
| Y41C | > 70 | > 70 | 64 | 0.5 | 20 | 0.25 |
| L34P | > 70 | 19 | 45 | 1.2 | 20 | n.d. |
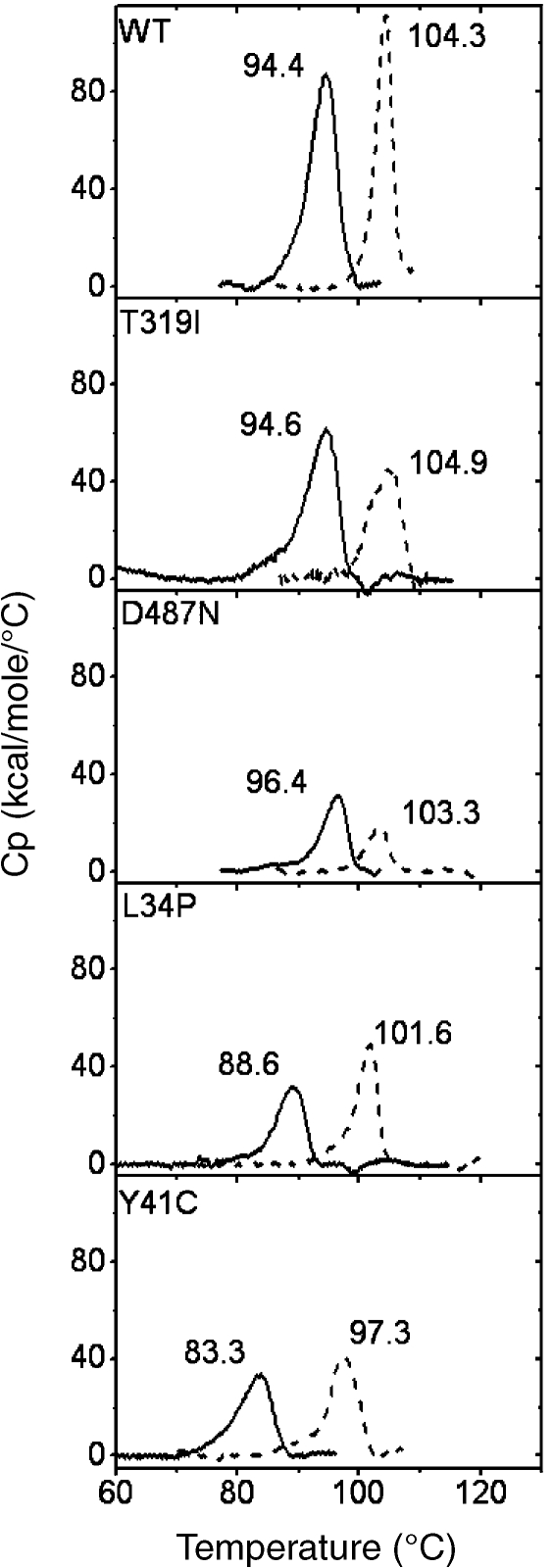
Thermal stability was also estimated by scanning microcalorimetry, which can reveal protein unfolding. Measurements were made at pH 4.0 (20 mM citrate buffer) and pH 8.0 (20 mM sodium phosphate buffer). In WT, T319I and D487N, dimers were predominant at pH 8.0, whereas octamers predominated at pH 4.0; in contrast, L34P and Y41C were dimeric at both pHs. Figure 3 shows that WT SSAM and the T319I and D487N mutants have similar Tm values and that these differ at pH 8.0 (94.4, 94.6 and 96.4 °C, respectively) and at pH 4.0 (104.3, 104.9 and 103.3 °C, respectively). These results suggest that in all three proteins dimers have a melting point about 10 °C lower than octamers. In the L34P and Y41C mutants, which are dimeric at both pH 4.0 and 8.0, the Tmvalues were lower at pH 8.0 (88.6 and 83.8 °C, respectively) than at pH 4.0 (101.6 and 97.3 °C, respectively). The finding that the Y41C mutant had the lowest Tm value is accounted for by the presence of the additional cysteine residue.
Figure 3.
Differential scanning calorimetry thermograms of Sulfolobus solfataricus recombinant WT amidase and its mutants at different pH values. Protein concentration expressed as moles of dimer. The buffer was 20 mM citrate at pH 4.0 (dashed lines) and 20 mM sodium phosphate at pH 8.0 (solid lines). Denaturation temperatures (Tm) are indicated above the corresponding peak.
Thermoactivity
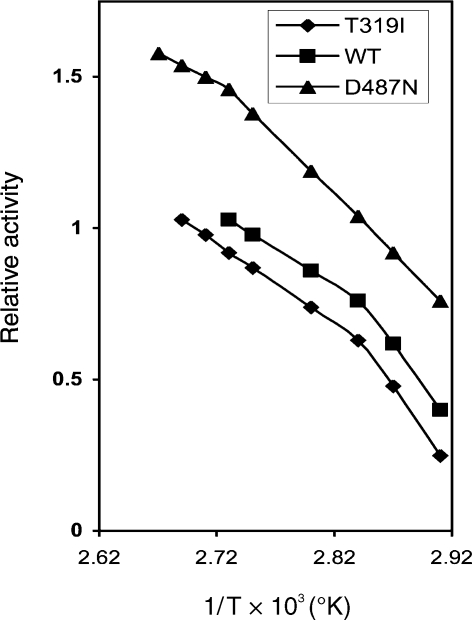
Activity–temperature relationships were assayed under standard conditions between 40 and 101 °C. Assays were performed in sealed plastic tubes. The optimal temperature was 95 °C for WT and Y41C; 98 °C for the T319I and L34P mutant proteins and 100 °C for the D487N mutant protein. An Arrhenius plot (Figure 4) yielded straight lines with a change of slope at 80 °C for WT, T319I, L34P and Y41C (for the sake of clarity only two representative plots are reported) and at 92 °C for D487N.
Figure 4.
Arrhenius plot for Sulfolobus solfataricus recombinant WT amidase and its D487N and T319I mutants. Activities were measured at pH 5.0 with the standard assay. WT amidase (j), T319I (r) and D487N (m) mutants.
The activation energy, calculated for temperatures between 80 and 90 °C and between 90 and 95 °C, was 13.8 kcal mol–1 for WT, 12.8 kcal mol–1 for T319I, and 18.8 kcal mol–1 for D487N. Between 90 and 95 °C the activation energy of D487N was 12.4 kcal mol–1, which was close to that for WT and T319I.
Effect of cyclodextrins
Oligomerization induced by a reduction in pH likely results from a conformational change leading to the exposure of nonpolar residue(s) on one dimer and to their interaction with a hydrophobic patch on an adjacent dimer. To test this hypothesis, the possible sequestration of such nonpolar residue(s) into the ring of α, β or γ cyclodextrins was assessed.
The WT protein was incubated overnight in 20 mM pH 8.0 phosphate buffer to induce dimerization. Thereafter, α, β or γ cyclodextrin in 1:20 and 1:40 molar excess over the dimer was added to the protein solution. To induce oligomerization, the pH was then adjusted to 5.0 by the addition of 0.5 M citrate buffer. After two hours at 20 °C, the mixture was injected onto a PWXL column and was eluted with 20 mM phosphate buffer pH 7.0, containing 150 mM NaCl. Treatment with α or β cyclodextrins resulted in a significant reduction in the amount of oligomer. Neither the amount of cyclodextrin nor an increase in temperature to 40 °C affected the dimer–octamer ratio. The samples treated with γ cyclodextrin remained unmodified (Table 5). Results with the T319I and D487N mutants were similar (results not shown).
Table 5.
Effects of 1:20 and 1:40 molar excess (me) of α, β and γ cyclodextrins (cd) on the acid pH-induced oligomerization of Sulfolobus solfataricus WT amidase. Results are reported as percentage of the area corresponding to the octamer in the elution patterns from the PWXL column.
| Octamer (%) | |
| WT | 100 |
| WT + α-cd (1:20 me) | 50 |
| WT + α-cd (1:40 me) | 50 |
| WT + β-cd (1:20 me) | 55 |
| WT + β-cd (1:40 me) | 55 |
| WT + γ-cd (1:20 me) | 100 |
| WT + γ-cd (1:40 me) | 100 |
Spectroscopic analyses
Scotto d’Abusco et al. (2005) showed that WT SSAM undergoes a conformational change between pH 5.0 and 8.0, which is evident from a decrease in intrinsic fluorescence and ellipticity, whereas no structural modifications were detected in the Y41C mutant. The circular dichroism (CD) and fluorescence spectra of T319I and D487N (results not shown) displayed the same pH dependence as the WT protein, whereas the L34P spectra were unaffected by change in pH.
Scotto d’Abusco et al. (2005) also showed that, following a change in pH from 8.0 to 5.0 in the presence of 1-anilino-8-naphthalene-sulfonate (ANS), the extrinsic fluorescence emission of WT SSAM almost doubled, reflecting increased dye binding, whereas that of the Y41C mutant changed little. Responses of the T319I and D487N mutants were similar to that of WT SSAM, whereas that of the L34P mutant was similar to that of Y41C (results not shown).
Kinetic parameters
To show whether the pH-induced structural changes observed in the WT T319I and D487N amidases are reflected in the active site structure, kinetic parameters were determined at pH 5.0 where the enzyme activity is at a maximum. Each of the enzymes investigated is active on many aliphatic and aromatic substrates (Table 6).
Table 6.
Kinetic properties of Sulfolobus solfataricus WT amidase (1strow), mutant T319I (2nd row), L34P (3rd row), Y41C (4th row). The turnover number (kcat) was obtained by dividing by the molecular mass of the monomer (55,784 Da); temperature was 70 °C; 1–10 µg of enzyme was present in a total volume of 200 µl; incubation times were 2–10 min in 50 mM citrate buffer pH 5.0. Values reported are representative from at least three independent experiments with different lots of enzymes. Abbreviation: Km = Michaelis constant.
| Substrate | Km (mM) | Kcat (s–1) | Kcat/Km (M–1 s–1) × 102 |
| Acetamide | 22.0 | 28.9 | 13.1 |
| 26.0 | 11.2 | 4.3 | |
| 21.0 | 10.3 | 4.9 | |
| 37.0 | 30.1 | 8.1 | |
| Propionamide | 10.0 | 82.9 | 82.9 |
| 25.0 | 214.0 | 85.6 | |
| 4.0 | 11.4 | 28.6 | |
| 10.0 | 99.4 | 99.4 | |
| Butyramide | 14.0 | 34.3 | 24.5 |
| 10.0 | 17.4 | 17.4 | |
| 7.0 | 10.7 | 15.3 | |
| 3.2 | 17.5 | 54.7 | |
| Isobutyramide | 7.0 | 48.9 | 69.9 |
| 1.4 | 27.6 | 197.2 | |
| 5.0 | 32.6 | 65.2 | |
| 6.5 | 58.4 | 89.8 | |
| Metacrylamide | 0.3 | 4.8 | 160.0 |
| 0.4 | 6.6 | 166.5 | |
| 1.0 | 8.6 | 86.0 | |
| 0.5 | 3.0 | 60.0 | |
| Benzamide | 0.9 | 6.7 | 74.7 |
| 0.4 | 7.8 | 195.3 | |
| 1.0 | 2.7 | 27.2 | |
| 0.6 | 7.2 | 120.0 | |
| o-Toluamide | 0.1 | 0.4 | 40.0 |
| 0.1 | 0.3 | 30.0 | |
| 0.1 | 0.2 | 20.0 | |
| 0.2 | 0.8 | 40.0 | |
| p-Toluamide | 0.2 | 12.2 | 610.0 |
| 0.3 | 13.1 | 436.8 | |
| 0.2 | 8.1 | 405.0 | |
| 0.3 | 14.2 | 473.3 | |
| 3-Phenyl-propionamide | 3.0 | 15.3 | 51.0 |
| 2.0 | 9.4 | 46.9 | |
| 1.8 | 8.6 | 47.6 | |
| 2.5 | 18.0 | 72.0 | |
| Indol-3-acetamide | 0.8 | 2.2 | 27.7 |
| 0.8 | 2.6 | 33.5 | |
| 2.5 | 2.2 | 8.8 | |
| 0.8 | 1.2 | 15.0 | |
To establish whether dimers are sensitive to pH, the turnover numbers of WT SSAM and the Y41C mutant were determined at pH 8.0, when both proteins were in the dimeric state, and at pH 5.0, when the WT SSAM was largely in the oligomerized form. At acid pHs, the Y41C dimers showed higher kcat values relative to the WT protein for some of the aliphatic and aromatic substrates tested (Table 7).
Table 7.
Turnover numbers (kcat, s–1) for Sulfolobus solfataricus WT amidase and Y41C determined at pH 5.0 and pH 8.0.
| Substrate | pH | WT | Y41C |
| Propionamide | 5.0 | 82.9 | 99.4 |
| 8.0 | 3.2 | 11.4 | |
| Isobutyramide | 5.0 | 48.9 | 58.4 |
| 8.0 | 21.9 | 40.8 | |
| Benzamide | 5.0 | 6.7 | 7.2 |
| 8.0 | 6.4 | 7.1 | |
| 3-Phenylpropionamide | 5.0 | 15.3 | 18.0 |
| 8.0 | 7.1 | 18.3 | |
Discussion
Recombinant amidase from S. solfataricus is a homodimer of 110 kDa that undergoes a fully reversible pH- and temperature-dependent association into octamers. Proton concentration and temperature act independently and additively on the process (Scotto d’Abusco et al. 2005). Thus, dimers are prevalent at alkaline pHs and at low temperatures. A decrease in pH to acid values primes the association of dimers to octamers such that oligomerization is complete at pH 3.0. Temperature-dependent oligomerization is detectable at 36 °C and increases up to 70–80 °C. Association of dimers into octamers is affected also by salt concentration, which favors octamer dissociation. At pH 7.0, the effect of salt can be detected at NaCl concentrations below 20 mM when temperature is low (4 °C). However, proton concentration appears to have the greatest influence on the dimer–octamer equilibrium. Marked pH sensitivity is attributable to conformational change induced by a reduction in pH, as revealed by UV CD spectra and increased ANS dye-binding. Scotto d’Abusco et al. (2005) observed that oligomerization was impaired by substitution of Y41, as in the Y41C mutant, and suggested that the conformational change induced by low pH results in exposure of hydrophobic residues that stabilize the octamer and that, among such residues, Y41 may trigger oligomerization.
The behavior of WT SSAM in the presence of cyclodextrins and the effect of site-specific mutations on oligomerization substantiate these ideas. Thus, when α or β cyclodextrin was added to the WT protein before acidification, the pH-induced oligomerization was reduced by more than 50%, whereas γ cyclodextrin had no such effect (Table 5). The Y41 side chain may become entrapped into the cyclohexaose or cycloheptaose ring of α or β cyclodextrin, but not in the large cyclomaltooctaose ring of γ cyclodextrin. As a consequence, when α or β but not γ cyclodextrin are in solution, the Y41 residue, though exposed on the dimer surface, cannot interact with a hydrophobic patch of the adjacent dimer and thereby trigger oligomerization.
The disparate oligomerization responses of the site-specific mutants to proton concentration and temperature provide further information on the portion of the dimer responsible for oligomer formation. The state of association of the T319I and D487N mutant proteins varied with pH and temperature in the same way as the WT protein. In contrast, the L34P and Y41C mutant proteins remained dimeric at every pH and temperature that we tested and, to judge by the CD spectra, they did not undergo conformational changes with change in pH or temperature. Moreover, they bound the same amount of ANS at pH 5.0 and pH 8.0, indicating no change with pH in dye-binding hydrophobic groups (Anderson and Weber 1966).
Thermal denaturation of the triad WT-T319I-D487N, as evaluated by microcalorimetry, was consistent with the observed thermal stability of the enzyme assessed by its half-life. Thus, the Tmvalues were comparable at pH 4.0, where octamers were prevalent, and at pH 8.0, where dimers were the dominant species (Figure 3) and the half-lives at pH 5.0 were all similarly high.
The WT protein and the mutants were characterized by high thermal unfolding temperatures at both pH 8.0 and 4.0 (Figure 3). The ΔTm values due to the pH change corresponded to 10 °C for the WT-T319I-D487N triad and to 13 °C for the L34P-Y41C couple. The increase in Tm value at low pH, particularly in L34P and Y41C, and the finding that the state of association of the protein was unaffected by pH, indicate that low pHs induce the most compact and favorable configuration of the active site as manifest by catalytic activity, which was maximal at pH 5.0.
The inferred induction of a more compact structure at pH 5.0 is supported by the high half-life of enzymatic activity at 70, 80 and 90 °C (Table 4). Such thermal stability is the highest reported for amidases. Compared with the L34P mutant, which has a 20 h half-life at 90 °C, amidase from Sulfolobus tokadai Strain 7 has a half-life of only 45 min at 85 °C (Suzuki and Ohta 2006).
All five SSAMs studied displayed exceptional thermal activities with values over 95 °C. The Arrhenius plot (Figure 4) displayed a downward curvature with a change in slope at 80 °C for WT, T319I, L34P and Y41C and at 92 °C for D487N. This feature has been reported for several thermophilic enzymes and is thought to reflect conformational flexibility (Wrba et al. 1990, Vihinen 1987). The change in slope is thought to indicate the temperature at which the conformational flexibility of the protein increases. The transition temperature is 12 °C higher in the D487N mutant than in the WT, T319I, L34P and Y41C proteins. Associated with the discontinuity in the Arrhenius plot, there were changes in activation energy, which was 13.8 and 12.8 kcal mol–1 between 80 and 95 °C for the WT and T319I proteins, respectively, and 18.8 kcal mol–1 between 80 and 92 °C for the D487N mutant (Figure 4). The latter value indicates that the compact mutant protein increases in flexibility above 92 °C and that between 92 an 95 °C the increase in flexibility is associated with a decrease in the activation energy to 12.4 kcal mol–1, a value resembling those of the other SSAMs examined. In principle, this behavior could be ascribed to a change in oligomerization state provided that the oligomeric forms differ in functionality. However this possibility can be excluded because the L34P and Y41C, which are always dimeric, behave similarly to the WT protein.
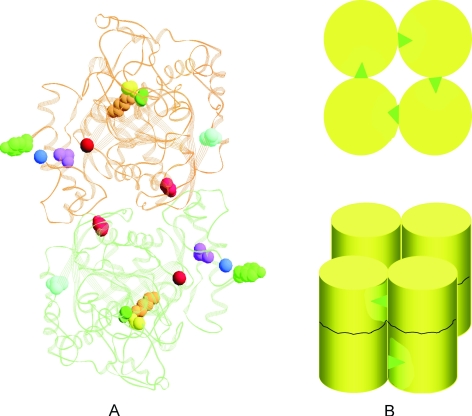
These results demonstrate that low pHs both increase protein compactness, resulting in greater thermal stability and a higher thermal unfolding temperature, and promote oligomerization in WT, T319I and D487N, but not in the other mutant proteins. Differences in pH response among the proteins must be related to the location of the mutation. The 3D structure of SSAM is as yet unavailable. Therefore the low resolution model of the protein based on the peptide amidase (PAM) from Stenotrophomonas maltophila (Labahn et al. 2002) computed by Scotto d’Abusco et al. (2005) was used to construct the SSAM dimer reported in Figure 5A, where putative locations of the mutations and of the catalytic triad at the active site are highlighted.
Figure 5.
Model of the Sulfolobus solfataricus recombinant WT amidase dimer, illustrating the mechanism of pH-, temperature- and ion-dependent oligomerization. In panel A, the residues of the dimer subjected to mutation are in blue (T319), in light red (D487), in magenta (L34), or in green (Y41). The amino- and carboxy-terminal residues of the protein are shown as blue and red spheres, respectively. The catalytic triad at the active site K96 (orange), S171 (yellow) and S195 (dark green) is also shown. Panel B provides a representation of the octamer: each cylinder represents a dimer. Tyrosine 41 residues are indicated by green triangles; hydrophobic patches on contiguous dimers are indicated in light yellow areas.
The L34P and Y41C mutations (highlighted in magenta and green, respectively) are located in the helical 33–48 region (LLKLQLESYERLDSLP) which is close to the amino- and carboxy-terminal residues of the protein (the blue and red spheres, respectively). The T319I mutation is located on the surface of the protein (in light blue) and the D487N mutation (in red) is in the core of the dimer, both far from and opposite to the amino-terminal segment and far from the active site where the catalytic triad K96 (orange), S171(yellow) and S196 (dark green) is shown. That the L34P and Y41C dimers do not form octamers suggests that the helical region of the protein close to the amino-terminus is involved in priming dimer association and plays a key role in this process.
The scheme reported in Figure 5B indicates a possible mechanism for the effects of pH and temperature on oligomerization. When the protein undergoes a pH- or temperature-induced conformational change, the tyrosine 41 residue (green triangle) is exposed and is thus able to interact with a hydrophobic area in the contiguous dimer (the light yellow area).
Because a large change in pH is necessary to cause a shift from one quaternary form to the other, the effect may not be due to the titration of a single residue. The role of the helical segment 33–48 in priming octamer formation is such that substitution with less hydrophobic amino acid, as inY41C and L34P, prevents oligomerization at all pHs and temperatures.
Having established that amino acid mutations in the protein region encompassing the 33–48 segment and those in other parts of the molecule affect the physico-chemical properties of SSAM differently, we examined whether the same is true of the enzyme activity. If so, the kinetic parameters and substrate specificity of the L34P and Y41C mutants should differ from those of the T319I and D479N ones.
The kinetic parameters showed that the WT protein and the four mutants studied were active on a large number of substrates, as is typical of signature amidases (Mayaux et al.1991, Chebrou et al. 1996), a family of proteins with a highly conserved core structure bearing the catalytic triad Ser-cisSer-Lys (Bracey et al. 2002, Labahn et al. 2002, Shin et al. 2003, Valina et al. 2004, Jiang and Cronan 2005), that are separated evolutionarily but diverged often to acquire a broad substrate specificity as in the case of mandelamide hydrolase from Pseudomonas putida (Gopalakrishna et al. 2004).
Scotto d’Abusco et al. (2005) showed that the activity curves of the WT protein and Y41C as a function of pH had a superimposable trend with a maximum at pH 5.0 demonstrating that octamers and dimers are both active. Despite the similarity in physico-chemical properties of the L34P and Y41C mutants, their kinetic behavior differed markedly. Thus, the Y41C kinetic parameters resemble those of WT SSAM, whereas those of L34P were all reduced. This finding indicates the occurrence of conformational change(s) at the active site that can be related to substitution of leucine with proline. Such conformational changes may be large even if not detectable by CD (Newman et al. 1989, Ward et al. 2000).
Although the kcat values for aromatic substrates were similar in all mutants, those for aliphatic substrates varied widely. The kcat for propionamide, which is one of the best aliphatic substrates for SSAM, varied from 82.9 s–1 for the WT protein, to 99.9 s–1 for Y41C and 214 s–1 for T319I. The KM values increased with kcat so that catalytic efficiency (kcat/Km) was unaffected. The observed variations in kinetic parameters may reflect a propagation of small mutation-induced conformational changes to the active site rendering it sensitive to the varying steric hindrance of aliphatic but not aromatic substrates.
In D487N, the slight conformational changes induced by the mutation resulted in fine modulation in the substrate binding, propionamide being used well (kcat 72.3 s–1), whereas butyramide was used poorly (kcat 10.5 s–1) and isobutyramide was not used at all; although all of these compounds are good substrates for the other mutants (Table 6). In contrast, aromatic substrates as benzamide and toluamide are utilized by D487N and by the other mutants in a similar fashion.
Selection of proteins in hyperthermophic organisms was believed to result in maximization of stability, not function. However, recent studies of active-site mutations have demonstrated the occurrence of “activity–stability trade-offs” in proteins of hyperthermophiles (Mukaiyama et al. 2006).
We obtained SSAM mutants in which the mutations were far from the active site, but which nevertheless affected optimization of function as indicated by the increase in kcat. However, the increase in kcat was not offset by reduced protein stability, which remained high, but by an increase of Km with a consequent unmodified efficiency (kcat/Km). The least efficient mutant, L34P, in which a leucine is substituted with a proline: showed low activity with both aliphatic and aromatic substrates.
Canonical inhibitors of serine proteases, such as phenylmethylsulfonyl fluoride, chymostatin and diisopropy1-fluorophosphate did not affect SSAM activity, demonstrating that these substances failed to reach the nucleophile. Other amidases, such ase PAM (Labahn et al. 2002), show a similar behavior. Phosphoramidates were found to be competitive inhibitors of SSAM. Although diethylphosphoroamidate showed no inhibitory activity, phenylphosphorodiamidate was a competitive inhibitor, the effect of which was fully reversed by an SSAM substrate such as benzamide, demonstrating the hydrophobicity of the active site.
Our results indicate that mutation-induced conformational changes influence the catalytic and physico-chemical properties differently, and confirm the importance of the 33–48 region in triggering oligomerization. The occurrence of oligomerization at low pHs and high temperatures, although an important feature of SSAM, is generally infrequent (e.g., Ralston 1991, Zeng et al. 1997, Calvete et al. 1999, Hochgrebe et al. 2000, Madern et al. 2001, Wah et al. 2001, Lew et al. 2003). A case similar to that presented here has been reported in prefoldin, a protein from the archaeon Thermococcus kodakaraensis KOD1 which is a tetramer at pH 8.0 and a dimer of tetramers at pH 6.8 (Kida et al. 2008), suggesting that acid-induced oligomerization may be common in archaea.
It is tempting to correlate the unusual behavior of SSAM to the physiology of S. solfataricus, which occupies a hot sulfur-rich environment (solfatara) with rapid changes in pH and temperature. Oligomerization may constitute an adaptation that limits protein denaturation under extremes of pH and temperature by dissipating thermal energy and excess protons.
Acknowledgments
This research was funded by Italian MIUR (Ministery of University and Research) to E.C. and R.S. and by ASI (Italian Space Agency).
References
- R1.Anderson S., Weber G. The reversibile acid dissociation and hybridization of lactic dehydrogenase. Arch. Biochem. Biophys. 1966;116:207–223. doi: 10.1016/0003-9861(66)90028-2. [DOI] [PubMed] [Google Scholar]
- R2.Bracey M.H., Hanson M.A., Masuda K.R., Stevens R.C., Cravatt B.F. Structural adaptation in a membrane enzyme that terminates endocannabinoid signaling. Science. 2002;296:1793–1796. doi: 10.1126/science.1076535. [DOI] [PubMed] [Google Scholar]
- R3.Cai G., Zhu S., Wang X., Jiang W. Cloning, sequence analysis and expression of the gene encoding a novel wide-spectrum amidase belonging to the amidase signature superfamily from Achromobacter xylosoxidans . FEMS Microbiol. Lett. 2005;249:15–21. doi: 10.1016/j.femsle.2005.05.038. [DOI] [PubMed] [Google Scholar]
- R4.Calvete J.J., Thole H.H., Raida M., Urbane C., Romero A. Molecular characterization and crystallization of Diocleinae lectins. Biochim. Biophys. Acta. 1999;1430:367–375. doi: 10.1016/s0167-4838(99)00020-5. [DOI] [PubMed] [Google Scholar]
- R5.Chebrou H., Bigey F., Arnaud A., Galzy P. Study of the amidase signature group. Biochim. Biophys. Acta . 1996;1298:285–293. doi: 10.1016/s0167-4838(96)00145-8. [DOI] [PubMed] [Google Scholar]
- R6.Chen R., Weng Z. Docking unbound protein using shape complementary, desolvation and electrostatics. Proteins. 2002;47:281–294. doi: 10.1002/prot.10092. [DOI] [PubMed] [Google Scholar]
- R7.Cravatt B.F., Giang D.K., Mayfield S.P., Boger D.L., Lerner R.A., Gilula N.B. Molecular characterization of an enzyme that degrades neuromodulatory fatty-acid amides. Nature. 1996;384:83–87. doi: 10.1038/384083a0. [DOI] [PubMed] [Google Scholar]
- R8.Curnow A.W., Hong K., Yuan R., Kim S., Martins O., Winkler W., Henkin T.M., Soll D. Glu-tRNAGln amidotransferase: a novel heterotrimeric enzyme required for correct decoding of glutamine codons during translation. Proc. Natl. Acad. Sci. USA . 1997;94:11819–11826. doi: 10.1073/pnas.94.22.11819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R9.Fariselli P., Pazos F., Valencia A., Casadio R. Prediction of protein–protein interaction sites in heterocomplexes with neural networks. Eur. J. Biochem. 2002;269:1356–1361. doi: 10.1046/j.1432-1033.2002.02767.x. [DOI] [PubMed] [Google Scholar]
- R10.Gaffney T.D., Da Costa E Silva O., Yamada T., Kosuge T. Indolacetic acid operon of Pseudomonas syringae subsp. savastanoi: transcription analysis and promoter identification. J. Bacteriol. 1990;172:5593–5601. doi: 10.1128/jb.172.10.5593-5601.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R11.Gill S., von Hippel P.H. Calculation of protein extinction coefficients from amino acid sequence data. Anal. Biochem. 1989;182:319–326. doi: 10.1016/0003-2697(89)90602-7. [DOI] [PubMed] [Google Scholar]
- R12.Gomi K., Kitamoto K., Kumagai C. Cloning and molecular characterization of the acetamidase-encoding gene (amdS) from Aspergillus orizae . Gene. 1991;108:91–98. doi: 10.1016/0378-1119(91)90491-s. [DOI] [PubMed] [Google Scholar]
- R13.Gopalakrishna K.N., Stewart B.H., Kneen M.M., Andricopulo A.D., Kenyon G.L., Mcleish M.J. Mandelamide hydrolase from Pseudomonas putida: characterization of a new member of the amidase signature family. Biochemistry. 2004;43:7725–7735. doi: 10.1021/bi049907q. [DOI] [PubMed] [Google Scholar]
- R14.Hochgrebe T., Pankhurst G.J., Wilce J., Easterbrook-Smith S.B. pH-Dependent changes in the in vitro ligand-binding properties and structure of human clusterin. Biochemistry. 2000;15:1411–1419. doi: 10.1021/bi991581b. [DOI] [PubMed] [Google Scholar]
- R15.Jiang Y., Cronan J.E. Expression cloning and demonstration of Enterococcus faecalis lipoamidase (pyruvate dehydrogenase inactivase) as a Ser-Ser-Lys triad amidohydrolase. J. Biol. Chem. 2005;280:2244–2256. doi: 10.1074/jbc.M408612200. [DOI] [PubMed] [Google Scholar]
- R16.Kida H., Sugano Y., Iizuka R., Fujihashi M., Yohda M., Miki K. Structural and molecular characterization of the prefoldin beta subunit from Thermococcus strain KS-1. J. Mol. Biol. 2008;383:465–474. doi: 10.1016/j.jmb.2008.08.041. [DOI] [PubMed] [Google Scholar]
- R17.Koutek B., Prestwich G.D., Howlett A.C., Chin S.A., Salehani D., Akhavan N., Deutsch D.G. Inhibitors of arachidonoyl ethanolamide hydrolysis. J. Biol. Chem. 1994;269:22937–22940. [PubMed] [Google Scholar]
- R18.Labahn J., Neumann S., Buldt G., Kula M.R., Granzin J. An alternative mechanism for amidase signature enzymes. J. Mol. Biol. 2002;322:1053–1064. doi: 10.1016/s0022-2836(02)00886-0. [DOI] [PubMed] [Google Scholar]
- R19.Leatherbarrows R. Cambridge, U.K.: Elsevier-Biosoft; 1987. ENZFITTER. A non linear regression data analysis program for the IBM-PC. [Google Scholar]
- R20.Lew S., Caputo G.A., London E. The effect of interactions involving ionizable residues flanking membrane-inserted hydrophobic helices upon helix–helix interactions. Biochemistry. 2003;42:10833–10842. doi: 10.1021/bi034929i. [DOI] [PubMed] [Google Scholar]
- R21.Madern D., Ebel C., Dale H.A., Lien T., Steen I.H., Birkeland N.K., Zaccai G. Differences in the oligomeric states of the LDH-like MalDH from the hyperthermophilic archaea Methanococcus jannaschii and Archaeoglobus fulgidus . Biochemistry . 2001;40:10310–10316. doi: 10.1021/bi010168c. [DOI] [PubMed] [Google Scholar]
- R22.Mayaux J.F., Cerebelaud E., Soubrier F., Yeh P., Blanche F., Petre D. Purification, cloning, and primary structure of new enantiomer-selective amidase from Rhodococcus strain: structural evidence for a conserved genetic coupling with nitrile hydratase. J. Bacteriol. 1991;173:6694–6704. doi: 10.1128/jb.173.21.6694-6704.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R23.McKinney M.K., Cravatt B.F. Structure and function of fatty acid amide hydrolase. Annu. Rev. Biochem. 2005;74:411–432. doi: 10.1146/annurev.biochem.74.082803.133450. [DOI] [PubMed] [Google Scholar]
- R24.Mukaiyama A., Haruki M., Ota M., Koga Y., Takano K. A hyperthermophilic protein acquires function at the cost of stability. Biochemistry. 2006;45:12673–12679. doi: 10.1021/bi060907v. [DOI] [PubMed] [Google Scholar]
- R25.Newman P.J., Derbes R.S., Aster R.H. The human platelet alloantigens, PIA1and PIA2, are associated with a leucine33/proline33 amino acid polymorphism in membrane glycoprotein IIIa, and are distinguishable by DNA typing. J. Clin. Invest. 1989;83:1778–1781. doi: 10.1172/JCI114082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R26.Patricelli M.P., Cravatt B.F. Clarifying the catalytic roles of conserved residues in the amidase signature family. J. Biol. Chem. 2000;275:19177–19180. doi: 10.1074/jbc.M001607200. [DOI] [PubMed] [Google Scholar]
- R27.Ralston G.B. Temperature and pH dependence of the self-association of human spectrin. Biochemistry. 1991;30:4179–4186. doi: 10.1021/bi00231a011. [DOI] [PubMed] [Google Scholar]
- R28.Sali A., Blundell T.L. Comparative protein modeling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- R29.Schon A., Kannangara C.G., Gough S., Soll D. Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature. 1998;331:187–190. doi: 10.1038/331187a0. [DOI] [PubMed] [Google Scholar]
- R30.Schuck P., Rossmanith P. Determination of the sedimentation coefficient distribution by least-squares boundary modeling. Biopolymers. 2000;54:328–341. doi: 10.1002/1097-0282(20001015)54:5<328::AID-BIP40>3.0.CO;2-P. [DOI] [PubMed] [Google Scholar]
- R31.Scotto d’Abusco A., Ammendola S., Scandurra R., Politi L. Molecular and biochemical characterization of the recombinant amidase from hyperthermophilic archaeon Sulfolobus solfataricus . Extremophiles. 2001;5:183–192. doi: 10.1007/s007920100190. [DOI] [PubMed] [Google Scholar]
- R32.Scotto d’Abusco A., Casadio R., Tasco G., et al. Oligomerization of Sulfolobus solfataricus signature amidase is promoted by acidic pH and high temperature. Archaea. 2005;1:411–423. doi: 10.1155/2005/543789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R33.Shin S., Yun Y.S., Koo H.M., Kim Y.S., Choi K.Y., Oh B.H. Characterization of a novel Ser-cisSer-Lys catalytic triad in comparison with the classical Ser-His-Asp triad. J. Biol. Chem. 2003;278:24937–24943. doi: 10.1074/jbc.M302156200. [DOI] [PubMed] [Google Scholar]
- R34.Suzuki Y., Ohta H. Identification of a thermostable and enantioselective amidase from the thermoacidophilic archaeon Sulfolobus tokadaii Strain 7. Protein Expr. Purif. 2006;45:368–373. doi: 10.1016/j.pep.2005.06.017. [DOI] [PubMed] [Google Scholar]
- R35.Valina A.L., Mazumder-Shivakumar D., Bruice T.C. Probing the Ser-Ser-Lys catalytic triad mechanism of peptide amidase: computational studies of the ground state, transition state, and intermediate. Biochemistry. 2004;43:15657–15672. doi: 10.1021/bi049025r. [DOI] [PubMed] [Google Scholar]
- R36.Vihinen M. Relationship of protein flexibility to thermostbility. Protein Eng. 1987;1:477–480. doi: 10.1093/protein/1.6.477. [DOI] [PubMed] [Google Scholar]
- R37.Wah D.A., Romero A., Gallego Del Sol F., Cavad B.S., Ramos M.V., Grangeiro T.B., Sampaio A.H., Calvete J.J. Crystal structure of native an Cd/Cd-substituted Dioclea guaianensis seed lectin. A novel manganese-binding site and structural basis of dimer-tetramer association. J. Mol. Biol. 2001;310:885–894. doi: 10.1006/jmbi.2001.4814. [DOI] [PubMed] [Google Scholar]
- R38.Ward C.M., Kestin A.S., Newman P.J. A Leu262Pro mutation in the integrin b3 subunit results in an aiib-b3 complex that binds fibrin but not fibrinogen. Blood. 2000;96:161–169. [PubMed] [Google Scholar]
- R39.Wei B.Q., Mikkelsen T.S., Mc Kinney M.K., Lander E.S., Cravatt B.F. A second fatty acid amide hydrolase with variable distribution among placental mammals. J. Biol. Chem. 2006;281:36569–36578. doi: 10.1074/jbc.M606646200. [DOI] [PubMed] [Google Scholar]
- R40.Wrba A., Schweiger A., Schultes V., Jaenicke R., Zbvodsky P. Extremely thermostable D-glyceraldeheyde-3-phosphate dehydrogenase from the eubacterium Thermotoga maritima . Biochemistry . 1990;29:7584–7592. doi: 10.1021/bi00485a007. [DOI] [PubMed] [Google Scholar]
- R41.Zeng X., Zhu H., Lashuel H.A., Hu J.C. Oligomerization properties of GCN4 leucine zipper e and g position mutants . Protein Sci. . 1997;6:2218–2226. doi: 10.1002/pro.5560061016. [DOI] [PMC free article] [PubMed] [Google Scholar]