Abstract
Various proteins with different biological activities have been observed to be translocated from the nucleus to the cytoplasm in an energy- and signal-dependent manner in eukaryotic cells. This nuclear export is directed by nuclear export signals (NESs), typically characterized by hydrophobic, primarily leucine, amino acid residues. Moreover, it has been shown that CRM1/exportin 1 is an export receptor for leucine-rich NESs. However, additional NES-interacting proteins have been described. In particular, eukaryotic initiation factor 5A (eIF-5A) has been shown to be a critical cellular cofactor for the nuclear export of the HIV type 1 (HIV-1) Rev trans-activator protein. In this study we compared the nuclear export activity of NESs of different origin. Microinjection of export substrates into the nucleus of somatic cells in combination with specific inhibitors indicated that specific nuclear export pathways exist for different NES-containing proteins. In particular, inhibition of eIF-5A blocked the nuclear export of NESs derived from the HIV-1 Rev and human T cell leukemia virus type I Rex trans-activators, whereas nucleocytoplasmic translocation of the protein kinase inhibitor-NES was unaffected. In contrast, however, inhibition of CRM1/exportin 1 blocked the nuclear export of all NES-containing proteins investigated. Our data confirm that CRM1/exportin 1 is a general export receptor for leucine-rich NESs and suggest that eIF-5A acts either upstream of CRM1/exportin 1 or forms a complex with the NES and CRM1/exportin 1 in the nucleocytoplasmic translocation of the HIV-1 Rev and human T cell leukemia virus type I Rex RNA export factors.
In nucleated cells, transport of macromolecules across the nuclear envelope is an active, bidirectional process mediated by the nuclear pore complex (NPC) in a temperature-, energy-, and signal-dependent manner (for reviews, see refs. 1–5). Protein import, in general, is specified by nuclear localization signals (NLSs), which are most commonly composed of a short stretch of basic amino acids and constitute the target of the dimeric importin-α/importin-β NLS–receptor complex (for reviews, see refs. 6 and 7). The process of protein import involves the importin-mediated binding of selected transport substrates to the cytoplasmic fibrils of the NPC, followed by their movement through the central gated channel and, finally, release of the import substrate into the nuclear interior. Multiple interactions between the soluble import protein cargo and components of the NPC are responsible for the orchestration of these events.
In comparison with protein import, the molecular mechanisms underlying the active nuclear export of proteins and RNAs are poorly understood. Nevertheless, it has become evident that RNA export is a receptor-mediated process involving the binding of the RNA cargo to specific export factors in the form of ribonucleoprotein complexes (RNPs). In addition, it generally is accepted that export of different classes of RNAs [e.g., mRNA, tRNA, rRNA] is mediated, at least in part, by specific transport factors (for reviews, see refs. 8–10). In analogy to import, nuclear export is specified by nuclear export signals (NESs), which reside in the protein moiety of RNPs and are characterized by a short stretch of hydrophobic, primarily leucine amino acids. This type of leucine-rich NES has been identified in various proteins, including proteins involved in viral RNA export, such as the HIV-1 Rev and type I human T-lymphotrophic virus (HTLV-I) Rex trans-activators (11–13), or in proteins with no obvious activity in nucleocytoplasmic RNA translocation, such as protein kinase inhibitor (PKI) (12, 14).
To date, the HIV-1 Rev protein is the most studied specific RNA export factor containing a leucine-rich NES (for a detailed review, see ref. 15). It was shown that different proteins are able to bind directly and specifically the HIV-1 Rev NES. These include eukaryotic initiation factor 5A (eIF-5A) (16, 17) and CRM1/exportin 1 (18, 19), which appears to be an essential export receptor for leucine-rich NESs (18–21). In contrast, however, the nucleoporin-like protein hRIP/Rab (22, 23) was shown to bind indirectly via CRM1/exportin 1 to the Rev NES (24). In the case of eIF-5A, distinct mutants of the factor have been described that inhibit the nuclear export of Rev protein and, thereby, HIV-1 replication (17, 25). Moreover, microinjection studies have demonstrated that antibodies directed against eIF-5A specifically block the nucleocytoplasmic translocation of HIV-1 Rev in somatic cells (26), indicating that eIF-5A is part of a specific nucleocytoplasmic export pathway.
In this work we compare the nuclear export activities of NESs derived from proteins with different biological activities in somatic cells. Glutathione S-transferase (GST)-fusion proteins containing the NES of the HIV-1 Rev or HTLV-I Rex trans-activators served as export cargoes that are involved in viral mRNA export, whereas the PKI NES represented a cellular, RNA-independent, nuclear export pathway in these studies. We demonstrate the existence of two different export pathways for leucine-rich NESs. Both types of pathways are blocked equally well by inhibitors of CRM1/exportin 1, confirming previous data that CRM1/exportin 1 is a general export factor for leucine-rich NESs. However, inhibitors of eIF-5A selectively blocked the nucleocytoplasmic translocation of NESs that were derived from the Rev or Rex retroviral RNA export factor. These data indicate that eIF-5A acts upstream of CRM1/exportin 1 in the regulation of nuclear export mediated by specific, leucine-rich NESs.
MATERIALS AND METHODS
Construction of Plasmids Encoding GST-Fusion Proteins.
Synthetic double-strand oligonucleotides specifying the amino acid positions 75–84 (NH2-LPPLERLTLD-COOH) in HIV-1 Rev wild-type (wt), amino acid positions 75–84 (NH2-LPPDLRLTLD-COOH) in the HIV-1 RevM10 mutant, amino acid positions 82–93 (NH2-LSAQLYSSLSLD-COOH) in HTLV-I Rex wt, and amino acid positions 37–46 (NH2-LALKLAGLDI-COOH) in PKI wt proteins, respectively, were fused to the carboxyl terminus of GST in the bacterial expression vector pGEX-3X (Pharmacia), generating the plasmids pGEX-Rev-NES, pGEX-RevM10-NES, pGEX-Rex-NES, and pGEX-PKI-NES. pGEX-eIF-5A and pGEX-eIF-5AM14 are bacterial vectors expressing the recently described eIF-5A wt and mutant M14 proteins (17).
Expression of Recombinant Proteins.
All GST-fusion proteins were expressed and purified from the Escherichia coli strain BL21 by affinity chromatography on glutathione-Sepharose 4B according to the manufacturer’s recommendations (Pharmacia). Eluted proteins were concentrated by ultrafiltration with a PM10 filter device (Amicon). Before microinjection, protein concentrations were determined by the method of Bradford (27). eIF-5A wt and eIF-5A-M14 proteins were expressed as GST fusions, and the GST moiety was removed by treatment with factor Xa and affinity chromatography on glutathione-Sepharose 4B according to the manufacturer’s protocol (Pharmacia).
Antibodies.
eIF-5A-specific IgG fraction from rabbits (16, 26) was affinity-purified on a column with recombinant eIF-5A coupled to Sepharose 4B according to the manufacturer’s recommendations (Pharmacia). The rabbit polyclonal anti-CRM1 antiserum, raised against the CRM1/exportin 1 carboxyl terminus (NH2- GIFNPHEIPEEMCD-COOH), was a kind gift of L. Gerace (The Scripps Research Institute).
Microinjections.
HeLa cells constitutively expressing the Rev response element (RRE) RNA (28) were cultured in DMEM supplemented with 10% FBS and antibiotics. These cells were microinjected to deliver fusion proteins into the nucleus by using a CompiC INJECT (Cellbiology Trading, Hamburg, Germany) computer-assisted injection system as described (17). In some experiments the respective GST-fusion proteins (1 mg/ml) were coinjected with rabbit IgG protein (1 mg/ml) as an internal injection control. Additional nuclear coinjections were performed with rabbit polyclonal anti-eIF-5A antibody (α-eIF-5A; 3 mg/ml) (26), rabbit polyclonal anti-CRM1 peptide antiserum (2 mg/ml), or eIF-5A wt or eIF-5A-M14 protein (using a 10-fold molar excess over the coinjected GST-fusion proteins). For antibody–antigen competition, the CRM1-derived peptide was included in the microinjection experiments at a concentration of 0.6 mg/ml. The cells were fixed 20 min after injection in 3% paraformaldehyde/1% BSA in PBS for 10 min, Triton X-100-treated for 10 min, and then labeled with a primary mouse monoclonal anti-GST antibody (Serotec) at a 1:50 dilution. The secondary antibodies, Cy2-conjugated goat anti-rabbit IgG and Cy3-conjugated goat anti-mouse IgG (Rockland, Gilbertsville, PA), were used at a dilution of 1:200. Immunofluorescence analyses were performed by using a Zeiss Axiovert-135 microscope. Images were recorded with a cooled MicroMax CCD camera (Princeton Instruments) and processed by using the IPLab spectrum and Adobe photoshop package.
RESULTS
Nucleocytoplasmic Translocation of GST-NES Fusion Proteins in Somatic Cells.
Nuclear export of proteins is mediated by NESs, which are short stretches of hydrophobic amino acids. The prototypic signal has been identified in the HIV-1 Rev trans-activator protein (11, 12) and, subsequently, in various proteins of different function, including HTLV-I Rex (13) and cAMP-dependent PKI (12, 14). The hallmark feature of this type of NES is a series of critically spaced leucine residues (29). However, functional investigation of a large number of randomly mutagenized NES regions also revealed a high diversity in allowable residues at a number of NES positions, including leucine residues that had been considered essential for function (30). Thus, a significant tolerance exists within the structural requirements of leucine-rich NESs. This sequence diversity also may reflect a diversity in the requirement of cofactors used for nuclear export. This hypothesis is strengthened further by the fact that proteins that contain a leucine-rich NES frequently have different biological activities. Thus, it is probable that specific levels of regulation exist for the nucleocytoplasmic translocation of potentially different classes of nuclear export cargoes. This fine regulation of nuclear export potentially could be regulated by modification of the respective NES, thereby allowing or preventing NES-CRM1/exportin 1 interaction. For example, phosphorylation of critical serine residues within the NES of cyclin B1 reduces its affinity for CRM1/exportin 1, subsequently reducing the nuclear export rate (31). Alternatively, specific regulation might be achieved by the action of small adapter molecules that target the NES-containing export cargo to CRM1/exportin 1, a mechanism similar to the importin-α/importin-β axis employed for the nuclear import of proteins containing a classical NLS (6, 7). Clearly, it also cannot be ruled out that, in some cases, the combination of both mechanisms might be used for specific regulation of nuclear export.
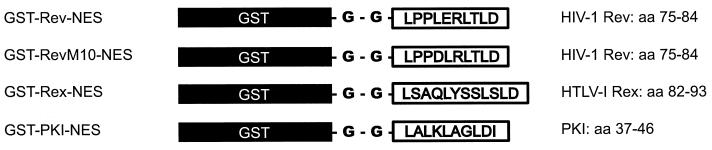
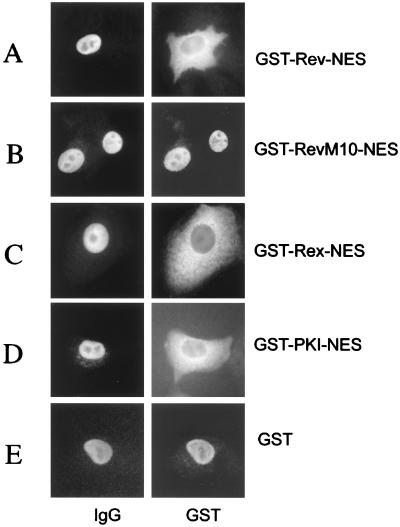
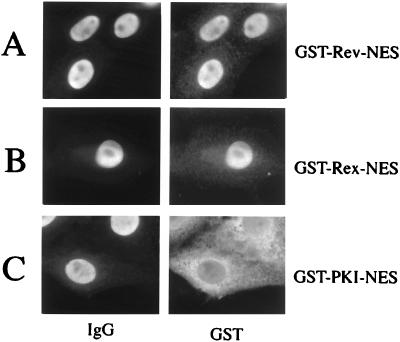
We generated a series of GST-fusion proteins to test NESs contained in different proteins for their capacity to translocate the heterologous GST protein moiety from the nucleus to the cytoplasm in mammalian cells. Short stretches of amino acids representing the NESs of the trans-activator proteins HIV-1 Rev and HTLV-I Rex, as well as of PKI, were fused via a linker consisting of two glycine residues to the GST carboxyl terminus (Fig. 1). In addition, wild-type GST as well as GST-RevM10-NES fusion protein, which is an NES mutant that specifies the trans-dominant phenotype of the RevM10 protein (32–34) and had been shown previously to be devoid of nuclear export function (35), served as export-deficient controls in our initial experiments. These proteins were microinjected into the nuclei of HeLa cells together with rabbit Igs. The comicroinjected IgG served as an internal control to confirm the correct injection site. Twenty minutes post-injection, the cells were fixed with paraformaldehyde and the injected proteins were visualized by indirect immunofluorescence microscopy. As shown in Fig. 2, GST-Rev-NES, GST-Rex-NES, and GST-PKI-NES proteins were readily exported from the HeLa cell nucleus, whereas the comicroinjected IgG protein was detected exclusively in the cell nucleus (Fig. 2 A, C, and D, respectively). As expected, the export-deficient GST-RevM10-NES and wild-type GST protein remained nuclear (Fig. 2 B and E). Notably, injection of GST-Rev-NES fusion protein, in which the structurally flexible glycine linker between the GST and NES element was omitted, resulted in significantly decreased nuclear export rates (J.H., unpublished results). These observations suggest that the NES must occupy a specific structural conformation to allow subsequent cofactor interaction. Notwithstanding, these data demonstrated that isolated NES elements of various origins are fully functional in the context of fusions to GST in somatic cells.
Figure 1.
Schematic depiction of the structure of the fusion proteins that were investigated for nuclear export. NESs (open boxes) of the HIV-1 Rev, HIV-1 RevM10, HTLV-I Rex, and PKI proteins, respectively, were fused via a flexible linker consisting of two glycine residues to the carboxyl terminus of GST (solid boxes). The amino acid position of the NESs within the respective proteins is indicated to the right. RevM10 is an export-deficient NES mutant (L78D and E79L) (11, 32).
Figure 2.
Nuclear export mediated by various NESs in human somatic cells. HeLa cells were comicroinjected into the nucleus with the indicated proteins and rabbit IgG (IgG, injection control). Cells were incubated for 20 min, fixed, and analyzed for GST- and rabbit IgG-specific localization by double-label, indirect immunofluorescence microscopy. The GST-fusion proteins were visualized by using a mouse monoclonal anti-GST antibody followed by Cy3-conjugated goat anti-mouse IgG (GST). The microinjected rabbit IgG was detected with a Cy2-conjugated goat anti-rabbit antibody. Injection of GST-RevM10-NES fusion protein (B) or wild-type GST (E) served as export-deficient controls.
Leptomycin B and Anti-CRM1/Exportin 1 Antibodies Inhibit NES-Mediated Nuclear Export.
Next, we wanted to investigate whether the nuclear export of our GST-NES proteins could be inhibited by inhibitors of CRM1/exportin 1. Leptomycin B has been shown to be a low-molecular-weight drug binding specifically to CRM1/exportin 1 and preventing the formation of stable NES-CRM1/exportin 1 complexes (19–21, 36). Furthermore, leptomycin B has been shown to block Rev-mediated RNA export as well as the nuclear export of Rev protein itself (19, 21, 37). We therefore pretreated our HeLa cell cultures with leptomycin B (20 nM) 45 min before microinjection. Fig. 3 summarizes the data, showing that leptomycin B completely abolishes the nuclear export of GST-Rev-NES, GST-Rex-NES, and GST-PKI-NES. These data confirmed the previous observations that leptomycin B is a general inhibitor of NES-mediated nuclear export.
Figure 3.
Inhibition of nuclear export by leptomycin B. HeLa cells were comicroinjected into the nucleus with the indicated fusion proteins and rabbit IgG (IgG, injection control) and analyzed as described above. Cell cultures were supplemented 45 min before microinjection with leptomycin B at a concentration of 20 nM.
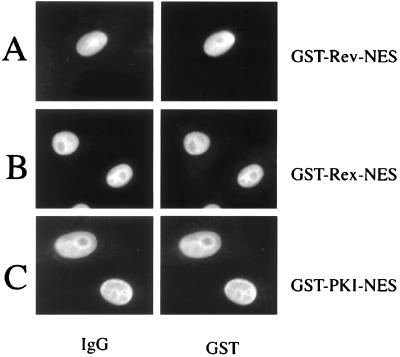
We were interested to know whether the inhibitory phenotype of leptomycin B on nuclear export also can be observed with antibodies to CRM1/exportin 1. For this, we comicroinjected our series of GST-NES fusion proteins together with a rabbit polyclonal antiserum raised against the carboxyl-terminal peptide (NH2-GIFNPHEIPEEMCD-COOH) of CRM1/exportin 1 (anti-CRM1). In agreement with the leptomycin B experiments and shown in Fig. 4, this anti-CRM1 antiserum completely blocked the nuclear export of all of our microinjected GST-NES fusion proteins (Fig. 4 A, C, and D). Importantly, the inhibitory effect of this antiserum was overcome when a large amount of the respective CRM1/exportin 1 peptide antigen was coinjected to saturate the anti-CRM1 antiserum (Fig. 4B). These data demonstrated that the observed block in Rev-NES-mediated nuclear export (Fig. 4A) indeed is due to inhibition of CRM1/exportin 1 by the microinjected antibody. Of note, nuclear export activities of all of our GST-NES fusion proteins were not affected in control experiments by comicroinjection of unrelated antibodies (not shown).
Figure 4.
Antibodies directed against CRM1/exportin 1 are inhibitors of nuclear export. The nuclei of HeLa cells were microinjected with the indicated fusion proteins together with anti-CRM1 antiserum. Cells were incubated for 20 min, fixed, and analyzed by double-label, indirect immunofluorescence microscopy as described above. The GST-fusion proteins were visualized by using a mouse monoclonal anti-GST antibody followed by Cy3-conjugated goat anti-mouse IgG (GST). The microinjected rabbit anti-CRM1 antiserum was detected with a Cy2-conjugated goat anti-rabbit antibody (IgG). Inhibition of GST-Rev-NES export by the anti-CRM1 antiserum could be released when the respective antigen was coinjected (B).
Antibodies to eIF-5A and eIF-5A-M14 Protein Are Specific Inhibitors of Export of Rev/Rex-Derived NESs.
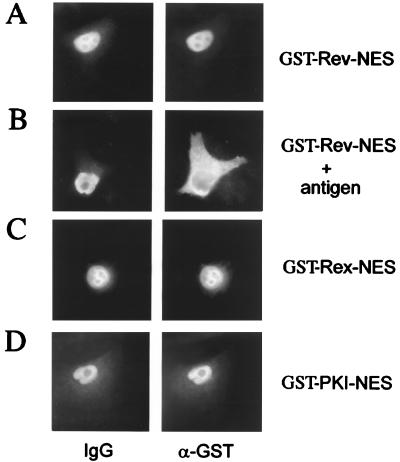
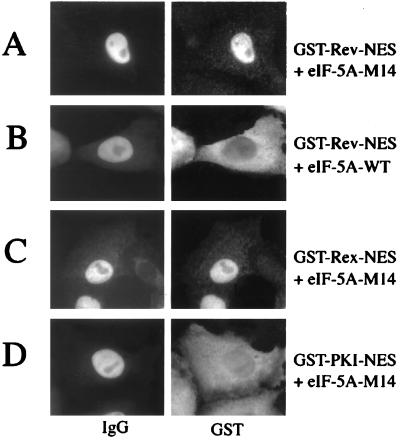
We have shown previously that the interaction of eIF-5A protein with Rev functionally mediates the Rev-dependent transport of unspliced or incompletely spliced HIV-1 mRNAs across the nuclear pore (16). Furthermore, we reported that mutants of eIF-5A (17) or antibodies raised against eIF-5A (26) specifically block the nucleocytoplasmic translocation of full-length HIV-1 Rev protein in somatic cells. To extend those studies, we tested these anti-eIF-5A antibodies for their ability to interfere with Rev-NES-, Rex-NES-, or PKI-NES-mediated nuclear export. As shown in Fig. 5, anti-eIF-5A antibodies effectively blocked the nuclear export of GST-Rev-NES and GST-Rex-NES (Fig. 5 A and B). Strikingly, the nuclear export of GST-PKI-NES was not affected by anti-eIF-5A antibodies in these microinjection experiments (Fig. 5C).
Figure 5.
Antibodies directed against eIF-5A block specifically the nuclear export of HIV-1 Rev and HTLV-I Rex NES. Comicroinjection of HeLa nuclei with the indicated export substrates and anti-eIF-5A antibodies (α-eIF-5A) (26). Cell were incubated for 20 min, fixed, and subjected to double-label, indirect immunofluorescence analysis. The GST-fusion proteins were visualized by using a mouse monoclonal anti-GST antibody followed by Cy3-conjugated goat anti-mouse IgG (GST). The microinjected rabbit antibodies directed against eIF-5A were detected with a Cy2-conjugated goat anti-rabbit antibody (IgG).
We next extended these antibody-inhibition studies by including eIF-5A wild-type and mutant protein (eIF-5A-M14). The eIF-5A-M14 protein previously has been shown to block the nuclear export of full-length GST-Rev protein and, hence, HIV-1 replication in trans (17, 25). In agreement with the previous antibody inhibition experiments and as shown in Fig. 6, comicroinjection of native eIF-5A-M14 protein specifically inhibited nuclear export of GST-Rev-NES and GST-Rex-NES, but not of GST-PKI-NES in somatic cells. Control experiments confirmed that the eIF-5A wt protein did not block nucleocytoplasmic translocation of GST-Rev-NES (Fig. 6B).
Figure 6.
An eIF-5A mutant protein blocks nuclear export mediated by HIV-1 Rev or HTLV-I Rex NES. HeLa cell nuclei were comicroinjected with the indicated GST-fusion proteins, rabbit IgG (injection control), and eIF-5A wild-type (B) or eIF-5A-M14 mutant protein (A, C, and D). Cells were incubated for 20 min, fixed, and subjected to double-label, indirect immunofluorescence analysis as described above.
Taken together, these data demonstrated that eIF-5A plays an essential role in the nuclear export pathway that is exploited particularly by the HIV-1 Rev and HTLV-I Rex trans-activator proteins.
DISCUSSION
The molecular export of macromolecules from the nucleus to the cytoplasm plays a fundamental role in orchestrating cellular activities. There is an increasing number of viral and cellular proteins with identified NESs. Because most of these proteins exert completely different biological functions, it is to be expected that their nuclear export is regulated specifically to avoid undesirable effects on the cell. In this report we demonstrate by microinjection experiments that different, specific nuclear export pathways exist for leucine-rich, NES-containing proteins, represented by the viral RNA export factors HIV-1 Rev and HTLV-I Rex, as opposed to the export pathway that is accessed by PKI.
In a first set of experiments we show (Fig. 2) that different NESs translocate the heterologous GST reporter protein from the nucleus into the cytoplasm of somatic cells. Inhibition experiments using leptomycin B, an inhibitor of CRM1/exportin 1 (19–21, 36, 37), and antibodies directed against CRM1/exportin 1 demonstrated that CRM1/exportin 1 is required for the nuclear export of Rev/Rex- and PKI-derived, leucine-rich NESs (Figs. 3 and 4). From previous studies (19–21, 38–40) and our own data presented here, it is reasonable to assume that CRM1/exportin 1 is a general export receptor for leucine-rich NESs. However, it is also conceivable that different nuclear export routes for proteins exist for accessing CRM1/exportin 1 for subsequent CRM1-mediated translocation through the NPC. Because it was shown previously that eIF-5A functionally interacts with the Rev- and Rex-NES (16, 41), we next assessed the effects of antibodies directed against eIF-5A on GST-Rev-NES, GST-Rex-NES, and GST-PKI-NES nuclear export. The results shown in Fig. 5 demonstrated that anti-eIF-5A antibodies specifically block the export of the Rev- and Rex-NES. In addition, a recently published eIF-5A mutant, eIF-5A-M14, which has been shown to effectively inhibit Rev nuclear export and HIV-1 replication (17, 25), displayed the same inhibitory phenotype on nucleocytoplasmic translocation of GST-Rev-NES and GST-Rex-NES, but not on GST-PKI-NES (Fig. 6), confirming the data collected with the anti-eIF-5A antibodies.
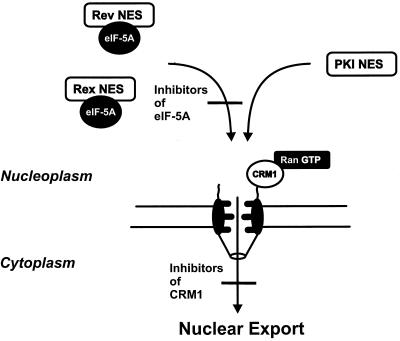
Considering the described cellular functions of proteins that were used to derive the NESs investigated in this study, it is obvious that HIV-1 Rev and HTLV-I Rex are involved in the nucleocytoplasmic transport of viral mRNAs (15, 42), whereas PKI binds and exports the catalytic subunit of protein kinase A from the nucleus and, therefore, has no role in RNA export (43). It therefore is reasonable to suggest different nuclear export pathways for leucine-rich, NES-containing proteins. The data shown in this study demonstrated that eIF-5A is specifically required for the nuclear export of the viral Rev/Rex RNA transport factors. These data also suggest that eIF-5A may act upstream of the general export receptor CRM1/exportin 1. Alternatively, it is also conceivable that eIF-5A may participate in a cooperative interaction of the respective NES and CRM1/exportin 1 or that eIF-5A binds to an already preformed NES-CRM1/exportin 1 complex. As depicted in Fig. 7, eIF-5A may target the Rev/Rex-NES-containing export cargo to the NPC for subsequent CRM1/exportin 1 interaction. In fact, several lines of evidence exist in favor of this hypothesis. Recently, it was demonstrated that eIF-5A accumulates at the nucleoplasmic surface of NPCs, which is assumed to act as the docking site for export substrates, and interacts with CRM1/exportin 1 (O.R. and J.H., unpublished results). CRM1/exportin 1 by itself has been shown to localize in the nucleus and at the NPC (44), where it associates with the nucleoporin Nup214/CAN (45). The necessity to localize particularly at the NPC has been demonstrated for the activity of CRM1/exportin 1 as a nuclear export factor, because displacement of CRM1/exportin 1 from the NPC by expression of a Nup214/CAN mutant (44, 46) also inhibits nuclear export and, thereby, function of HIV-1 Rev and HTLV-I Rex (47, 48). Thus, it is conceivable that eIF-5A may be active in the intranuclear transport of Rev/Rex-bound viral mRNAs from specific subnuclear compartments to the NPC. Of note, current evidence suggests that active gene transcription and RNA processing take place at distinct sites within the nucleus, which appear to be clearly separated from the nuclear envelope (for a recent review see ref. 49). Whether eIF-5A indeed targets Rev and Rex to the NPC will be investigated in future studies. Nevertheless, eIF-5A is clearly dispensable for PKI-NES-mediated nuclear export (Figs. 5 and 6). Thus, the PKI NES may interact in the cell nucleus directly with CRM1/exportin 1 or via an as yet uncharacterized adapter molecule. In fact, a PKI-NES-binding factor distinct from CRM1/exportin 1 was reported recently (50). In every case, however, CRM1/exportin 1 is considered to act as general export receptor that mediates, together with RanGTP (21, 51), the translocation of all leucine-rich NESs across the nuclear envelope (Fig. 7). This notion is in perfect agreement with the data shown above and also with all previous studies in which the effect of leptomycin B on nucleocytoplasmic translocation was investigated. In fact, inhibition of CRM1/exportin 1 by leptomycin B always resulted in perfect inhibition of nuclear export, irrespective of the origin of the investigated leucine-rich NES (19–21, 38–40, 52). This observation also implies that inhibition of CRM1/exportin 1 activity leads to a general block in nuclear export, which, in turn, then should result in toxic side-effects on cellular metabolism. This is indeed the case, because cellular toxicity has been observed in tissue cultures that were exposed for extended periods of time to leptomycin B (37). Furthermore, inhibition of CRM1/exportin 1 by overexpression of a Nup214/CAN mutant has been reported recently to be toxic to the cell by inhibiting cell growth and inducing apoptosis (53).
Figure 7.
Multiple cellular factors are involved in nuclear export. eIF-5A is required for nuclear export mediated by the leucine-rich NES of the HIV-1 Rev and HTLV-I Rex trans-activator proteins, but not for nucleocytoplasmic translocation mediated by the PKI NES. CRM1/exportin 1 is a general export receptor that mediates together with RanGTP the translocation of all export cargoes containing leucine-rich NESs through the NPC. The data presented in this work suggest that eIF-5A either acts before CRM1/exportin 1 in the nuclear export of specific export substrates or, alternatively, forms a complex with the respective NES and CRM1/exportin 1 (for details, see text).
Taken together, our data provide evidence that nuclear export is orchestrated by the concerted action of multiple cellular proteins. The existing data indicate that CRM1/exportin 1 acts as a general nuclear export receptor for all leucine-rich NESs whereas, in contrast, eIF-5A acts before or simultaneously with CRM1/exportin 1 in nuclear export of the retroviral Rev and Rex RNA transport factors. It remains to be seen whether eIF-5A also plays a role in the nuclear export of other viral and cellular proteins and whether additional cellular proteins are involved in nucleocytoplasmic transport of other leucine-rich, NES-containing export substrates.
Acknowledgments
We thank Dr. Larry Gerace (The Scripps Research Institute) for the kind gift of anti-CRM1 antibodies and Dr. Ivan Lindley for his comments on the manuscript. This work was supported, in part (O.R. and J.H.), by the Deutsche Forschungsgemeinschaft (SFB466).
ABBREVIATIONS
- GST
glutathione S-transferase
- NLS
nuclear localization signal
- NES
nuclear export signal
- NPC
nuclear pore complex
- PKI
protein kinase inhibitor
- eIF-5A
eukaryotic initiation factor 5A
Footnotes
This paper was submitted directly (Track II) to the Proceedings Office.
References
- 1.Nigg E A. Nature (London) 1997;386:779–787. doi: 10.1038/386779a0. [DOI] [PubMed] [Google Scholar]
- 2.Corbett A H, Silver P A. Microbiol Mol Biol Rev. 1997;61:193–211. doi: 10.1128/mmbr.61.2.193-211.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ohno M, Fornerod M, Mattaj I W. Cell. 1998;92:327–336. doi: 10.1016/s0092-8674(00)80926-5. [DOI] [PubMed] [Google Scholar]
- 4.Görlich D. EMBO J. 1998;17:2721–2727. doi: 10.1093/emboj/17.10.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Melchior F, Gerace L. Trends Cell Biol. 1998;8:175–179. doi: 10.1016/s0962-8924(98)01252-5. [DOI] [PubMed] [Google Scholar]
- 6.Dingwall C, Laskey R A. Trends Biochem Sci. 1991;16:478–481. doi: 10.1016/0968-0004(91)90184-w. [DOI] [PubMed] [Google Scholar]
- 7.Boulikas T. Crit Rev Eukaryotic Gene Expression. 1993;3:193–227. [PubMed] [Google Scholar]
- 8.Gerace L. Cell. 1995;82:341–344. doi: 10.1016/0092-8674(95)90420-4. [DOI] [PubMed] [Google Scholar]
- 9.Izaurralde E, Mattaj I W. Cell. 1995;81:153–159. doi: 10.1016/0092-8674(95)90323-2. [DOI] [PubMed] [Google Scholar]
- 10.Nakielny S, Fischer U, Michael W M, Dreyfuss G. Annu Rev Neurosci. 1997;20:269–301. doi: 10.1146/annurev.neuro.20.1.269. [DOI] [PubMed] [Google Scholar]
- 11.Fischer U, Huber J, Boelens W C, Mattaj I W, Lührmann R. Cell. 1995;82:475–483. doi: 10.1016/0092-8674(95)90436-0. [DOI] [PubMed] [Google Scholar]
- 12.Wen W, Meinkoth J L, Tsien R Y, Taylor S S. Cell. 1995;82:463–473. doi: 10.1016/0092-8674(95)90435-2. [DOI] [PubMed] [Google Scholar]
- 13.Palmeri D, Malim M H. J Virol. 1996;70:6442–6445. doi: 10.1128/jvi.70.9.6442-6445.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fridell R A, Bogerd H P, Cullen B R. Proc Natl Acad Sci USA. 1996;93:4421–4424. doi: 10.1073/pnas.93.9.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pollard V W, Malim M H. Annu Rev Microbiol. 1998;52:491–532. doi: 10.1146/annurev.micro.52.1.491. [DOI] [PubMed] [Google Scholar]
- 16.Ruhl M, Himmelspach M, Bahr G M, Hammerschmid F, Jaksche H, Wolff B, Aschauer H, Farrington G K, Probst H, Bevec D, et al. J Cell Biol. 1993;123:1309–1320. doi: 10.1083/jcb.123.6.1309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bevec D, Jaksche H, Oft M, Wöhl T, Himmelspach M, Pacher A, Schebesta M, Koettnitz K, Dobrovnik M, Csonga R, et al. Science (Washington, DC) 1996;271:1858–1860. doi: 10.1126/science.271.5257.1858. [DOI] [PubMed] [Google Scholar]
- 18.Stade K, Ford C S, Guthrie C, Weis K. Cell. 1997;90:1041–1050. doi: 10.1016/s0092-8674(00)80370-0. [DOI] [PubMed] [Google Scholar]
- 19.Fornerod M, Ohno M, Yoshida M, Mattaj I W. Cell. 1997;90:1051–1060. doi: 10.1016/s0092-8674(00)80371-2. [DOI] [PubMed] [Google Scholar]
- 20.Ossareh-Nazari B, Bachelerie F, Dargemont C. Science (Washington, DC) 1997;278:141–144. doi: 10.1126/science.278.5335.141. [DOI] [PubMed] [Google Scholar]
- 21.Fukuda M, Asano S, Nakamura T, Adachi M, Yoshida M, Yanagida M, Nishida E. Nature (London) 1997;390:308–311. doi: 10.1038/36894. [DOI] [PubMed] [Google Scholar]
- 22.Bogerd H P, Fridell R A, Madore S, Cullen B R. Cell. 1995;82:485–494. doi: 10.1016/0092-8674(95)90437-9. [DOI] [PubMed] [Google Scholar]
- 23.Fritz C C, Zapp M L, Green M R. Nature (London) 1995;376:530–533. doi: 10.1038/376530a0. [DOI] [PubMed] [Google Scholar]
- 24.Neville M, Stutz F, Lee L, Davies L I, Rosbash M. Curr Biol. 1997;7:767–775. doi: 10.1016/s0960-9822(06)00335-6. [DOI] [PubMed] [Google Scholar]
- 25.Junker U, Bevec D, Barske C, Kalfoglou C, Escaich S, Dobrovnik M, Hauber J, Böhnlein E. Hum Gene Ther. 1996;7:1861–1869. doi: 10.1089/hum.1996.7.15-1861. [DOI] [PubMed] [Google Scholar]
- 26.Schatz O, Oft M, Dascher C, Schebesta M, Rosorius O, Jaksche H, Dobrovnik M, Bevec D, Hauber J. Proc Natl Acad Sci USA. 1998;95:1607–1612. doi: 10.1073/pnas.95.4.1607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradford M M. Anal Biochem. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 28.Zimmermann K, Weber S, Dobrovnik M, Hauber J, Böhnlein E. Hum Gene Ther. 1992;3:155–161. doi: 10.1089/hum.1992.3.2-155. [DOI] [PubMed] [Google Scholar]
- 29.Bogerd H P, Fridell R A, Benson R E, Hua J, Cullen B R. Mol Cell Biol. 1996;16:4207–4214. doi: 10.1128/mcb.16.8.4207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang M J, Dayton A I. Biochem Biophys Res Commun. 1998;243:113–116. doi: 10.1006/bbrc.1997.8070. [DOI] [PubMed] [Google Scholar]
- 31.Yang J, Bardes E S G, Moore J D, Brennan J, Powers M A, Kornbluth S. Genes Dev. 1998;12:2131–2143. doi: 10.1101/gad.12.14.2131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Malim M H, Böhnlein S, Hauber J, Cullen B R. Cell. 1989;58:205–214. doi: 10.1016/0092-8674(89)90416-9. [DOI] [PubMed] [Google Scholar]
- 33.Malim M H, Freimuth W W, Liu J, Boyle T J, Lyerly H K, Cullen B R, Nabel G J. J Exp Med. 1992;176:1197–1201. doi: 10.1084/jem.176.4.1197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bevec D, Dobrovnik M, Hauber J, Böhnlein E. Proc Natl Acad Sci USA. 1992;89:9870–9874. doi: 10.1073/pnas.89.20.9870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meyer B E, Malim M H. Genes Dev. 1994;8:1538–1547. doi: 10.1101/gad.8.13.1538. [DOI] [PubMed] [Google Scholar]
- 36.Kudo N, Wolff B, Sekimoto T, Schreiner E P, Yoneda Y, Yanagida M, Horinouchi S, Yoshida M. Exp Cell Res. 1998;242:540–547. doi: 10.1006/excr.1998.4136. [DOI] [PubMed] [Google Scholar]
- 37.Wolff B, Sanglier J J, Wang Y. Chem Biol. 1997;4:139–147. doi: 10.1016/s1074-5521(97)90257-x. [DOI] [PubMed] [Google Scholar]
- 38.Engel K, Kotlyarov A, Gaestel M. EMBO J. 1998;17:3363–3371. doi: 10.1093/emboj/17.12.3363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wada A, Fukuda M, Mishima M, Nishida E. EMBO J. 1998;17:1635–1641. doi: 10.1093/emboj/17.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Freedman D A, Levine A J. Mol Cell Biol. 1998;18:7288–7293. doi: 10.1128/mcb.18.12.7288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Katahira J, Ishizaki T, Sakai H, Adachi A, Yamamoto K, Shida H. J Virol. 1995;69:3125–3133. doi: 10.1128/jvi.69.5.3125-3133.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gitlin S D, Dittmer J, Reid R L, Brady J N. In: Human Retroviruses. Cullen B R, editor. Oxford: IRL; 1993. pp. 159–192. [Google Scholar]
- 43.Wen W, Harootunian A T, Adams S R, Feramisco J, Tsien R Y, Meinkoth J L, Taylor S S. J Biol Chem. 1994;269:32214–32220. [PubMed] [Google Scholar]
- 44.Fornerod M, van Deursen J, van Baal S, Reynolds A, Davis D, Murti K G, Fransen J, Grosveld G. EMBO J. 1997;16:807–816. doi: 10.1093/emboj/16.4.807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fornerod M, Boer J, van Baal S, Morreau H, Grosveld G. Oncogene. 1996;13:1801–1808. [PubMed] [Google Scholar]
- 46.Fornerod M, Boer J, van Baal S, Jaegle M, von Lindern M, Murti K G, Davis D, Bonten J, Buijs A, Grosveld G. Oncogene. 1995;10:1739–1748. [PubMed] [Google Scholar]
- 47.Bogerd H P, Echarri A, Ross T M, Cullen B R. J Virol. 1998;72:8627–8635. doi: 10.1128/jvi.72.11.8627-8635.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zolotukhin A S, Felber B K. J Virol. 1999;73:120–127. doi: 10.1128/jvi.73.1.120-127.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Lamond A I, Earnshaw W C. Science (Washington, DC) 1998;280:547–553. doi: 10.1126/science.280.5363.547. [DOI] [PubMed] [Google Scholar]
- 50.Holaska J M, Paschal B M. Proc Natl Acad Sci USA. 1998;95:14739–14744. doi: 10.1073/pnas.95.25.14739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Askjaer P, Jensen T H, Nilsson J, Englmeier L, Kjems J. J Biol Chem. 1998;273:33414–33422. doi: 10.1074/jbc.273.50.33414. [DOI] [PubMed] [Google Scholar]
- 52.Toyoshima F, Moriguchi T, Wada A, Fukuda M, Nishida E. EMBO J. 1998;17:2728–2735. doi: 10.1093/emboj/17.10.2728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Boer J, Bonten-Surtel J, Grosveld G. Mol Cell Biol. 1998;18:1236–1247. doi: 10.1128/mcb.18.3.1236. [DOI] [PMC free article] [PubMed] [Google Scholar]