Abstract
In part due to the existence of simple methods for its cultivation and genetic manipulation, Haloferax volcanii is a major archaeal model organism. It is the only archaeon for which the whole set of post-transcriptionally modified tRNAs has been sequenced, allowing for an in silico prediction of all RNA modification genes present in the organism. One approach to check these predictions experimentally is via the construction of targeted gene deletion mutants. Toward this goal, an integrative “Gateway vector” that allows gene deletion in H. volcanii uracil auxotrophs was constructed. The vector was used to delete three predicted tRNA modification genes: HVO_2001 (encoding an archaeal transglycosyl tranferase or arcTGT), which is involved in archeosine biosynthesis; HVO_2348 (encoding a newly discovered GTP cyclohydrolase I), which catalyzes the first step common to archaeosine and folate biosynthesis; and HVO_2736 (encoding a member of the COG1444 family), which is involved in N4-acetylcytidine (ac4C) formation. Preliminary phenotypic analysis of the deletion mutants was conducted, and confirmed all three predictions.
Keywords: Archaea, GTP-cyclohydrolase I, halophile, tRNA-modification
Introduction
The Archaea encompass a diverse set of organisms and represent the third domain of life. Although they share similarities to both prokaryotes and eukaryotes they have evolved many unique ecological and life-style adaptations. Archaea have become models for the characterization of specific cellular machineries they share with eukaryotes such as those involved in replication (Grabowski and Kelman 2003) and protein degradation (Maupin-Furlow et al. 2006). Despite the increasing availability of genome sequences, the use of archaea as model organisms has been slow, mainly because most archaeal organisms cannot be cultured under standard laboratory conditions and few archaeal genetic systems have been developed (Allers and Mevarech 2005). Three archaeons are emerging as “user friendly” organisms: the crenarchaeum, Sufolobus solafataricus and the euryarchaeal halophiles, H. volcanii and Halobacterium salinarium (Allers and Mevarech 2005). The latter are obligate halophiles, which are aerobic, genetically stable, able to grow on complex and minimal media and which have been genetically sequenced (Ng et al. 2000, Schneider et al. 2006). Transformation systems (Cline et al. 1989), shuttle vectors (Holmes et al. 1991) and a reporter system for these organisms (Holmes and Dyall-Smith 2000) are available. Current gene deletion strategy relies mainly on the use the pyrE2 selective/counter-selective marker (Bitan-Banin et al. 2003). An inducible promoter to generate conditional lethal mutants and to study complementation or depletion of essential proteins is also on hand (Large et al. 2007). The availability of these tools facilitates the genetic manipulation of H. volcanii and H. salinarium;however, because the direct transformation and recombination of a linear PCR product is extremely inefficient (Allers and Mevarech 2005), gene disruption by homologous recombination in these organisms remains a slow process. The most efficient gene-knockout method available requires inserting 0.5–1 kb of the regions upstream and downstream of the gene to be deleted into a suicide vector carrying pyrE (Bitan-Banin et al. 2003), requiring two restriction-digest/ligation-dependent steps. This technique has recently been shortened by the use of inverse PCR (Guangyin et al. 2008), and “fusion PCR/restriction selection cloning” (Hammelmann and Soppa 2008), both requiring only one restriction-digest/ligation-dependent step.
The general focus of our work is to use comparative genomics to uncover the biochemical activities of genes of unknown function, particularly in the areas of coenzyme biosynthesis (Daugherty et al. 2002, de Crécy-Lagard et al. 2007) and RNA modification (Reader et al. 2004, Van Lanen et al. 2005, de Crécy-Lagard 2007, Phillips et al. 2008). We have recently predicted the existence of over thirty putative H. volcanii RNA modification genes (Grosjean et al. 2008), and are now in the process of testing these predictions. To streamline the design of deletion cassettes and insure robustness we wanted to avoid several steps used previously. To this end we established an archaeal Gateway platform based on the Invitrogen (Carlsbad, CA) “Gateway system,” obviating the need to: (1) generate a restriction map of the region targeted for deletion; (2) choose appropriate endonucleases for cloning into multiple cloning sites (MCS) of H. volcanii suicide vector; (3) include such sites into polymerase chain reaction (PCR) primers used to amplify fragments to be cloned; (4) treat PCR products with restriction enzymes to generate MCS compatible cohesive ends (often inefficient); and (5) perform blunt-end ligations which are often inefficient. In this work we describe a facile method for the construction of targeted gene deletions via use of a H. volcanii Gateway suicide destination vector. We have employed this system to delete three genes: HVO_2736 and HVO_ 2001 encoding proteins COG1444 and COG0343, respectively, which are predicted to be involved in the synthesis of two tRNA modifications; and HVO_2348, which encodes COG1469 and is predicted to be involved in both the biosynthesis of a tRNA modification and folate.
Archaeosine (G+) is an archaea specific 7-deazaguanosine nucleoside present at position 15 (dihydrouridine loop or D-loop) in the majority of tRNA species (Gregson et al. 1993, Watanabe et al. 1997, Iwata-Reuyl 2003). The function of archaeosine in vivo has yet to be investigated but its location has led to the hypothesis that it may function in stabilizing the tertiary structure of the tRNA through global interactions between the D-loop and the T-stem and loop (Gregson et al. 1993). The archaeosine biosynthetic pathway is complex and poorly characterized (Oliva et al. 2007). It has been shown that 7-cyano-7-deazaguanine (preQ0) is an intermediate in the pathway and is inserted into tRNA by a tRNA-guanine transglycosylase (TGT, EC 2.4.2.29) encoded by the tgtA gene (Bai et al. 2000, Watanabe et al. 2001). This enzyme has been extensively investigated biochemically and structurally (Ishitani et al. 2002), but no deletion mutant has ever been constructed in any archaeal organism. By deleting the corresponding HVO_2001 gene, we will address the essentiality of a protein family that is found in most archaeal genomes sequenced thus far.
We have recently shown that the synthesis of deazapurines derived from preQ0, such as G+, share a common first step with the folate pathway, catalyzed by the folE gene encoding the canonical GTP cyclohydrolase enzyme IA (Katzenmeier et al. 1990, McCarty and Bandarian 2008, Phillips et al. 2008). Haloferax volcanii is one of the rare archaea with both active folate and G+ pathways (Gupta 1984, Gupta 1986, Ortenberg et al. 2000); however, no folE gene has been identified in its genome. Instead, H. volcanii possesses a folE2 gene (HVO_2348) encoding a newly discovered GTP cyclohydrolase enzyme IB that is involved in archaeosine biosynthesis (El Yacoubi et al. 2006, Phillips et al. 2008). A role in folate biosynthesis has yet to be demonstrated.
Finally, the N4-acetylcytidine (ac4C) modification is found at position 34 in many H. volcanii tRNAs and has also been detected in ribosomal RNA in certain archaea and eukaryotes (Gupta 1984, Gupta 1986, Grosjean et al. 2008). The COG1444 member YpfI (now called TmcA) has been shown responsible for ac4C formation in the tRNA initiator of E. coli (Ikeuchi et al. 2008). In yeast, the homolog (YNL132W) is essential (Giaever et al. 2002), whereas the E. coli ypfI gene is not (Baba et al. 2006). We predicted the enzyme family COG1444 as a potential candidate for ac4C (Grosjean et al. 2008) and that constructing a gene deletion mutant would address both the validity of the functional prediction in archaea and the essentiality of this modification in H. volcanii.
Materials and methods
Strains, media and transformation protocols.
All strains and plasmids used are listed in Table 1. Escherichia coli derivatives were routinely grown in LB medium (BD Diagnostic System) at 37 °C. Growth media were solidified with 15 g l–1 agar (BD Diagnostic System) for the preparation of plates. Transformations of E. coli were performed following standard procedures (Miller 1972, Sambrook et al. 2001). Ampicillin (Ap, 100 µg ml–1), chloramphenicol (Cm, 20 µg ml–1), spectinomycin (50 µg ml–1) and kanamycin (Kn, 50 µg ml–1) were added as required. Haloferax volcanii derivatives were grown at 45 °C in Hv-YPC rich medium (Kauri et al. 1990) (125 g l–1 NaCl, 50g l–1 MgCl2·6H2O, 2.5 g l–1 KSO4, 0.134 g l–1 CaCl2·2H2O, 5 g l–1 tryptone, and 5 g l–1 yeast extract) or a chemically defined medium Hv-CA (Kauri et al. 1990) (125g l–1 NaCl, 50g l–1 MgCl2·6H2O, 2.5 g l–1 KSO4, 0.134 g l–1 CaCl2·2H2O and 0.5% casamino acids) supplemented when needed with 40 µg ml–1 dT, 40 µg ml–1 hypoxanthine, 40 µg ml–1 pantothenic acid, 50 or 10 µg ml–1 uracil, and 50 µg ml–1 5-fluoroorotic acid (FOA). Transformations of H. volcanii were performed essentially as described by Cline et al. (1989).
Table 1.
List of strains and plasmids used in this study.
| Relevant features | Reference | |
| Strains | ||
| E. coli DB3.1 | F- gyrA462 endA1 (sr1-recA) mcrB mrr hsdS20(rB-, mB-) supE44 ara-14 galK2 lacY1 | Invitrogen (Calsbad, CA) |
| proA2 rpsL20(SmR) xyl-5 - leu mtl1 | ||
| E. coli TOP 10 | F- mcrA Δ(mrr-hsdRMS-mcrBC) ϕ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(araleu) 7697 | Invitrogen (Calsbad, CA) |
| galU galK rpsL (StrR) endA1 nupG | ||
| E. coli INV110 | F´ {traΔ36 proAB lacIq lacZΔM15} rpsL (StrR) thr leu endA thi-1 lacY galK galT ara tonA | Invitrogen (Calsbad, CA) |
| tsx dam dcm supE44 <>Δ(lac-proAB)Δ(mcrC-mrr)102::Tn10 (TetR) | ||
| H. volcanii DS70 | (Wendoloski et al. 2001) | |
| H. volcanii H26 | DS70 ΔpyrE2 | (Allers et al. 2004) |
| VDC5273 | DB3.1/pBY158 | This study |
| VDC3241 | HVO_2001 deletion mutant clone 1; H26 derivative; | This study |
| VDC3242 | HVO_2001 deletion mutant clone 2; H26 derivative | This study |
| VDC3234 | HVO_2348 deletion mutant clone 1; H26 derivative | This study |
| VDC3235 | HVO_2348 deletion mutant clone 2; H26 derivative | This study |
| VDC3236 | HVO_2348 deletion mutant clone 3; H26 derivative | This study |
| VDC2177 | HVO_2736 deletion mutant clone 1; H26 derivative | This study |
| VDC2178 | HVO_2736 deletion mutant clone 2; H26 derivative | This study |
| Plasmids | ||
| pYES-DEST52 | Cloning vector; ApR and G418 | Invitrogen (Calsbad, CA) |
| pGEM-T Easy | Cloning vector; ApR | Promega (Madison, WI) |
| pTA131 | pBluescript II with BamHI-XbaI fragment from pGB70 containing pyrE2 under ferredoxin promoter; ApR | (Allers et al. 2004) |
| pENTR/D | Standard cloning vector; KnR | Invitrogen (Carlsbad, CA) |
| pCR8 | Standard cloning vector; SpR | Invitrogen (Carlsbad, CA) |
| pBY156.14 | pGEMTez derivative containing the attCmRCcdR cassette, T/A cloned | This study |
| pBY158 | pTA131 derivative containing the attCmRCcdR cassette from pBY158 as a HindIII, XhoI fragment | This study |
| pNAB144 | pENTR/D derivative containing HVO_2001 with 1 Kb of upstream and 1 Kb of down-stream sequence | This study |
| pBY163 | pCR8 derivative containing HVO_2348 with 1 Kb of upstream and 1 Kb of down-stream sequence | This study |
| pGPP005 | pNAB144 derivative containing a deletion of HVO_2001; result of reverse PCR | This study |
| pGPP032 | pBY163 derivative containing a deletion of HVO_2348; result of reverse PCR | This study |
| pBY165.31 | pBY158 derivative containing Up_ΔHVO_2001_Dn; result of recombination between pBY158 and pGPP005 | This study |
| pGPP092 | pBY158 derivative containing Up_ΔHVO_2348_Dn; result of recombination between pBY158 and pGPP032 | This study |
| pGPP018.1 | pCR8 derivative containing HVO_2736 with 1 Kb of upstream and 1 Kb of down-stream sequence | This study |
| pGPP037 | pGPP18.1 derivative containing a deletion of HVO_2736; result of reverse PCR | This study |
| pGPP066 | pBY158 derivative containing Up_ΔHVO_2736_Dn; result of recombination between pBY158 and pGPP037 | This study |
| pIKB103 | pBY158 derivative containing Up_ΔHVO_Dn; result of recombination between pBY158 and pIKB153 | This study |
| pIKB152 | pENTR derivative containing Up_HVO_2736_DN | This study |
| pIKB153 | pIKB152 derivative containing UP_ΔHVO_2736_DN | This study |
Molecular biology
PCR
All PCRs were performed using Phusion™ Hot Start (Finnzymes, Espoo, Finland).
LR reactions
All LR reactions were performed following the Gateway LR protocols (Invitrogen, Carlsbad, CA) using the Gateway LR clonase enzyme mix (Invitrogen, Carlsbad, CA).
Reverse PCR
Oligonucleotides were designed to anneal within the gene to be deleted and to amplify the remaining DNA fragment of the plasmid. The PCR was performed using Phusion according to the manufactures directions in a volume of 100 µl. The<> subsequent PCR product was column purified (QIAquick, Qiagen) and eluted with 50 µl of elution buffer. The purified PCR product was ligated according to the manufacturer’s directions (NEB; New England Biolabs, IPSWICH, MS). Briefly, reactions included 1 µl of T4 DNA ligase buffer, 6 µl of purified vector (product of the reverse PCR), 2.5 µl of water, 0.5 µl of T4 DNA ligase. Reactions were performed at 16 °C overnight. The ligase was inactivated according to the manufacturer’s protocol and the ligation mix treated with DpnI to remove template DNA. For that purpose, 2 µl of DpnI, 3 µl of NEB buffer 1, and 5 µl of water were added to the ligation mixture and incubated at 37 °C for 3 hours.
pBY158 construction
The Gateway destination H. volcanii suicide vector pBY158 (Table 1) was constructed as follows; the region comprising the attR1 recombination sequence, the CmR gene (the positive selection marker), the ccdB gene (the negative selection marker) and the attR2 recombination sequence was amplified from pYES-DEST52 (Invitrogen, Carlsbad, CA) with primers AttCmCcd.ol1 and AttCmCcd.ol2 that contained HindIII and XhoI restriction sites, respectively (see Table 2 for a list of all primers used). After A-addition using Taq polymerase, the product was ligated into the T/A cloning vector, pGEMT-Easy (Promega, Madison, WI) and the ligation product transformed into E. coli One Shot® Library Efficiency® DB3.1 ™ chemically competent cells (DB3.1 carries a mutation in gyrA that confers resistance to CcdB). The resulting plasmid, pBY156.14 was digested with HindIII and XhoI to liberate the attR1CmRCcdBattR2 cassette that was then ligated into pTA131 (Allers et al. 2004) linearized with the corresponding enzymes. The ligation product was then transformed into DB3.1 cells. Plasmid preparations were recovered and verified by restriction profiling, sequencing of the attccdBCmR region, and phenotyping (Cm resistance and lethality to cells not carrying the ccdB gene).
Table 2.
List of primers used in this study.
| Primer name | Sequence |
| UpDn_HVO2001_fwd | CCGCGGCAACCGCAGACG |
| UpDn_HVO2001_rev | CTCTCTTTCGCCCGTCGTTA |
| RevPCR_HVO2001_fwd | CACCCCATCGGCGACTACTTCTTC |
| RevPCR_HVO2001_rev | CGACGCAATCTCCTCGATGG |
| HVO2001_Check-in_fwd | AACTCCTACATCATCAAGACG |
| HVO2001_Check-in_rev | TCGTTGGAGGCGTAGAAGAA |
| HVO2001_Check-out_fwd | GAGGCGGTCTCCACGGTGAC |
| HVO2001_Check-out_rev | GCGCTCGCCTGCGTTCGCG |
| HVO2001_DIG_fwd | GGTGACAGCTCGTCATCGG |
| HVO2001_DIG_rev | GTGTGACGCCACGATTATGG |
| UpDn_HVOB2348_fwd | ATGGAGTGGCGTTTCCCGA |
| UpDn_HVO2348_rev | TCGGTCGGTCGCGTCCG |
| RevPCR_HVO2348_fwd | AGTGCCGTTGCTGATTAGTG |
| RevPCR_HVO2348_rev | AGCGCGAGGTGACGCTGG |
| HVO2348_Check-in_fwd | CGGCCATCGAGCGGACGC |
| HVO2348_Check-in_rev | GAGGACATGTGCGGCGACGC |
| HVO2348_Check-out_fwd | CGGGCATCGACATCCACC |
| HVO2348_Check-out_rev | GACGCGACTCGCGAGCAA |
| HVO2348_DIG_fwd | CTGGCCTGCGTTCGCGGTTC |
| HVO2348_DIG_rev | GAGCTGTTTGAGCGCGGAGA |
| UpDn_HVOB2736_fwd | CTG GAG GAT GTC GCC CTC GC |
| UpDn_HVO2736_rev | GAC GGT CAG GTG CGC CCC |
| RevPCR_HVO2736_fwd | GACGCGGCCCGCGAGGAA |
| RevPCR_HVO2736_rev | TTCGGTCATGGGATGGATAC |
| HVO2736_Check-in_fwd | GTT TCT GGT GGT CGA GAG GT |
| HVO2736_Check-in_rev | TAC CGA CAG GGA ATC AGC TT |
| HVO2736_Check-out_fwd | TCTCCGACCCTGATTTTCTG |
| HVO2736_Check-out_rev | CGTGATGAAGTCCGACGAGAC |
| HVO2736_DIG_fwd | TCG TTA CCG GAG ATG AGG TC |
| HVO2736_DIG_rev | AGT AGC TTC ACC GGC TAC GA |
H. volcanii DNA preparation for PCR
The DNA preparations were performed according to The Halohandbook (Dyall-Smith 2007). In brief, a single colony of H. volcanii was used to inoculate 5 ml of Hv-YPC and incubated at 45 °C overnight. Two hundred µl of the overnight culture was centrifuged at 6000 g for 5 minutes, the supernatant decanted, and the cells resuspended in 400 µl of water. A 0.5 µl aliquot of the cell suspension was used in a 20-µl PCR reaction. The PCR reaction included 200 µM primers, 3% DMSO and 0.4 units of Phusion Hot-Start polymerase. The cycling conditions routinely used were: 98 °C for 30 s; 98 °C for 10 s, annealing temperature for 10 s, 72 °C for 30 s per kb (30 cycles); 72 °C for 5 min.
Southern hybridization
Genomic DNA preparations were performed according to The Halohandbook (Dyall-Smith 2007). After digestion with the appropriate restriction enzymes, the DNA was transferred by capillary transfer to a positively charged nylon membrane using the alkaline transfer method according to Sambrook et al. (2001). The membrane was processed according to Roche Biosciences DIG protocols (ROCHE Molecular Biochemical, Indianapolis, IN). The probes were designed to hybridize outside the 5′ and 3′ flanking regions. Stringency washes were performed according to Roche Biosciences (ROCHE Molecular Biochemical, Indianapolis, IN) except for the second stringency wash which was performed in 0.5 × SSC at 65 °C.
H. volcanii tRNA extraction and analysis
Bulk tRNA was prepared, hydrolyzed and analyzed by liquid chromatography-tandem mass spectrometry (LC-MS-MS) asdescribed by Phillips et al. (2008). All tRNA purifications and analyses were performed at least twice independently.
Databases
The H. volcanii genome sequence was accessed through the UCSC archaeal genome browser (Schneider et al. 2006).
Results
Construction of a gateway destination vector to allow subsequent deletions of H. volcanii genes
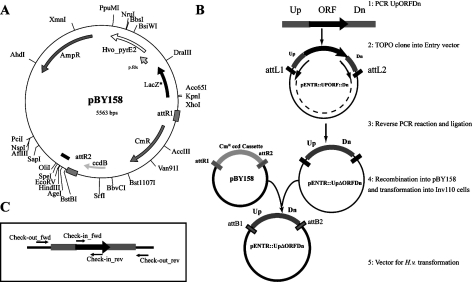
The gateway destination H. volcanii suicide vector pBY158 (Table 1) was constructed as described in the Materials and methods. The resulting destination plasmid, pBY158 (Figure 1A), is an H. volcanii suicide vector that can recombine DNA fragments from Gateway compatible entry plasmids.
Figure 1.
Haloferax volcanii deletion cassette construction using pBY158, an H. volcanii gateway suicide vector. A. Map of pBY158. B. Schematic of deletion cassette construction using pBY158. C. PCR verification scheme for checking H. volcanii deletion mutants.
A generalized strategy for constructing gene deletions with pBY158 is summarized in Figure 1B. Briefly, a fragment comprising the gene of interest flanked by 500 to 1000 bp on both the 5′ and the 3′ sides (Up-ORF-Dn) is amplified using a blunt-end generating polymerase (e.g. Phusion™ Hot Start). The Up-ORF-Dn fragment is then cloned into one of the entry plasmids, pENTR/D (for blunt directional cloning) or pCR8/GW (for non-directional T/A cloning). Reverse PCR (see Figure 1B) is performed using a high-fidelity polymerase resulting in a deletion of the target ORF. The deletion is either exact (i.e., from ATG to the stop codon) or designed to retain the integrity of the flanking genes and operon organization. In the reverse PCR, phosphorylated oligonucleotides are used to allow for ligation of the product. After ligation of the reverse PCR product, DpnI treatment is used to eliminate the methylated template plasmid before transformation into E. coli competent cells. The resulting plasmid contains the Up-ΔORF-Dn cassette in an entry plasmid. The Up-ΔORF-Dn region is then recombined into pBY158 and the recombination mix (pBY158::Up-ΔORF-Dn) is transformed into INV110 competent cells. These cells are gyrA+, and are therefore killed by the presence of the ccdB allele (this counter-selection allele eliminates plasmids that have not recombined) and dam dcm, which for plasmid preparations that will not be cleaved by endogenous H. volcanii restriction enzymes (Holmes et al. 1991). Transformants are plated onto LB agar supplemented with ampicillin. Individual clones are then replica-plated onto LB supplemented with ampicillin and chloramphenicol and ampicillin alone. For clones showing chloramphenicol sensitivity and ampicillin resistance, diagnostic of successful replacement of the ccdB-CmR cassette by the Up-ΔORF-Dn casette, plasmid DNA was extracted and verified by restriction enzyme profiling and DNA sequencing before transformation into H. volcanii H26.
Use of pBY158 for targeted disruption of H. volcanii genes
With the procedure described here, three H. volcanii deletion strains were constructed. The first carrying a deletion of HVO_2736 (1551 bp) was predicted to lack the N4-acetylcytidine (ac4C) modification in tRNA. The second and third with deletions of HVO_2001 (2265 pb) and HVO_2348 (927 bp) genes were predicted to be deficient in archaeosine (G+) and G+ and folate biosynthesis, respectively.
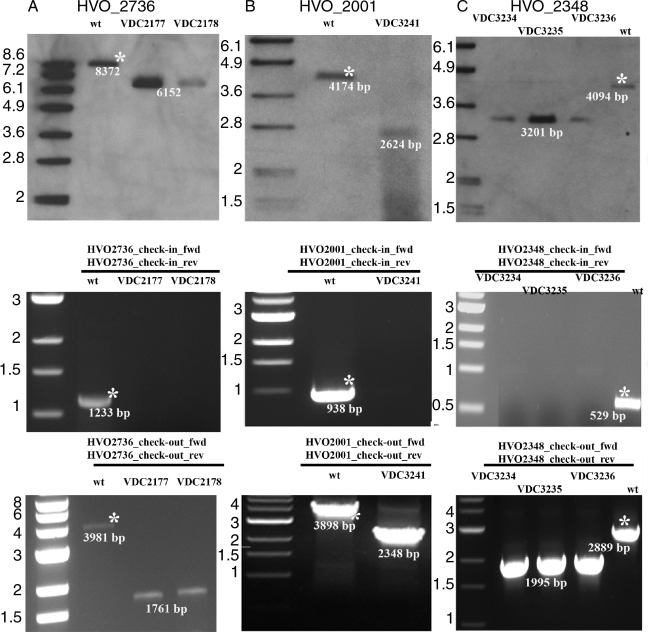
Deletion plasmids were constructed as described in Figure 1B, using the primers listed in Table 2, and transformed into H. volcanii H26 (pyrE2). Single crossover events were first selected using uracil prototrophy. Subsequent double crossover events leading to elimination of the genes (in 50% of the cases) were selected based on 5-fluoroorotic acid resistance. For the deletion of the predicted folE2 gene (HVO_2348), thymidine and hypoxanthine were added to all selection media. For all three ORFs, candidate H. volcanii colonies were first screened by colony PCR using primers designed to anneal within the target gene (Check-in primers) (Figure 1C), thus amplifying a fragment internal to the ORF targeted for deletion. The colonies that yielded no product in the first screen were tested in a second PCR screen using one oligonucleotide priming at a region 5′ of the upstream homology sequence and another priming 3′ of the downstream homology sequence (Check-out primers) (Figure 1C). Clones that yielded a PCR product of the predicted size were analyzed by Southern hybridization. For all mutants both PCR and Southern hybridization analyses confirmed the deletion of the targeted gene and total disappearance of the wild-type copy (Figure 2). Therefore, HVO_2736, HVO_2001 and HVO_2348 were found to be non essential under the condition tested.
Figure 2.
Southern blot analyses and PCR verifications of the three H. volcanii mutants constructed. Deletion strain for HVO_2736 (A), deletion strain for HVO_2001 (B) and deletion strain for HVO_2348 (C). Upper panels are Southern hybridization profiles, middle panel are inside primers PCR checks and lower panels are outside primers PCR checks. Relevant DNA marker sizes (kb) are indicated on the right side of each panel and wild type bands (bp) are indicated by an asterisk (*), expected sizes are indicated under each band.
Phenotype of deletion mutants
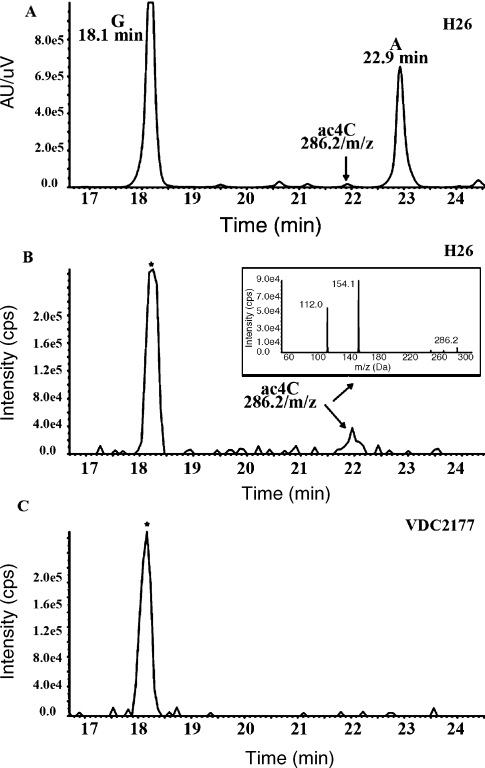
Ribonucleosides released by digestion of the bulk tRNA extract from WT strain H26 and the HVO_2736 deletion strain (VDC2177), were analyzed by LC-MS/MS (Figure 3). The peak eluting at 21.9 min on the UV trace of H26 (Figure 3A), corresponding to ac4C as showed by the extraction ion chromatogram (XIC) and the tandem mass fragmentation pattern of 286.2 m/z (Figure 3B), is absent in the deletion mutant (Figure 3C). Because other modified bases elute at the same position (Ikeuchi et al. 2008), only the XIC showed the absence of the ac4C modification in the deletion strain under the 21.9 min peak. Quantification of the N2,N2-dimethyguanosine (m22G) content was used as an internal standard to estimate the variation caused by the loading amount of tRNA analyzed in the mutant strain. The ratio of m22G between the mutant and WT was 88%. These results strongly suggest that COG1444 is involved in introducing the ac4C modification in H. volcanii tRNA.
Figure 3.
Analysis of LC-MS/MS tRNA extracted from H. volcanii strains showing the presence or absence of the ac4C peak on the UV trace at 254 nm for H26 (A) and the corresponding extraction ion chromatogram for the H26 and VDC2177 strains (B).
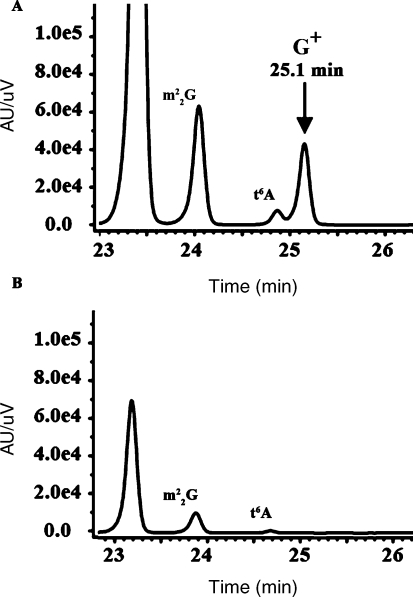
Similarly, we analyzed the G+ content of tRNA extracted and digested from the WT and the HVO_2001 deletion strain (VDC3241). As shown in Figure 4, the peak eluting at 25.1 min, which was present in the WT strain, and confirmed by the XIC of 325 m/z ion to be G+ (Phillips et al. 2008; data not shown), was absent in the mutant. Although the tRNA concentration was lower in the mutant preparation (m22G ratio of 32%), the N6-threonylcarbamoyladenosine (t6A) modification (413 m/z) eluting at 24.7 min was nevertheless detectable (confirmed by the XIC, data not shown). Therefore the absence of the G+ peak in the mutant phenotype is unlikely due to the lower amount of tRNA analyzed, but rather to the deletion of HVO_2001.
Figure 4.
Analysis by LC-MS of tRNA extracted from H. volcanii strains showing the presence or absence of the G+ absorbance peak at 254 nm for H26 (A and B) and VDC3242 strains (C). Two additional modification are indicated, m22G and t6A.
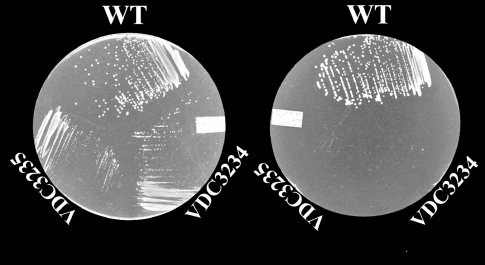
As in H. volcanii, tetrahydrofolate derivatives are essential methyl donors for several metabolic reactions, the predicted phenotype for HVO_2348 deletion strains (VDC3234, VDC 3235 and VDC 3236) is the requirement for dT, hypoxanthine and pantothenate on Hv-CA (minimal medium supplemented with casaminoacids (Levin et al. 2004)). As predicted, the mutant strains were auxotrophic for dT (Figure 5). Hypoxanthine auxotrophy was also observed and the absence of pantothenate led only to a slow-growth phenotype, likely because of trace amounts of the vitamin in the casaminoacid mix (data not shown).
Figure 5.
Haloferax volcanii folE2 deletion mutant phenotype analysis. H26, VDC3234 and VDC3235 (two HVO_2348 independent deletion mutants) growing on Hv-CA supplemented with dT/hypoxanthine (left plate) or hypoxanthine only (right plate) (pantothenate was added to both plates). Cells were grown at 45 °C for 72 hours.
Discussion
We have constructed and demonstrated the utility of an H. volcanii Gateway suicide vector (pBY158) for constructing deletion cassettes. The major advantage of the platform presented here is the combination of a simple and efficient cloning technique, specifically the Gateway platform, with a simple and rapid design strategy applicable to high throughput mutant generation in any gene. With this protocol, restriction/ligation dependent steps are eliminated (there is one straightforward blunt ligation, which allows for re-circularizing the LR reaction product). Also, large fragments often needed for deleting large genes can easily be accommodated (see deletion cassette construction for COG1444; Up (1 kb)-ORF (2.2 kb)-Dn (1 kb) with a total of 4.2 kb). This deletion platform offers the following advantages over existing methods: (1) it requires at most one post-PCR gel purification step (after the initial PCR if more than one fragment is obtained, see Figure 1), (2) it requires a simple PCR primer design, i.e., no endonuclease restriction sites need be incorporated in the primer sequences, and (3) fragment cloning is independent of the DNA and vector sequences, which obviates the need for restriction map construction and analysis of DNA sequences. The method can easily be extended to generate complementation test plasmids carrying H. volcanii ORFs under their native promoters without additional designs. In this application, UN-ORF-DN fragments could be recombined into replicative destination vectors to allow for transgene expression driven by the natural promoter, a feature often needed for accurate complementation studies.
The platform was successfully used to confirm the function of three proteins out of a list of 30 genes predicted to encode proteins involved in RNA metabolism (Grosjean et al. 2008). Deletion of the folE2 gene (HVO_2348) predicted to encode a novel family of GTP cyclohydrolase I in H. volcanii yielded a strain with the predicted phenotypes, i.e., auxotrophy for both thymidine and hypoxanthine and tRNAs devoid of archaeosine. It is unlikely that these phenotypes are due to polarity effects as the folE2 gene is upstream of HV_2349, which is transcribed in the opposite orientation. This is the first folE 2 deletion mutant in any prokaryote, and its phenotypic analysis confirms our prediction that this gene family is involved in both G+ (Phillips et al. 2008) and folate biosynthesis in archaea. We have shown elsewhere that this strain displays a growth rate comparable to H26 in supplemented medium (Phillips et al. 2008). The folE2 gene can therefore be used as a positive auxotrophic selectable marker adding to the few already available (Ortenberg et al. 2000, Bitan-Banin et al. 2003, Allers et al. 2004).
Deletion of HVO_2736 and analysis of the tRNA profile of the resulting mutant confirmed the prediction that this family is involved in ac4C synthesis in tRNA. This modification is found at position 34 in five H. volcanii tRNAs (Gupta 1984, Gupta 1986), but its function is unknown. It has been postulated that the conformational rigidity of this nucleoside could stabilize RNA molecules of thermophilic archaea (Kawai et al. 1992, Bruenger et al. 1993, Oliva et al. 2007). Our data indicate that an H. volcanii strain with tRNA lacking ac4C remains viable. Similarly, we have constructed the first Δtgt archaeal strain and showed that its tRNA is lacking G+. The strong conservation of the pathway for G+ synthesis across archaea suggests an essential role for this modification. We are currently testing the influence of growth conditions on the phenotypes of both G+ and ac4C deficient mutants. The possibility that both the ac4Cand archaeosine deficiency phenotypes are due to a polar effect on downstream genes (less than 100 bp downstream of the stop codon) has not been ruled out by classical genetics means, i.e., complementation test. However, this is unlikely because independent in vitro biochemical data (Watanabe et al. 1997, Ikeuchi et al. 2008) corroborate our predications based on comparative genomics for these genes (Grosjean et al. 2008).
In summary, use of the TOPO/Gateway platform in combination with PCR to screen and verify deletion mutants provides an efficient method for simultaneous processing of large numbers of clones, enabling the generation of deletion libraries. These libraries could provide the basis for high-throughput functional genomics in archaea.
Acknowledgments
This work was supported by The National Science foundation (MCB-05169448). The authors thank Julie Maupin and Thorsten Allers for advice on H. volcanii manipulations and Sophie Alvarez for LC/MS analyses.
References
- R1.Allers T., Mevarech M. Archaeal genetics—the third way. Nat. Rev. Genet. 2005;6:58–73. doi: 10.1038/nrg1504. [DOI] [PubMed] [Google Scholar]
- R2.Allers T., Ngo H., Mevarech M., Lloyd R.G. Development of additional selectable markers for the halophilic archaeon Haloferax volcanii based on the leuB and trpA genes. Appl. Environ. Microbiol. 2004;70:943–953. doi: 10.1128/AEM.70.2.943-953.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R3.Baba T., Ara T., Hasegawa M., Takai Y., Okumura Y., Baba M., Datsenko K.A., Tomita M., Wanner B.L., Mori H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006;2:1–11. doi: 10.1038/msb4100050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R4.Bai Y., Fox D.T., Lacy J.A., Van Lanen S.G., Iwata-Reuyl D. Hypermodification of tRNA in thermophilic archaea. Cloning, overexpression, and characterization of tRNA-guanine transglycosylase from Methanococcus jannaschii. J. Biol. Chem. 2000;275:28731–28738. doi: 10.1074/jbc.M002174200. [DOI] [PubMed] [Google Scholar]
- R5.Bitan-Banin G., Ortenberg R., Mevarech M. Development of a gene knockout system for the halophilic archaeon Haloferax volcanii by use of the pyrE gene. J. Bacteriol. 2003;185:772–778. doi: 10.1128/JB.185.3.772-778.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R6.Bruenger E., Kowalak J.A., Kuchino Y., Mccloskey J.A., Mizushima H., Stetter K.O., Crain P.F. 5S rRNA modification in the hyperthermophilic archaea Sulfolobus solfataricus and Pyrodictium occultum . FASEB J. 1993;7:196–200. doi: 10.1096/fasebj.7.1.8422966. [DOI] [PubMed] [Google Scholar]
- R7.Cline S.W., Lam W.L., Charlebois R.L., Schalkwyk L.C., Doolittle W.F. Transformation methods for halophilic archaebacteria. Can. J. Microbiol. 1989;35:148–152. doi: 10.1139/m89-022. [DOI] [PubMed] [Google Scholar]
- R8.Daugherty M., Polanuyer B., Farrell M., Scholle M., Lykidis A., De Crécy-Lagard V., Osterman A. Complete reconstitution of the human coenzyme A biosynthetic pathway via comparative genomics. J. Biol. Chem. 2002;277:21431–21439. doi: 10.1074/jbc.M201708200. [DOI] [PubMed] [Google Scholar]
- R9.de Crécy-Lagard V. Identification of genes encoding tRNA modification enzymes by comparative genomics. Methods Enzymol. 2007;425:153–183. doi: 10.1016/S0076-6879(07)25007-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R10.de Crécy-Lagard V., El Yacoubi B., De La Garza R.D., Noiriel A., Hanson A.D. Comparative genomics of bacterial and plant folate synthesis and salvage: Predictions and validations. BMC Genomics. 2007;8:245. doi: 10.1186/1471-2164-8-245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R11.Dyall-Smith M. The halohandbook: Protocols for halobacterial genetics. http://www.haloarchaea.com/resources/halohandbook/index.html. 2007 [Google Scholar]
- R12.El Yacoubi B., Bonnett S., Anderson J.N., Swairjo M.A., Iwata-Reuyl D., De Crécy-Lagard V. Discovery of a new prokaryotic type I GTP cyclohydrolase family. J. Biol. Chem. 2006;281:37586–37593. doi: 10.1074/jbc.M607114200. [DOI] [PubMed] [Google Scholar]
- R13.Giaever G., Chu A.M., Ni L., et al. Functional profiling of the Saccharomyces cerevisiae genome. Nature. 2002;418:387–391. doi: 10.1038/nature00935. [DOI] [PubMed] [Google Scholar]
- R14.Grabowski B., Kelman Z. Archaeal DNA replication: Eukaryal proteins in a bacterial context. Annu. Rev. Microbiol. 2003;57:487–516. doi: 10.1146/annurev.micro.57.030502.090709. [DOI] [PubMed] [Google Scholar]
- R15.Gregson J.M., Crain P.F., Edmonds C.G., Gupta R., Hashizume T., Phillipson D.W., Mccloskey J.A. Structure of the archaeal transfer RNA nucleoside G*–15 (2-amino-4,7- dihydro-4-oxo-7-beta-D-ribofuranosyl-1H-pyrrolo[2,3-d]pyrimidine-5-carboximi dam ide (archaeosine)) J. Biol. Chem. 1993;268:10076–10086. [PubMed] [Google Scholar]
- R16.Grosjean H., Marck C., Gaspin C., Decatur W., De Crécy Lagard V. RNomics and Modomics in the halophilic archaea Haloferax volcanii: identification of RNA modification genes. BMC Genomics. 2008;9:470–496. doi: 10.1186/1471-2164-9-470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R17.Guangyin Z., Kowalczyk D., Humbard M.A., Rohatgi S., Maupin-Furlow J.A. Proteasomal components required for cell growth and stress responses in the haloarchaeon Haloferax volcanii . J. Bacteriol. 2008;190:8096–8105. doi: 10.1128/JB.01180-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R18.Gupta R. Halobacterium volcanii tRNAs. Identification of 41 tRNAs covering all amino acids, and the sequences of 33 class I tRNAs. J. Biol. Chem. 1984;259:9461–9471. [PubMed] [Google Scholar]
- R19.Gupta R.C. Transfer RNAs of Halobacterium volcanii: Sequences of five leucine and three serine tRNAs. Syst. Appl. Microbiol. 1986;7:102–105. [Google Scholar]
- R20.Hammelmann M., Soppa J. Optimized generation of vectors for the construction of Haloferax volcanii deletion mutants. J. Microbiol. Methods. 2008;75:201–204. doi: 10.1016/j.mimet.2008.05.029. [DOI] [PubMed] [Google Scholar]
- R21.Holmes M.L., Dyall-Smith M.L. Sequence and expression of a halobacterial β-galactosidase gene. Mol. Microbiol. 2000;36:114–122. doi: 10.1046/j.1365-2958.2000.01832.x. [DOI] [PubMed] [Google Scholar]
- R22.Holmes M.L., Nuttall S.D., Dyall-Smith M.L. Construction and use of halobacterial shuttle vectors and further studies on Haloferax DNA gyrase. J. Bacteriol. 1991;173:3807–3813. doi: 10.1128/jb.173.12.3807-3813.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R23.Ikeuchi Y., Kitahara K., Suzuki T. The RNA acetyltransferase driven by ATP hydrolysis synthesizes N4-acetylcytidine of tRNA anticodon. EMBO J. 2008;27:2194–2203. doi: 10.1038/emboj.2008.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R24.Ishitani R., Nureki O., Fukai S., Kijimoto T., Nameki N., Watanabe M., Kondo H., Sekine M., Okada N., Nishimura S., Yokoyama S. Crystal structure of archaeosine tRNA-guanine transglycosylase. J. Mol. Biol. 2002;318:665–677. doi: 10.1016/S0022-2836(02)00090-6. [DOI] [PubMed] [Google Scholar]
- R25.Iwata-Reuyl D. The Biosynthesis of the 7-deazaguanosine modified nucleosides of transfer RNA. Bioorg. Chem. 2003;31:24–43. doi: 10.1016/s0045-2068(02)00513-8. [DOI] [PubMed] [Google Scholar]
- R26.Katzenmeier G., Schmid C., Bacher A. Cloning and expression of the putative gene coding for GTP cyclohydrolase I from Escherichia coli . FEMS Microbiol. Lett. 1990;54:231–234. doi: 10.1016/0378-1097(90)90288-2. [DOI] [PubMed] [Google Scholar]
- R27.Kauri T., Wallace R., Kushner D.J. Nutrition of the halophilic archaebacterium, Haloferax volcanii . Syst. Appl. Microbiol. 1990;13:14–18. [Google Scholar]
- R28.Kawai G., Yamamoto Y., Kamimura T., Masegi T., Sekine M., Hata T., Iimori T., Watanabe T., Miyazawa T., Yokoyama S. Conformational rigidity of specific pyrimidine residues in tRNA arises from posttranscriptional modifications that enhance steric interaction between the base and the 2′-hydroxyl group. Biochemistry. 1992;31:1040–1046. doi: 10.1021/bi00119a012. [DOI] [PubMed] [Google Scholar]
- R29.Large A., Stamme C., Lange C., Duan Z., Allers T., Soppa J., Lund P.A. Characterization of a tightly controlled promoter of the halophilic archaeon Haloferax volcanii and its use in the analysis of the essential cct1 gene. Mol. Microbiol. 2007;66:1092–1106. doi: 10.1111/j.1365-2958.2007.05980.x. [DOI] [PubMed] [Google Scholar]
- R30.Levin I., Giladi M., Altman-Price N., Ortenberg R., Mevarech M. An alternative pathway for reduced folate biosynthesis in bacteria and halophilic archaea. Mol. Microbiol. 2004;54:1307–1318. doi: 10.1111/j.1365-2958.2004.04339.x. [DOI] [PubMed] [Google Scholar]
- R31.Maupin-Furlow J.A., Humbard M.A., Kirkland P.A., Li W., Reuter C.J., Wright A.J., Zhou G., Gerald P.S. Proteasomes from structure to function: perspectives from archaea. Curr. Top. Dev. Biol. 2006;75:125–169. doi: 10.1016/S0070-2153(06)75005-0. [DOI] [PubMed] [Google Scholar]
- R32.McCarty R.M., Bandarian V. Deciphering deazapurine biosynthesis: Pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem. Biol. 2008;15:790–798. doi: 10.1016/j.chembiol.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R33.Miller J.H. Cold Spring Harbor: CSHL Press; 1972. Experiments in molecular genetics.468 [Google Scholar]
- R34.Ng W.V., Kennedy S.P., Mahairas G.G., et al. From the cover: Genome sequence of Halobacterium species NRC-1. Proc. Natl. Acad. Sci. USA. 2000;97:12176–12181. doi: 10.1073/pnas.190337797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R35.Oliva R., Tramontano A., Cavallo L. Mg2+ binding and archaeosine modification stabilize the G15 C48 Levitt base pair in tRNAs. RNA. 2007;13:1427–1436. doi: 10.1261/rna.574407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R36.Ortenberg R., Rozenblatt-Rosen O., Mevarech M. The extremely halophilic archaeon Haloferax volcanii has two very different dihydrofolate reductases. Mol. Microbiol. 2000;35:1493–1505. doi: 10.1046/j.1365-2958.2000.01815.x. [DOI] [PubMed] [Google Scholar]
- R37.Phillips G., El Yacoubi B., Lyons B., Alvarez S., Iwata-Reuyl D., De Crécy-Lagard V. The biosynthesis of the 7-deazaguanosine modified tRNA nucleosides: A new role for GTP cyclohydrolase I. J. Bacteriol. 2008;190:7876–7884. doi: 10.1128/JB.00874-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R38.Reader J.S., Metzgar D., Schimmel P., De Crécy-Lagard V. Identification of four genes necessary for biosynthesis of the modified nucleoside queuosine. J. Biol. Chem. 2004;279:6280–6285. doi: 10.1074/jbc.M310858200. [DOI] [PubMed] [Google Scholar]
- R39.Sambrook J., Fritsch E.F., Maniatis T. Cold Spring Harbor: CSHL Press; 2001. Molecular cloning: A laboratory manual.745 [Google Scholar]
- R40.Schneider K.L., Pollard K.S., Baertsch R., Pohl A., Lowe T.M. The UCSC archaeal genome browser. Nucleic Acids Res. 2006;34:D407–D410. doi: 10.1093/nar/gkj134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R41.Van Lanen S.G., Reader J.S., Swairjo M.A., De Crécy-Lagard V., Lee B., Iwata-Reuyl D. From cyclohydrolase to oxidoreductase: Discovery of nitrile reductase activity in a common fold. Proc. Natl. Acad. Sci. USA. 2005;102:4264–4269. doi: 10.1073/pnas.0408056102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- R42.Watanabe M., Matsuo M., Tanaka S., Akimoto H., Asahi S., Nishimura S., Katz J.R., Hashizume T., Crain P.F., Mccloskey J.A., Okada N. Biosynthesis of archaeosine, a novel derivative of 7-deazaguanosine specific to archaeal tRNA, proceeds via a pathway involving base replacement of the tRNA polynucleotide chain. J. Biol. Chem. 1997;272:20146–20151. doi: 10.1074/jbc.272.32.20146. [DOI] [PubMed] [Google Scholar]
- R43.Watanabe M., Nameki N., Matsuo-Takasaki M., Nishimura S., Okada N. tRNA recognition of tRNA-guanine transglycosylase from a hyperthermophilic archaeon, Pyrococcus horikoshii . J. Biol. Chem. 2001;276:2387–2394. doi: 10.1074/jbc.M005043200. [DOI] [PubMed] [Google Scholar]
- R44.Wendoloski D., Ferrer C., Dyall-Smith M.L. A new simvastatin (mevinolin)-resistance marker from Haloarcula hispanica and a new Haloferax volcanii strain cured of plasmid pHV2. Microbiology. 2001;147:959–964. doi: 10.1099/00221287-147-4-959. [DOI] [PubMed] [Google Scholar]