Abstract
Munc18-1 and soluble NSF attachment protein receptors (SNAREs) are critical for synaptic vesicle fusion. Munc18-1 binds to the SNARE syntaxin-1 folded into a closed conformation and to SNARE complexes containing open syntaxin-1. Understanding which steps in fusion depend on the latter interaction and whether Munc18-1 competes with other factors such as complexins for SNARE complex binding is critical to elucidate the mechanisms involved. In this study, we show that lentiviral expression of Munc18-1 rescues abrogation of release in Munc18-1 knockout mice. We describe point mutations in Munc18-1 that preserve tight binding to closed syntaxin-1 but markedly disrupt Munc18-1 binding to SNARE complexes containing open syntaxin-1. Lentiviral rescue experiments reveal that such disruption selectively impairs synaptic vesicle priming but not Ca2+-triggered fusion of primed vesicles. We also find that Munc18-1 and complexin-1 bind simultaneously to SNARE complexes. These results suggest that Munc18-1 binding to SNARE complexes mediates synaptic vesicle priming and that the resulting primed state involves a Munc18-1–SNARE–complexin macromolecular assembly that is poised for Ca2+ triggering of fusion.
Introduction
The release of neurotransmitters by Ca2+-triggered synaptic vesicle exocytosis is a key event in interneuronal communication. Release involves a series of steps that include vesicle docking to the plasma membrane, priming to a release-ready state, and Ca2+-triggered membrane fusion (Südhof, 2004). The protein machinery that governs these steps contains components that have homologues in most types of intracellular membrane traffic and are thus believed to underlie a conserved mechanism of membrane fusion. Particularly important for fusion are proteins from the Sec1/Munc18 (SM) and SNARE families, which, in neuronal synapses, are represented by Munc18-1 and the SNAREs syntaxin-1, SNAP-25, and synaptobrevin/vesicle-associated membrane protein (Rizo and Südhof, 2002; Toonen and Verhage, 2003; Jahn and Scheller, 2006). The SNAREs function by forming tight four-helix bundles called SNARE complexes through sequences known as SNARE motifs (Sollner et al., 1993; Poirier et al., 1998; Sutton et al., 1998); assembly of these complexes brings the two membranes together (Hanson et al., 1997) and is key for membrane fusion (Brunger, 2005; Rizo and Rosenmund, 2008). The function of SM proteins is less clear.
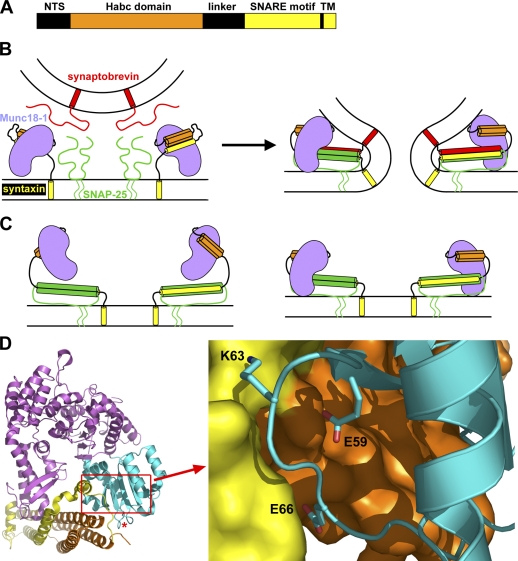
Munc18-1 was identified and linked to synaptic vesicle fusion by virtue of its tight binding to syntaxin-1 (Hata et al., 1993). In addition to a SNARE motif preceding a C-terminal transmembrane region, syntaxin-1 contains a flexible N-terminal sequence, a three-helix bundle called the Habc domain, and a flexible linker (Fig. 1 A; Fernandez et al., 1998). The Habc domain and SNARE motif bind intramolecularly, forming a “closed conformation” that is crucial for tight binding to Munc18-1 (Fig. 1 B; Dulubova et al., 1999; Misura et al., 2000). Sso1p, the yeast plasma membrane syntaxin, also adopts a closed conformation (Nicholson et al., 1998; Fiebig et al., 1999), but this feature is not generally conserved in syntaxins (Dulubova et al., 2001, 2002), and the yeast SM protein Sec1p binds to assembled SNARE complexes rather than to isolated Sso1p (Carr et al., 1999). Moreover, the syntaxins from the ER, Golgi, TGN, and early endosomes of yeast and mammals bind tightly to their cognate SM proteins through the short N-terminal sequence (Bracher and Weissenhorn, 2002; Dulubova et al., 2002, 2003; Yamaguchi et al., 2002). The apparently diverging picture emerging from these early findings was partially unified by evidence showing that SM proteins generally bind to their cognate SNARE complexes and that most of these interactions involve syntaxin N-terminal sequences (Peng and Gallwitz, 2002; Carpp et al., 2006; Latham et al., 2006; Stroupe et al., 2006; Togneri et al., 2006; Dulubova et al., 2007; Shen et al., 2007). These data suggest that Munc18-1 interacts with the SNAREs in at least two different modes: a binary interaction with the syntaxin-1 closed conformation that is not universal and likely meets specific requirements of regulated exocytosis (Gerber et al., 2008) and a multifaceted interaction with the SNARE complex that likely underlies the general function of SM proteins (Fig. 1 B; Dulubova et al., 2007; Shen et al., 2007).
Figure 1.
Design of mutations to disrupt Munc18-1–SNARE interactions. (A) Domain diagram of syntaxin-1. NTS, N-terminal sequence; TM, transmembrane region. (B) Diagrams of the binary complex between Munc18-1 and closed syntaxin-1 (left) and the Munc18-1–SNARE complex assembly (right). Munc18-1 is purple, synaptobrevin is red, SNAP-25 is green, and syntaxin-1 is orange (Habc domain) and yellow (SNARE motif and transmembrane region). The model of the binary complex is based on its crystal structure (Misura et al., 2000; Burkhardt et al., 2008). The model of the Munc18-1–SNARE complex assemblies is based on NMR data suggesting a multifaceted interaction and illustrates the overall notion that these assemblies are critical for membrane fusion (Dulubova et al., 2007; and for a concrete physical model of how these assemblies can induce membrane fusion, see Rizo et al. [2006]). (C) Models of potential interactions between Munc18-1 and open syntaxin-1 within syntaxin-1–SNAP-25 heterodimers. The model on the left is based on the finding that Munc18-1 can bind to the isolated syntaxin-1 N-terminal region (Khvotchev et al., 2007; Burkhardt et al., 2008), whereas the model on the right also incorporates interactions with the SNARE motifs (Weninger et al., 2008). (D) Ribbon diagram of the binary Munc18-1–syntaxin-1 complex with Munc18-1 colored in purple except for the N-terminal domain, which is in cyan, and syntaxin-1, which is in orange (Habc domain) and yellow (linker and SNARE motif). The red asterisk indicates the position where cerulean was inserted for the rescue experiments. A close-up of the interface showing the mutated residues is shown on the right. The diagrams were prepared with Pymol (DeLano Scientific).
The physiological relevance of the binary Munc18-1–syntaxin-1 complex was suggested by diverse evidence (Wu et al., 1998; Verhage et al., 2000; Rizo and Südhof, 2002) and has been demonstrated through analysis of knockin mice bearing an LE (L165A, E166A) mutation that destabilizes the syntaxin-1 closed conformation and impairs Munc18-1 binding, leading to an increase in vesicle release probability (Gerber et al., 2008). This phenotype likely arises because the LE mutation facilitates SNARE complex formation, as the SNARE complex is incompatible with the closed conformation (Dulubova et al., 1999; Misura et al., 2000), and Munc18-1 binding stabilizes this conformation (Chen et al., 2008), thus hindering SNARE complex assembly (Yang et al., 2000).
Munc18-1–SNARE complex assemblies were proposed to form the core of the membrane fusion machinery (Fig. 1 B; Dulubova et al., 2007), and their importance for exocytosis was supported by transfection assays and peptide injection experiments in the calyx of Held synapse (Khvotchev et al., 2007). Moreover, reconstitution assays suggested that Munc18-1 stimulates the rate of lipid mixing between SNARE proteoliposomes, interacting with both syntaxin-1 and synaptobrevin (Shen et al., 2007; Rodkey et al., 2008). However, a recent study concluded that Munc18-1–SNARE complex binding is mediated only by the syntaxin-1 Habc domain and N-terminal sequence (Burkhardt et al., 2008), and Munc18-1 was shown to also bind to syntaxin-1–SNAP-25 heterodimers (Guan et al., 2008; Weninger et al., 2008). Although the results of these latter studies do not truly contradict the model of Fig. 1 B, they do suggest that Munc18-1 can interact with open syntaxin-1 within more than one type of complex (Fig. 1, B and C) and emphasize that we are still far from understanding how these interactions control release.
To reach such an understanding, it is crucial to determine which of the steps leading to release depends on the binding of Munc18-1 to open syntaxin-1. Moreover, because several components of the release machinery also bind to SNARE complexes (Südhof, 2004; Brunger, 2005), elucidating whether such interactions are compatible with Munc18-1 binding is critical to dissect the order of the molecular events leading to release. Particularly important in this context is to unravel whether Munc18-1 competes for SNARE complex binding with complexins because it is well established that these small soluble proteins function in the Ca2+-triggering step of release (Reim et al., 2001; Tang et al., 2006), and they are generally believed to be bound to the SNARE complex before Ca2+ influx (Rizo and Rosenmund, 2008). In this study, we have addressed these questions using a combination of biophysical experiments and a Munc18-1 knockout (KO) rescue approach. This approach has been hindered because Munc18-1–deficient neurons die early (Verhage et al., 2000) and are thus difficult to analyze. We have overcome this problem through lentiviral expression of Munc18-1 in neurons from Munc18-1 KO mice. We have designed mutations in Munc18-1 that impair binding to SNARE complexes containing open syntaxin-1 while still retaining tight binding to closed syntaxin-1 and the ability to rescue survival in Munc18-1 KO neurons. Importantly, these mutations cause a selective disruption of synaptic vesicle priming without altering the efficiency of release of primed vesicles. Moreover, we show that Munc18-1 and complexin-1 bind simultaneously to the SNARE complex. Our data show that interactions of Munc18-1 with open syntaxin-1 are critical for priming vesicles to a release-ready state that likely involves macromolecular assemblies comprising Munc18-1, SNAREs, and complexins.
Results
Design of mutations to distinguish Munc18-1–SNARE interactions
The x-ray structure of the syntaxin-1–Munc18-1 complex revealed that Munc18-1 has an arch shape with a cavity where the syntaxin-1 closed conformation binds (Fig. 1 D; Misura et al., 2000). The N-terminal domain of Munc18-1 (Fig. 1 D, cyan) plays a key role in the interaction, making extensive contacts with the Habc domain and the SNARE motif of syntaxin-1 (Fig. 1 D, orange and yellow, respectively). The syntaxin-1 N-terminal sequence was not observable in the initial crystal structure but also participates in binding (Khvotchev et al., 2007; Burkhardt et al., 2008). Although no high resolution structure of the Munc18-1–SNARE complex is available, nuclear magnetic resonance (NMR) data showed that formation of the complex involves the syntaxin-1 N-terminal sequence and Habc domain as well as the four-helix bundle formed by the SNARE motifs (Fig. 1 B, right; Dulubova et al., 2007). These observations suggest that the interactions of Munc18-1 with the syntaxin-1 C terminus must change drastically in the transition between the two complexes, whereas interactions with the N-terminal sequence and Habc domain may involve similar residues in both complexes. However, the energetic contributions of individual interactions to binding are likely to change during the transition between the two complexes because of the syntaxin-1 C-terminal rearrangements, particularly if the interactions involve residues near the interface between the Habc domain and SNARE motif in the closed conformation. Thus, replacing residues near this interface is likely to have differential disruptive effects on binding of Munc18-1 to syntaxin-1 or the SNARE complex. Based on these considerations, three residues of the Munc18-1 N-terminal domain that contact the syntaxin-1 Habc domain (E59), the SNARE motif (K63), or both (E66; Fig. 1 D) were selected for mutagenesis, and three point mutants of Munc18-1 bearing substitutions in one of these three residues were prepared. Two of the substitutions (E59K and K63E) were charge reversals to try to enhance their disruptive effects, whereas the third (E66A) only neutralized the charge to aim for more moderate effects.
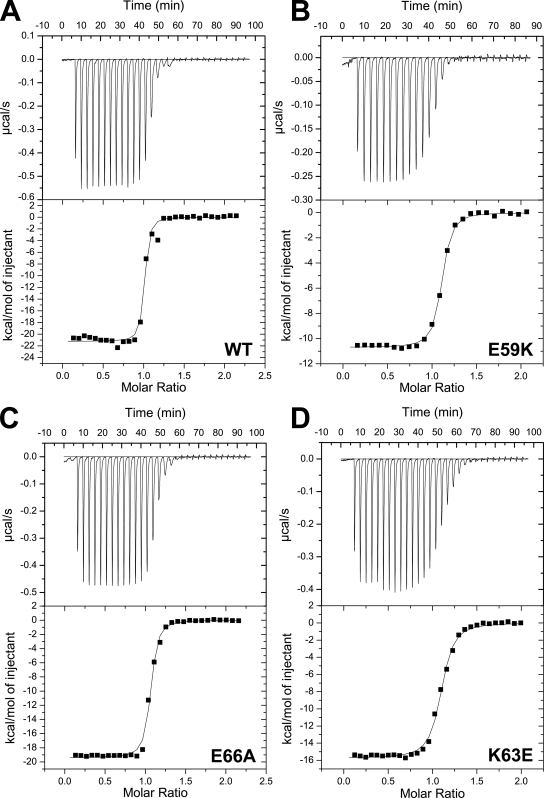
The effects of the three mutations on the binary Munc18-1–syntaxin-1 interaction were investigated by isothermal titration calorimetry (ITC; Fig. 2). For this purpose, we used a syntaxin-1 fragment encompassing residues 2–243, which include all of the sequences that make contact with Munc18-1 in the crystal structure of the binary complex (ITC experiments using a slightly longer fragment, syntaxin-1[2–253], yielded comparable but less consistent results because of the tendency of this fragment to oligomerize; Chen et al., 2008). Triplicate experiments with syntaxin-1(2–243) and the wild-type (WT) or mutant Munc18-1s yielded the following mean Kd and standard deviations: WT, 7.5 ± 2.7 nM; E59K, 12.0 ± 5.6 nM; K63E, 20.5 ± 11.7 nM; and E66A, 11.3 ± 4.1 nM. The Kd measured for WT Munc18-1 is comparable with the 10–20-nM Kd values measured previously by other methods (Pevsner et al., 1994; Khvotchev et al., 2007) and the 2.7-nM Kd reported in a recent ITC study (Burkhardt et al., 2008). The differences between these values can be attributed to differences in experimental conditions and/or the protein fragments used as well as to intrinsic difficulties in obtaining accurate measurements for such high affinities, which also underlie the relatively large standard deviations in our measurements. Despite these difficulties, it is clear from our ITC data that the three Munc18-1 mutants retain tight binding to syntaxin-1. Each of the mutations do appear to impair the binary interaction slightly, particularly the K63E mutation, but none of the differences between the Kd measured for WT Munc18-1 and the mutants is statistically significant.
Figure 2.
ITC analysis of binding of WT and mutant Munc18-1 to syntaxin-1. (A–D) Illustrative examples of the ITC data obtained for binding of WT Munc18-1 (A) and E59K (B), E66A (C), and K63E (D) Munc18-1 mutants to syntaxin-1(2–243) are shown. A polynomial baseline correction was applied to remove a slight drift in the initial points of each titration before fitting the data to a single-site binding model. This correction did not substantially alter the Kd values obtained.
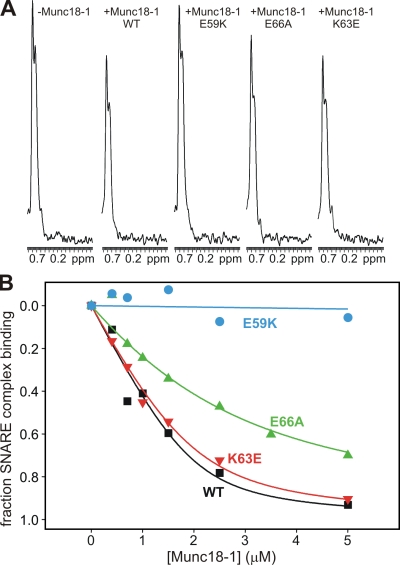
ITC experiments with WT Munc18-1 and the SNARE complex yielded very small binding enthalpies (Burkhardt et al., 2008; and unpublished data), which suggests that binding is entropically driven and hinders quantitative comparisons by ITC. To measure the effects of the mutations in Munc18-1 on its interaction with open syntaxin-1 within the SNARE complex, we turned to an NMR method that we used previously to demonstrate this interaction (Dulubova et al., 2007). The method is based on the observation of a decrease in the intensity of the strongest methyl resonance (SMR) in 1D 13C-edited 1H-NMR spectra of a 13C-labeled protein (or complex) upon binding to an unlabeled protein as a result of the broadening caused by formation of a larger species (Arac et al., 2003). To apply this method to study Munc18-1–SNARE complex binding, we prepared SNARE complexes containing 13C-labeled syntaxin-1 (hereafter referred to as 13C-labeled SNARE complex for simplicity) and added WT or mutant Munc18-1s. Addition of 2.5 µM WT Munc18-1 induced a moderate but reproducible decrease in the SMR intensity of 2 µM 13C-labeled SNARE complex (Fig. 3 A), which reflects formation of the Munc18-1–SNARE complex assembly. Interestingly, the K63E Munc18-1 mutant had a similar effect as WT Munc18-1, whereas the E59K mutant did not significantly decrease the SMR intensity of the 13C-labeled SNARE complex, and the E66A mutant had an intermediate effect (Fig. 3 A). Multiple titrations in which we measured the decrease in the SMR intensity of the 13C-labeled SNARE complex as a function of WT Munc18-1 concentration (Fig. 3 B) yielded a Kd of 266 ± 41 nM, which is consistent with the values of 100–300 nM that we estimated previously (Dulubova et al., 2007). Titrations with the Munc18-1 mutants yielded a Kd of 310 ± 82 nM for K63E, which is not significantly different from WT, and a Kd of 1.61 ± 0.35 µM for E66A, which reveals a considerable disruption of Munc18-1 binding to the SNARE complex. Titrations with the E59K mutant consistently showed that this mutation strongly impairs SNARE complex binding, although there appeared to be some residual binding at the higher concentrations (Fig. 3 B). Based on the sensitivity of the method, we estimate a Kd of >30 µM for this mutant. Thus, these results show that the three mutations in Munc18-1 have markedly different effects on SNARE complex binding and that two of them (E66A and E59K) disrupt this interaction much more strongly than the binary interaction with the syntaxin-1 closed conformation.
Figure 3.
Differential disruption of Munc18-1 binding to the SNARE complex by Munc18-1 point mutations. (A) Sample traces of the methyl regions of 1D 13C-edited 1H-NMR spectra of 2 µM SNARE complex containing uniformly 13C-labeled syntaxin-1(2–243) in the absence or presence of 2.5 µM of unlabeled WT or mutant Munc18-1s. ppm, parts per million. (B) Binding curves obtained from the SMR intensities observed in 1D 13C-edited 1H-NMR spectra of 2 µM SNARE complex containing uniformly 13C-labeled syntaxin-1(2–243) in the presence of increasing amounts of unlabeled WT or mutant Munc18-1s. The data were fit to a standard single-site binding model and normalized to the percentage of binding using as limit values the initial intensity in the absence of Munc18-1 (0% binding) and the intensity extrapolated to infinite Munc18-1 concentration (100% binding).
Rescue of survival and neurotransmitter release in Munc18-1 KO neurons
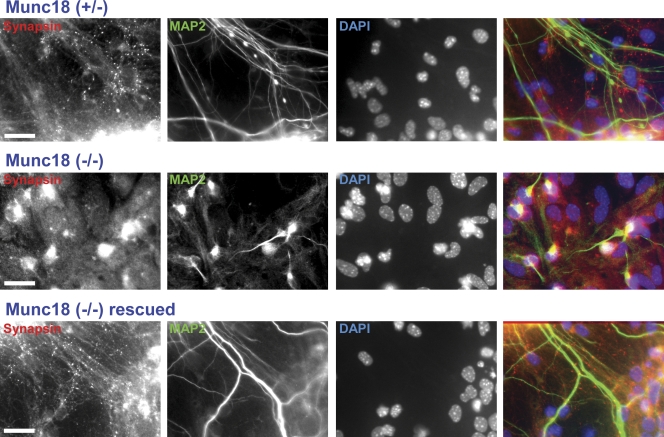
Munc18-1 KO mice die immediately at birth and exhibit a total abrogation of spontaneous, hypertonic sucrose-induced and Ca2+-triggered neurotransmitter release (Verhage et al., 2000). To explore whether we could rescue release in neurons from these mice by overexpression of WT Munc18-1, we used primary cortical cultures from mouse embryos at embryonic day (E) 16.5. During the first week in vitro, Munc18-1–deficient neurons exhibited apparently normal neurite outgrowth and synapse formation as judged by immunocytochemistry and electron microscopy (unpublished data). Subsequently, neurons from Munc18-1 KO mice degenerated rapidly, and cultures did not survive >10 d in vitro (DIV; Fig. 4). To overcome this problem, we used lentiviral expression of Munc18-1. In these experiments, we aimed to visualize the expressed Munc18-1 to make it easier to monitor the levels of WT and mutant Munc18-1. Thus, we tagged Munc18-1 with the cerulean variant of GFP. We initially explored a C-terminal cerulean fusion protein and three fusion proteins in which cerulean was inserted into loops of Munc18-1. The three loops were chosen in exposed surface locations of Munc18-1 that, based on the crystal structure of the Munc18-1–syntaxin-1 complex (Misura et al., 2000), were close to syntaxin-1 (for future fluorescence resonance energy transfer studies in vivo) and were predicted to be able to harbor the insertion of cerulean without disrupting folding and/or binding. Although no systematic experiments were performed, preliminary experiments indicated that the insertion of cerulean between residues 24 and 25 (Fig. 1 D, red asterisk) allowed efficient rescue of the survival and the neurotransmitter release phenotypes in Munc18-1–deficient neurons (Fig. S1, A and B, available at http://www.jcb.org/cgi/content/full/jcb.200812026/DC1) and that the rescue was better than that observed with the other fusion proteins. Therefore, we performed all of our functional experiments with this cerulean fusion protein of Munc18-1.
Figure 4.
Rescue of neuronal survival in cortical cultures by Munc18-1 expression. Representative images of cortical synapses from littermate heterozygotes (top), homozygote KOs for Munc18-1 (middle), or Munc18-1 KOs infected with Munc18-1-containing lentivirus (bottom). Cells were maintained in culture for 11 d before being labeled with antibodies against the presynaptic marker synapsin (first column), the neurofilament marker MAP2 (second column), and the nuclear DAPI marker (third column). The last column shows the combined image of the three labeling procedures with colors that match the relevant labels in the other columns. Bars, 20 µm.
We infected cultured Munc18-1–deficient neurons at 1 DIV with lentiviruses expressing WT or E59K, E66A, or K63E mutant Munc18-1 that was fused to cerulean and monitored the survival and morphology of the neurons and expression levels using fluorescence microscopy at 11 DIV. All Munc18-1 proteins rescued neuronal survival, and no significant difference was observed between the number of synapses in WT neurons and in neurons rescued with WT or mutant Munc18-1 proteins (Fig. S1 C). Similar expression levels were observed for the WT and mutant Munc18-1 proteins as monitored from cerulean fluorescence intensities (Fig. S2 A, available at http://www.jcb.org/cgi/content/full/jcb.200812026/DC1), although Western blots indicated lower levels of E59K mutant (25–37% compared with WT; Fig. S2 B). The reason for this discrepancy is unclear, but these data suggest that the electrophysiological results described below for this mutant need to be interpreted with caution.
Next, we used patch-clamp recordings to test whether the cerulean-tagged WT Munc18-1 rescues neurotransmitter release in Munc18-1 KO neurons. As expected, no evoked or spontaneous activity was observed in untreated KO cultures at 6–7 DIV, when there are still some surviving neurons (unpublished data), which confirmed previous findings (Verhage et al., 2000). Lentiviral expression of WT Munc18-1 rescued spontaneous neurotransmitter release (“minis”) and also restored release evoked by field stimulation at low frequency (0.4 Hz), as monitored by recordings at 12–18 DIV (Fig. 5). Similarly, release stimulated at higher frequencies (10 Hz; Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200812026/DC1) or by hyperosmotic sucrose (Fig. 6) was rescued by WT Munc18-1. These results show that lentiviral expression of WT Munc18-1 is efficient enough to confer WT electrophysiological responses on Munc18-1–deficient neurons.
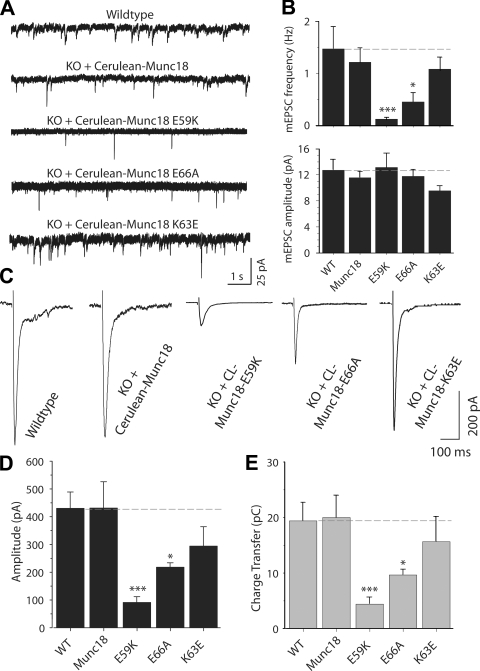
Figure 5.
Synaptic release depends on Munc18-1 binding to the SNARE complex. (A) Analysis of spontaneous synaptic release upon rescue with WT and mutant Munc18-1s. Representative 10-s segments from 10-min-long traces of spontaneous excitatory synaptic activity, which was recorded at a holding potential of −70 mV in the presence of 1 µM tetrodotoxin and 50 µM picrotoxin. (B) Bar diagram describing the frequency (top) and amplitude (bottom) of spontaneous release (WT, n = 13; Munc18, n = 19; E59K, n = 10; K63E and E66A, n = 9). (C) Representative traces of field stimulation (at 0.4 Hz) evoked excitatory responses from neurons of WT (n = 14) or Munc18-1 KO infected with WT (n = 9), E59K (n = 7), E66A (n = 16), or K63E (n = 11) Munc18-1–24-cerulean. Note that only the first 400 ms of the traces are shown for clarity. (D) Bar diagram summarizing the amplitudes of evoked responses for cultures rescued with the WT Munc18-1 and different Munc18-1 mutants. (E) Synaptic responses characterized as the amount of transferred charge. Asterisks in the bar diagrams mark statistical significance of the difference between the WT and mutant rescues (*, P < 0.05; ***, P < 0.005). (B, D, and E) Data are shown as means ± SEMs. Dashed lines indicate WT values.
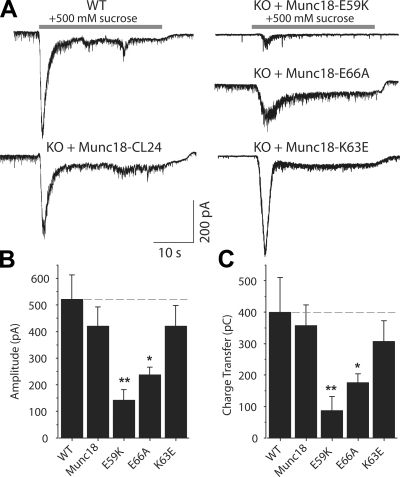
Figure 6.
Munc18-1 binding to the SNARE complex is critical for release readiness of synaptic vesicles. (A) Representative traces of synaptic excitatory responses to hypertonic solution (+500 mM sucrose to the bath) in WT neurons from littermate controls and Munc18-1 KO neurons rescued with lentivirus-expressing WT Munc18-1–24-cerulean or Munc18-1–24-cerulean with the E59K, K63E, or E66A mutations. (B) Bar diagram depicting the amplitudes of responses to hypertonic sucrose solution for WT cultures or Munc18-1 KO cultures rescued with the WT and mutant Munc18-1s (WT, n = 5; Munc18-1, K63E, and E66A, n = 8; E59K, n = 5). (C) Readily releasable synaptic excitatory transmission characterized as the amount of charge transfer induced by hypertonic sucrose. Asterisks in the bar diagrams mark statistical significance of the difference between the WT and mutant rescues (*, P < 0.05; **, P < 0.01). (B and C) Data are shown as means ± SEMs. Dashed lines indicate WT values.
Mutations in Munc18-1 impair vesicle priming
We next investigated the effects of the three mutations in Munc18-1 on its ability to rescue neurotransmitter release. We first examined spontaneous release (minis) and found that the K63E mutation had no significant effect, but the E66A mutation strongly decreased the mini frequency (>60%) while leaving the mini amplitude unaltered (Fig. 5, A and B). The E59K mutation decreased the mini frequency even more strongly (90%). We then measured the effects of the Munc18-1 mutations on evoked neurotransmitter release (Fig. 5, C–E). The K63E mutation had no statistically significant effect, although there was a trend for less release. The E66A mutation again caused a considerable impairment of release (50%), which was even more pronounced for the E59K mutant (80%). Note that the finding that the E66A mutation markedly impairs spontaneous and evoked release, whereas the K63E mutation has little or no effect, clearly correlates with the effects of these mutations on SNARE complex binding. The E59K mutant data further extends this correlation, although we cannot rule out the possibility that the disruption of release caused by this mutation arises in part from decreased protein levels.
The decrease in evoked release caused by the mutations could in principle arise from a reduction in the size of the readily releasable pool (RRP) of vesicles and/or in the vesicular release probability. To distinguish between these possibilities, we analyzed evoked responses at 10-Hz stimulation frequency. In these experiments, a reduced release probability is expected to lead to synaptic facilitation. The effects of the mutations on the amplitude of the first response of the train paralleled those observed at low frequency stimulation, but, importantly, all mutants exhibited strong synaptic depression during the stimulus train (Fig. S3). We then determined the size of the RRP by measuring synaptic response to 0.5 M hypertonic sucrose. We found that the E66A mutation led to a marked decrease in the size of the RRP (∼50% decrease), whereas the E59K mutation decreased the size of the RRP even more strongly (76%), and the K63E mutation caused no significant effect (Fig. 6). Thus, the effects of the mutations on the RRP parallel those observed in the spontaneous and evoked responses (Fig. 5) as well as in the Munc18-1–SNARE complex binding assays (Fig. 3). Correspondingly, the ratio between the synaptic charge transfer in evoked release (Fig. 5 E) and the sucrose-induced charge transfer (Fig. 6 C) was very similar for the WT and mutant Munc18-1 rescues (Fig. S4, available at http://www.jcb.org/cgi/content/full/jcb.200812026/DC1), suggesting that, for the vesicles that are primed, Ca2+ triggering of fusion is normal. These results show that the impairment in release caused by the Munc18-1 mutations occurs at the vesicle-priming step and suggest that the interaction of Munc18-1 with the Habc domain in open syntaxin-1 is critical for this step but not for the downstream events that lead to release.
A Munc18-1–SNARE–complexin macromolecular assembly
A fundamental question to understand the mechanism of action of Munc18-1 in neurotransmitter release is to characterize the relation between its binding to the SNARE complex and other interactions that have been described for this complex (Südhof, 2004; Brunger, 2005). It is particularly crucial to determine whether Munc18-1 and complexins can bind simultaneously to the SNARE complex because complexins function at the Ca2+-triggering step of release (Reim et al., 2001) and they are generally believed to be bound to the SNARE complex before Ca2+ influx (Rizo and Rosenmund, 2008). Thus, such simultaneous binding would suggest that Munc18-1 is bound to the SNAREs within the release-ready state that results after vesicle priming.
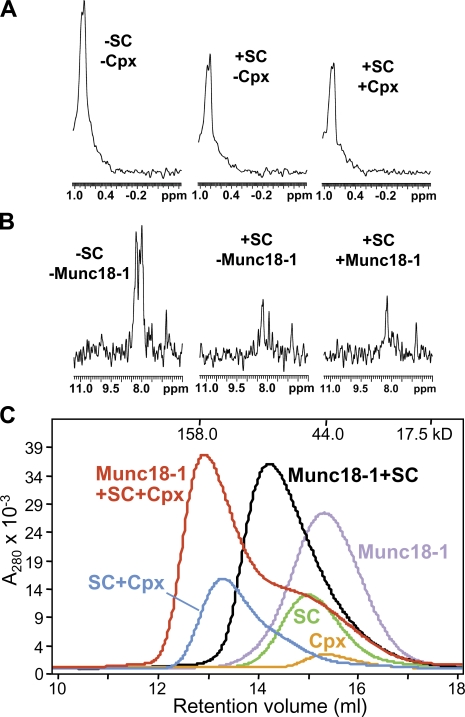
To address this question, we first used 1D isotope-edited 1H-NMR assays (Arac et al., 2003; Dulubova et al., 2005) that rely on the same principles as the experiments of Fig. 3. Thus, the SMR intensity of 1D 13C-edited 1H-NMR spectra of 2 µM Munc18-1 (65 kD) was considerably reduced upon addition of unlabeled SNARE complex (55 kD), reflecting the formation of the Munc18-1–SNARE complex assembly (Fig. 7 A). Addition of 15N-labeled complexin-1 (16 kD) led to a further decrease in the SMR intensity of Munc18-1, which can be attributed to binding of complexin-1 to the Munc18-1–SNARE complex assembly (note that if complexin-1 displaced Munc18-1 from the SNARE complex, the SMR intensity would have been restored to that of isolated Munc18-1). The formation of a Munc18-1–SNARE–complexin macromolecular assembly was confirmed by 1D 15N-edited 1H-NMR spectra of 2 µM 15N-labeled complexin-1 (Fig. 7 B). Many complexin-1 signals in these spectra are broadened beyond detection upon SNARE complex binding and are still not observable upon addition of Munc18-1, showing that Munc18-1 does not displace complexin-1 from the SNARE complex (signals that remain observable correspond to complexin-1 regions that are flexible regardless of the presence or absence of Munc18-1). In particular, note that the spectra containing 13C-labeled Munc18-1, unlabeled SNARE complex, and 15N-labeled complexin-1 (Fig. 7, A and B, right) were acquired on the same sample and unambiguously demonstrate that Munc18-1 and complexin-1 can bind simultaneously to the SNARE complex.
Figure 7.
Munc18-1 and complexin-1 can bind simultaneously to the SNARE complex. (A) Sample traces of the methyl regions of 1D 13C-edited 1H-NMR spectra of 2 µM Munc18-1 in the absence or presence of 2 µM of unlabeled SNARE complex or 2 µM of unlabeled SNARE complex plus 2 µM 15N-labeled complexin-1. (B) Sample traces of 1D 15N-edited 1H-NMR spectra of 2 µM 15N-labeled complexin-1 in the absence or presence of 2 µM of unlabeled SNARE complex or 2 µM of unlabeled SNARE complex plus 2 µM 13C-labeled Munc18-1. (A and B) The spectra on the right were acquired with the same sample. ppm, parts per million. (C) Gel filtration on a Superdex S200 (10/300GL) column of Munc18-1, complexin-1, SNARE complex (SC), and mixtures of the SNARE complex with Munc18-1, complexin (Cpx), or both. The SNARE complexes used for all of these experiments contained syntaxin-1(2–253), synaptobrevin-2(29–93), SNAP-25(11–82), and SNAP-25(141–203).
To confirm this conclusion by a different method, we used gel filtration experiments (Fig. 7 C). A mixture of complexin-1 and SNARE complex coeluted at smaller volumes than those of separate samples of complexin-1 and SNARE complex, as expected from the high affinity of their interaction (McMahon et al., 1995). Similar results were obtained with samples of Munc18-1 and SNARE complex, as described previously (Dulubova et al., 2007). Importantly, addition of complexin-1 further decreased the elution volume of the Munc18-1–SNARE complex assembly (Fig. 7 C), demonstrating that complexin-1 binds to this assembly. It is worth noting in these profiles that the elution of the SNARE complex is shifted to smaller volumes by complexin-1 than by Munc18-1 despite the much smaller size of complexin-1. We attribute this difference to the formation of a more compact structure in the SNARE complex upon Munc18-1 binding as the result of interactions of Munc18-1 with the four-helix bundle and the N-terminal region of syntaxin-1 (Dulubova et al., 2007) and to the fact that complexin-1 contains large unfolded regions even after SNARE complex binding (Pabst et al., 2000). To rule out the possibility that the coelution of Munc18-1 with complexin-1 and the SNARE complex might arise from the binding of Munc18-1 to these unfolded regions of complexin-1, we performed additional gel filtration experiments with a shorter complexin-1 fragment (residues 26–83), which spans the region that becomes structured upon SNARE binding (Pabst et al., 2000; Chen et al., 2002). Analogous results were obtained with this fragment (Fig. S5, available at http://www.jcb.org/cgi/content/full/jcb.200812026/DC1), again confirming the formation of a Munc18-1–SNARE–complexin assembly.
Discussion
The importance of Munc18-1 and the SNAREs for neurotransmitter release is well established, but it is still unclear how their functions are coupled. The binary interaction initially identified between Munc18-1 and the closed conformation of syntaxin-1 (Hata et al., 1993; Dulubova et al., 1999), which stabilizes both proteins and gates the entry of syntaxin-1 into SNARE complexes (Verhage et al., 2000; Gerber et al., 2008), does not appear to be general and may have emerged to meet specific requirements of regulated secretion. Munc18-1 binding to SNARE complexes containing open syntaxin-1 does seem to be universal (Fig. 1 B; Dulubova et al., 2007; Shen et al., 2007; Rodkey et al., 2008), and functional data provided evidence for the physiological relevance of SM protein–SNARE complex interactions in diverse systems (Carr et al., 1999; Grote et al., 2000; Yamaguchi et al., 2002; Collins et al., 2005; Khvotchev et al., 2007; Shen et al., 2007). However, the point of action of Munc18-1–open syntaxin-1 interactions in release was unknown, and the relationship between these interactions and those of the SNAREs with other key proteins such as complexins was unclear. Our data now provide fundamental insights into these questions, showing that binding of Munc18-1 to the Habc domain of open syntaxin-1 is critical for synaptic vesicle priming but not for the release step and that Munc18-1 and complexin-1 can bind simultaneously to the SNARE complex.
The development of a strategy to rescue survival and neurotransmitter release in neurons from Munc18-1 KO mice, which is strongly hindered by the deleterious effects arising from deletion of this protein, was key for this study. In addition, we wanted to perform the rescue with fluorescently tagged Munc18-1 to ensure that we could monitor the proper localization of the expressed protein. After extensive efforts, we overcame these difficulties by identifying a Munc18-1–cerulean fusion that rescues the Munc18-1 deficiency phenotype when expressed with a lentivirus, which allowed us to study the functional effects of the three Munc18-1 mutations with the precision of electrophysiology and correlate them with our in vitro binding data.
Although the energetic contributions of individual residues to protein–protein interactions are difficult to predict from 3D structures, the observed biochemical effects of the Munc18-1 mutations can be rationalized according to general knowledge on the energetics of protein–protein interactions and to the model used to design these mutations. In this model, the syntaxin-1 SNARE motif contributes strongly to the binary interaction with Munc18-1 but needs to be released upon formation of the Munc18-1–SNARE complex assembly, leading to a different interaction of Munc18-1 with the four-helix bundle (Fig. 1 B). In contrast, the Habc domain–Munc18-1 interface is likely similar in both complexes (Khvotchev et al., 2007). The latter assumption implies that, in the Munc18-1–SNARE complex assembly, E59 contacts the Habc domain extensively, E66 makes fewer contacts, and K63 does not interact with the Habc domain (in the binary complex, E66 is between the Habc domain and the SNARE motif, whereas K63 only contacts the SNARE motif; Fig. 1 D); this prediction correlates very well with the relative impairments in Munc18-1–SNARE complex binding caused by the E59K and E66A mutations and the lack of an effect by the K63E mutation. A key prediction of the model was that, because of the proximity of E59, K63, and E66 to the Habc domain–SNARE motif interface in the binary Munc18-1–syntaxin-1 complex and because of the large rearrangement of the syntaxin-1 SNARE motif upon formation of the Munc18-1–SNARE complex assembly, the energetic contributions of these residues to binding would be different in the two complexes. The different effects of the mutations on the binary Munc18-1–syntaxin-1 interaction compared with those caused on Munc18-1–SNARE complex binding agree with this prediction. It is not surprising that none of the mutations strongly reduced Munc18-1 binding to syntaxin-1 given the adaptability of protein–protein interfaces upon introduction of point mutations (Atwell et al., 1997), particularly for complexes of high affinity and involving large interfaces such as that in the Munc18-1–syntaxin-1 complex (>4,000 Å of buried surface area; Misura et al., 2000; Burkhardt et al., 2008). Although not reaching statistical significance, the K63E mutation did appear to have an effect on Munc18-1–syntaxin-1 binding, which is consistent with the notion that interactions with the SNARE motif contribute strongly to the affinity of the binary complex; such a contribution likely facilitates toleration of the E59K and E66A mutations.
The considerable impairment of Ca2+-evoked, sucrose-induced, and spontaneous release caused by the E66A mutation and the little functional consequences of the K63E mutation (Figs. 5 and 6) correlate very well with the effects of these mutations on binding of Munc18-1 to SNARE complexes containing open syntaxin-1 (Fig. 3) but not with their effects on binding of Munc18-1 to closed syntaxin-1 (Fig. 2). Although the functional data obtained for the E59K mutant needs to be interpreted with caution, the data appear to extend the correlation between impairment of release and disruption of Munc18-1–SNARE complex binding but not Munc18-1–syntaxin-1 binding. Note that the expression of this mutant is sufficient to rescue survival in Munc18-1 KO neurons and that decreased Munc18-1 levels in syntaxin-1B LE mutant mice lead to a much more moderate decrease in the RRP (Gerber et al., 2008) than that caused by the E59K mutation (Fig. 6 C). Thus, it seems likely that the strong impairment of release caused by this mutation arises at least in part from disruption of the interaction between Munc18-1 and open syntaxin-1. Importantly, the decreases in spontaneous and Ca2+-evoked release caused by the E66A and E59K mutations mirror the corresponding RRP reductions. This finding is in sharp contrast with the phenotype observed in the syntaxin-1 LE mutant mice, in which the RRP decreased (likely because of the lower protein levels), but spontaneous release and release of primed vesicles were enhanced (Gerber et al., 2008). Because the LE mutation impairs Munc18-1 binding to closed syntaxin-1 but not to SNARE complexes (Gerber et al., 2008), this contrast further supports the conclusion that the functional effects of the E66A and E59K mutations arise from the disruption of interactions of Munc18-1 with open syntaxin-1 rather than closed syntaxin-1.
Synaptic vesicle docking was not affected in Munc18-1 KO mice (Verhage et al., 2000). Thus, it is unlikely that the E66A and E59K mutations alter docking. This observation together with the finding that these mutations cause parallel decreases in spontaneous, sucrose-induced, and Ca2+-triggered release, resulting in evoked release/RRP ratios that are similar to WT (Fig. S4), strongly suggest that the mutations selectively disrupt synaptic vesicle priming but not the downstream events that lead to evoked release. Immediate questions that arise are do Munc18-1–SNARE interactions play any role after priming, and does Munc18-1 form part of the macromolecular complex that results after priming and is poised for Ca2+-triggered release? As a first step to address these questions, we examined whether complexin-1 and Munc18-1 compete for SNARE complex binding. Complexins play a role in the Ca2+-triggering step of release (Reim et al., 2001) and bind tightly to SNARE complexes (McMahon et al., 1995), interacting with the SNARE four-helix bundle (Chen et al., 2002). These observations strongly suggest that complexins are key components of the primed macromolecular assembly that is ready for release (Rizo and Rosenmund, 2008), and our demonstration that Munc18-1 and complexin-1 can bind simultaneously to the SNARE complex suggests that Munc18-1 likely forms part of this assembly as well. Structural studies will be required to characterize in detail the resulting Munc18-1–SNARE–complexin assembly, but it is noteworthy that Munc18-1 binding barely shifts the elution profile of the complexin-1–SNARE complex despite doubling its molecular mass (Fig. 7 C). This observation suggests that such binding leads to a more compact shape caused by interactions of Munc18-1 with both the N-terminal region of syntaxin-1 and the four-helix bundle, as proposed for the Munc18-1–SNARE complex assembly (Fig. 1 B, right; Dulubova et al., 2007).
In contrast, a recent study concluded that binding of Munc18-1 to the SNARE complex involves interactions with only the syntaxin-1 N-terminal region (residues 1–179) based on the similar affinities of Munc18-1 for the SNARE complex and syntaxin-1(1–179) (Burkhardt et al., 2008). However, this conclusion ignores the possibility that the energy gained from Munc18-1–four-helix bundle interactions may be offset by release of interactions contributing to the affinity of Munc18-1 for syntaxin-1(1–179) (e.g., involving the syntaxin-1 linker region), and abundant evidence has demonstrated interactions of Munc18-1 with the SNARE motifs (Dulubova et al., 2007; Shen et al., 2007; Rodkey et al., 2008; Weninger et al., 2008). Nevertheless, the similar affinities suggest that there is little cooperativity between interactions of Munc18-1 with the four-helix bundle and the syntaxin-1 N-terminal region. Conversely, Munc18-1–membrane interactions seem to cooperate with binding to the four-helix bundle, as the syntaxin-1 N-terminal region is required for binding of Munc18-1 to soluble SNARE complexes but not for binding to membrane-anchored SNARE complexes (Shen et al., 2007).
These observations, together with our data, suggest a model whereby synaptic vesicle priming involves the opening of syntaxin-1, and binding of Munc18-1 to the syntaxin-1 Habc domain is critical for the opening reaction but not for downstream events leading to release. In this model, transition from the Munc18-1–closed syntaxin-1 complex to the Munc18-1–SNARE complex assembly (Fig. 1 B) involves a series of intermediate states. Thus, release of the SNARE motif from closed syntaxin-1 to bind to SNAP-25 (likely assisted by Munc13-1; Guan et al., 2008; Weninger et al., 2008) may involve a transient state in which Munc18-1 is only interacting with the syntaxin-1 N-terminal region (Fig. 1 C, left). This interaction might keep Munc18-1 near the site of action to facilitate the establishment of new interactions with the syntaxin-1/SNAP-25 SNARE motifs (Fig. 1 C, right), forming an acceptor complex for synaptobrevin binding. The resulting Munc18-1–SNARE complex assembly (Fig. 1 B, right) may involve cooperative interactions of Munc18-1 with the four-helix bundle and one or both membranes, which might be key for membrane fusion but might shift the energetic balance so that the interactions of Munc18-1 with the syntaxin-1 N-terminal region become dispensable. These features can explain why binding of Munc18-1 to the Habc domain of open syntaxin-1 is crucial for priming but not for the downstream events leading to fusion. The existence of the proposed intermediate states is supported by the finding that Munc18-1 can bind to isolated syntaxin-1 N-terminal fragments (Khvotchev et al., 2007; Burkhardt et al., 2008) and to syntaxin-1–SNAP-25 heterodimers (Guan et al., 2008; Weninger et al., 2008). The hypothesis that Munc18-1 binds to the four-helix bundle and the two apposed membranes correlates with the role proposed for the homotypic fusion and vacuole protein-sorting complex (which includes the Munc18-1 homologue Vps33p) in discriminating trans- from cis-SNARE complexes in yeast vacuolar fusion (Starai et al., 2008). Our model is also consistent with evidence suggesting that Munc18-1 plays multiple roles in the different steps that lead to release (Gulyas-Kovacs et al., 2007), but, clearly, further research will be required to test this model and to elucidate how the function of Munc18-1 is coupled to those of other SNARE-binding proteins such as complexins, Munc13s, and synaptotagmin-1.
Materials and methods
Constructs
Bacterial expression vectors to express full-length rat Munc18-1 and full-length rat complexin-1 and fragments corresponding to the cytoplasmic region of rat syntaxin-1A (residues 2–243 or 2–253), the SNARE motifs of rat synaptobrevin-2(29–93) and human SNAP-25B (11–82 and 141–203), or residues 26–83 of rat complexin-1 as GST fusion proteins were described previously (McMahon et al., 1995; Chen et al., 2002; Dulubova et al., 2007). Analogous vectors to express full-length rat Munc18-1 point mutants (E59K, K63E, and E66A) were generated from the WT construct using the QuikChange Site-Directed Mutagenesis kit (Agilent Technologies) and custom-designed primers and were verified by sequencing. For the construction of Munc18-1–24-cerulean constructs, a Munc18-1 (aa 1–24) fragment was first generated by PCR to introduce EcoRI and SseI cloning sites at the N and C termini, and then a Munc18-1 (aa 25–601) fragment was produced by PCR to introduce BsrGI and XbaI cloning sites at the N and C termini. These two PCR products were then inserted to the N terminus and C terminus of cerulean, respectively, on a PCMV5-cerulean vector. The cDNAs of WT and mutant Munc18-1–24-cerulean versions were subcloned between the EcoRI and BamHI sites into the pFUGW shuttle vector for virus production.
Preparation of recombinant proteins and SNARE complexes
Syntaxin-1A(2–243), syntaxin-1A(2–253), synaptobrevin-2(29–93), SNAP-25(11–82), SNAP-25(141–203), complexin-1, complexin-1(26–83), and WT and mutant full-length Munc18-1 were expressed in bacteria as GST fusion proteins, isolated by affinity chromatography, cleaved with thrombin, and purified by ion exchange or gel filtration chromatography as described previously (Dulubova et al., 1999; Chen et al., 2002; Dulubova et al., 2007). Mass spectrometry analysis showed that the proteolysis of the syntaxin-1A N terminus reported in a recent study (Burkhardt et al., 2008) does not occur using our protocols. Uniform 13C or 15N labeling was accomplished by expression in bacteria using 13C6-glucose as the sole carbon source or 15NH4Cl as the sole nitrogen source, respectively. SNARE complexes were prepared by overnight incubation of the four purified SNARE fragments (with either syntaxin-1A[2–243] or syntaxin-1A[2–253] plus synaptobrevin-2[29–93], SNAP-25[11–82], and SNAP-25[141–203]), and further purification was performed by gel filtration on a column (Superdex 200; GE Healthcare) in PBS, pH 7.4, containing 2 mM Tris(2-carboxyethyl)phosphine (Dulubova et al., 2007).
ITC
ITC experiments were performed using a VP-ITC system (GE Healthcare) at 20°C in PBS, pH 7.4, containing 2 mM Tris(2-carboxyethyl)phosphine with samples of 5–10 µM WT or mutant Munc18-1s in the sample cell and successive injections of 100–150 µM syntaxin-1A(2–243). All proteins were purified by gel filtration on a Superdex 200 column in the same buffer before the experiments. After polynomial baseline correction to remove a slight drift of the initial data points, the data were fitted with a nonlinear least squares routine using a single-site binding model with Origin for ITC version 5.0 (GE Healthcare). The baseline correction did not substantially alter the Kd values obtained.
NMR experiments
1D 13C-edited or 15N-edited 1H-NMR spectra were acquired on a spectrometer (INOVA600; Varian) at 25°C in 20 mM sodium phosphate, pH 7.1, 150 mM NaCl, and 2 mM DTT, acquiring the first trace of 1H-13C or 1H-15N heteronuclear single quantum correlation spectra (1,500 scans and 30-min total acquisition time). For the titrations of Fig. 3 B, samples contained 2 µM SNARE complex (with uniformly 13C-labeled syntaxin-1A[2–243]) and the desired concentration of unlabeled WT or mutant Munc18-1s; a separate sample was prepared for each concentration. The data were fit to a single-site binding model using SigmaPlot (SPSS, Inc.; Arac et al., 2003).
Cortical cultures
Homozygote Munc18-1 KO mice were bred by crossing heterozygous mutant Munc18-1 KO mice (Verhage et al., 2000). Cortical neurons from littermate mice at E16 were dissociated by trypsin (5 mg/ml for 5 min at 37°C), triturated with a siliconized Pasteur pipette, and plated onto 12-mm coverslips coated with Matrigel (∼12 coverslips/cortex). Neurons were cultured at 37°C in a humidified incubator with 95% air and 5% CO2 in minimal essential media containing 5 g/liter glucose, 0.1 g/liter transferrin, 0.25 g/liter insulin, 0.3 g/liter glutamine, 5–10% heat-inactivated FCS, 2% B-27 supplement, and 1 µM cytosine arabinoside and were used after 5–22 DIV.
Lentiviral infection
Constructs were cotransfected with plasmids for viral enzymes and envelope proteins into HEK 293 cells using a transfection system (FuGENE6; Roche) according to the manufacturer's specifications, and lentivirus-containing culture medium was harvested 2 d later, filtered through 0.45-µm pores, and immediately used for infection or frozen in liquid nitrogen and stored at −80°C. Cortical cultures were infected at 1 DIV by adding 300 µl of viral suspension to each well.
Immunocytochemistry
Neurons attached to the glass coverslips were rinsed once in PBS and fixed for 15 min on ice in 4% formaldehyde and 4% sucrose in PBS. After fixation, the neurons were washed with PBS twice and then incubated for 30 min in blocking solution, PBS containing 3% milk, 0.1% saponin, and in PBS followed by 1-h incubation with primary and rhodamine- and FITC-conjugated secondary antibodies diluted in blocking solution. The coverslips were then mounted on glass slides with Aqua-Poly/Mount medium (Polysciences, Inc.) and analyzed at room temperature (22–24°C) with a confocal microscope (DMIRE2; Leica) equipped with a confocal system (TCS SPX; Leica) using a 63× NA 1.32 oil immersion objective. The images were collected using confocal software (Leica) and processed using Photoshop software (Adobe). All digital manipulations were equally applied to the entire image.
Electrophysiology
Synaptic responses were recorded from pyramidal cells in modified Tyrode bath solution in the whole-cell patch configuration. The solution routinely contained 50–100 µM picrotoxin to block inhibitory synaptic currents via γ-aminobutyric acid receptors. For spontaneous release experiments (Deák et al., 2006), 1 µM tetrodotoxin was added to inhibit voltage-gated sodium channels and action potential propagation. For evoked responses, tetrodotoxin was omitted from the bath. Data were acquired with an amplifier (Axopatch 200B; MDS Analytical Technologies) and Clampex 8.0 software (MDS Analytical Technologies), filtered at 2 kHz, and sampled at 200 µs. The internal pipette solution was set to 300 mosM and included 115 mM Cs-MeSO3, 10 mM CsCl, 5 mM NaCl, 0.1 mM CaCl2, 10 mM Hepes, 4 mM Cs-BAPTA, 20 mM triethylamine-Cl, 4 mM Mg-ATP, 0.3 mM Na2-GTP, and 10 mM lidocaine N-ethyl-bromide, pH 7.35. A hypertonic solution, which was prepared by the addition of 500 mM sucrose to the nominally Ca2+-free Tyrode solution, was perfused directly into the close vicinity of the cell from which the recording was made. Field stimulations (24-mA pulses of 0.6 ms) were applied with parallel platinum electrodes immersed into the perfusion chamber.
Data analysis
Evoked responses were adjusted with baseline subtraction for each stimulus. Synchronized responses were determined as those within 100 ms of the stimulus, and transferred charge was also calculated for this period. Normal distribution of data was analyzed with the Shapiro-Wilk's W test. Paired Student's t test or variance analysis followed with Tukey's test was used to determine significance, which is marked on the figures as asterisks (*, P < 0.05; **, P < 0.01; and ***, P < 0.005 levels of significance).
Gel filtration binding assays
Samples contained 5 µM Munc18-1, 7.5 µM complexin-1, 5 µM SNARE complex (formed with syntaxin-1A[2–253], synaptobrevin-2[29–93], SNAP-25[11–82], and SNAP-25[141–203]), or different combinations of these proteins at the same concentrations and were dissolved in 400 µl of 20 mM Tris-HCl, pH 7.4, and 120 mM NaCl. The samples were incubated for 30 min, injected in a column (Superdex S200 10/300 GL; GE Healthcare), and eluted with the same buffer.
Online supplemental material
Fig. S1 shows a comparison of Ca-dependent synaptic release from WT neurons and Munc18-1 KO neurons rescued with WT Munc18-1 or Munc18-1–24-cerulean, as monitored by FM2-10 destaining, and the quantification of synaptic densities in the rescue experiments with WT and mutant Munc18-1s. Fig. S2 shows the protein expression levels in the rescue experiments with WT and mutant Munc18-1s as assessed by fluorescence and Western blotting. Fig. S3 shows the synaptic depression observed for WT Munc18-1 and Munc18-1 mutants at 10-Hz field stimulation. Fig. S4 shows the ratios between charge transfer induced by an action potential and by hypertonic sucrose in the rescue experiments with WT and mutant Munc18-1s. Fig. S5 shows gel filtration profiles of Munc18-1, complexin-1(26–83), SNARE complex, and mixtures of them. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200812026/DC1.
Acknowledgments
We would like to thank Siqi Liu for preparing bacterial expression vectors for Munc18-1 mutants, Andrea Roth, Iza Kornblum, and Ewa Borowicz for excellent technical assistance, and Lin Fan and Jason Mitchell for help with mouse husbandry.
This work was supported by grant I-1304 from the Welch Foundation and by grant NS37200 (to J. Rizo) from the National Institutes of Health.
Footnotes
Abbreviations used in this paper: DIV, day in vitro; ITC, isothermal titration calorimetry; KO, knockout; NMR, nuclear magnetic resonance; RRP, readily releasable pool; SM, Sec1/Munc18; SMR, strongest methyl resonance; WT, wild type.
References
- Arac D., Murphy T., Rizo J. 2003. Facile detection of protein-protein interactions by one-dimensional NMR spectroscopy.Biochemistry. 42:2774–2780 [DOI] [PubMed] [Google Scholar]
- Atwell S., Ultsch M., De Vos A.M., Wells J.A. 1997. Structural plasticity in a remodeled protein-protein interface.Science. 278:1125–1128 [DOI] [PubMed] [Google Scholar]
- Bracher A., Weissenhorn W. 2002. Structural basis for the Golgi membrane recruitment of Sly1p by Sed5p.EMBO J. 21:6114–6124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunger A.T. 2005. Structure and function of SNARE and SNARE-interacting proteins.Q. Rev. Biophys. 38:1–47 [DOI] [PubMed] [Google Scholar]
- Burkhardt P., Hattendorf D.A., Weis W.I., Fasshauer D. 2008. Munc18a controls SNARE assembly through its interaction with the syntaxin N-peptide.EMBO J. 27:923–933 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carpp L.N., Ciufo L.F., Shanks S.G., Boyd A., Bryant N.J. 2006. The Sec1p/Munc18 protein Vps45p binds its cognate SNARE proteins via two distinct modes.J. Cell Biol. 173:927–936 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carr C.M., Grote E., Munson M., Hughson F.M., Novick P.J. 1999. Sec1p binds to SNARE complexes and concentrates at sites of secretion.J. Cell Biol. 146:333–344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X., Tomchick D.R., Kovrigin E., Arac D., Machius M., Südhof T.C., Rizo J. 2002. Three-dimensional structure of the complexin/SNARE complex.Neuron. 33:397–409 [DOI] [PubMed] [Google Scholar]
- Chen X., Lu J., Dulubova I., Rizo J. 2008. NMR analysis of the closed conformation of syntaxin-1.J. Biomol. NMR. 41:43–54 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins K.M., Thorngren N.L., Fratti R.A., Wickner W.T. 2005. Sec17p and HOPS, in distinct SNARE complexes, mediate SNARE complex disruption or assembly for fusion.EMBO J. 24:1775–1786 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deák F., Shin O.H., Kavalali E.T., Südhof T.C. 2006. Structural determinants of synaptobrevin 2 function in synaptic vesicle fusion.J. Neurosci. 26:6668–6676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Sugita S., Hill S., Hosaka M., Fernandez I., Südhof T.C., Rizo J. 1999. A conformational switch in syntaxin during exocytosis: role of munc18.EMBO J. 18:4372–4382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Yamaguchi T., Wang Y., Südhof T.C., Rizo J. 2001. Vam3p structure reveals conserved and divergent properties of syntaxins.Nat. Struct. Biol. 8:258–264 [DOI] [PubMed] [Google Scholar]
- Dulubova I., Yamaguchi T., Gao Y., Min S.W., Huryeva I., Südhof T.C., Rizo J. 2002. How Tlg2p/syntaxin 16 “snares” Vps45.EMBO J. 21:3620–3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Yamaguchi T., Arac D., Li H., Huryeva I., Min S.W., Rizo J., Südhof T.C. 2003. Convergence and divergence in the mechanism of SNARE binding by Sec1/Munc18-like proteins.Proc. Natl. Acad. Sci. USA. 100:32–37 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Lou X., Lu J., Huryeva I., Alam A., Schneggenburger R., Südhof T.C., Rizo J. 2005. A Munc13/RIM/Rab3 tripartite complex: from priming to plasticity? EMBO J. 24:2839–2850 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dulubova I., Khvotchev M., Liu S., Huryeva I., Südhof T.C., Rizo J. 2007. Munc18-1 binds directly to the neuronal SNARE complex.Proc. Natl. Acad. Sci. USA. 104:2697–2702 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernandez I., Ubach J., Dulubova I., Zhang X., Südhof T.C., Rizo J. 1998. Three-dimensional structure of an evolutionarily conserved N-terminal domain of syntaxin 1A.Cell. 94:841–849 [DOI] [PubMed] [Google Scholar]
- Fiebig K.M., Rice L.M., Pollock E., Brunger A.T. 1999. Folding intermediates of SNARE complex assembly.Nat. Struct. Biol. 6:117–123 [DOI] [PubMed] [Google Scholar]
- Gerber S.H., Rah J.C., Min S.W., Liu X., de Wit H., Dulubova I., Meyer A.C., Rizo J., Arancillo M., Hammer R.E., et al. 2008. Conformational switch of syntaxin-1 controls synaptic vesicle fusion.Science. 321:1507–1510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote E., Carr C.M., Novick P.J. 2000. Ordering the final events in yeast exocytosis.J. Cell Biol. 151:439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan R., Dai H., Rizo J. 2008. Binding of the Munc13-1 MUN domain to membrane-anchored SNARE complexes.Biochemistry. 47:1474–1481 [DOI] [PubMed] [Google Scholar]
- Gulyas-Kovacs A., de Wit H., Milosevic I., Kochubey O., Toonen R., Klingauf J., Verhage M., Sorensen J.B. 2007. Munc18-1: sequential interactions with the fusion machinery stimulate vesicle docking and priming.J. Neurosci. 27:8676–8686 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanson P.I., Roth R., Morisaki H., Jahn R., Heuser J.E. 1997. Structure and conformational changes in NSF and its membrane receptor complexes visualized by quick-freeze/deep-etch electron microscopy.Cell. 90:523–535 [DOI] [PubMed] [Google Scholar]
- Hata Y., Slaughter C.A., Südhof T.C. 1993. Synaptic vesicle fusion complex contains unc-18 homologue bound to syntaxin.Nature. 366:347–351 [DOI] [PubMed] [Google Scholar]
- Jahn R., Scheller R.H. 2006. SNAREs–engines for membrane fusion.Nat. Rev. Mol. Cell Biol. 7:631–643 [DOI] [PubMed] [Google Scholar]
- Khvotchev M., Dulubova I., Sun J., Dai H., Rizo J., Südhof T.C. 2007. Dual modes of Munc18-1/SNARE interactions are coupled by functionally critical binding to syntaxin-1 N terminus.J. Neurosci. 27:12147–12155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham C.F., Lopez J.A., Hu S.H., Gee C.L., Westbury E., Blair D.H., Armishaw C.J., Alewood P.F., Bryant N.J., James D.E., Martin J.L. 2006. Molecular dissection of the Munc18c/syntaxin4 interaction: implications for regulation of membrane trafficking.Traffic. 7:1408–1419 [DOI] [PubMed] [Google Scholar]
- McMahon H.T., Missler M., Li C., Südhof T.C. 1995. Complexins: cytosolic proteins that regulate SNAP receptor function.Cell. 83:111–119 [DOI] [PubMed] [Google Scholar]
- Misura K.M., Scheller R.H., Weis W.I. 2000. Three-dimensional structure of the neuronal-Sec1-syntaxin 1a complex.Nature. 404:355–362 [DOI] [PubMed] [Google Scholar]
- Nicholson K.L., Munson M., Miller R.B., Filip T.J., Fairman R., Hughson F.M. 1998. Regulation of SNARE complex assembly by an N-terminal domain of the t-SNARE Sso1p.Nat. Struct. Biol. 5:793–802 [DOI] [PubMed] [Google Scholar]
- Pabst S., Hazzard J.W., Antonin W., Südhof T.C., Jahn R., Rizo J., Fasshauer D. 2000. Selective interaction of complexin with the neuronal SNARE complex. Determination of the binding regions.J. Biol. Chem. 275:19808–19818 [DOI] [PubMed] [Google Scholar]
- Peng R., Gallwitz D. 2002. Sly1 protein bound to Golgi syntaxin Sed5p allows assembly and contributes to specificity of SNARE fusion complexes.J. Cell Biol. 157:645–655 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pevsner J., Hsu S.C., Braun J.E., Calakos N., Ting A.E., Bennett M.K., Scheller R.H. 1994. Specificity and regulation of a synaptic vesicle docking complex.Neuron. 13:353–361 [DOI] [PubMed] [Google Scholar]
- Poirier M.A., Xiao W., Macosko J.C., Chan C., Shin Y.K., Bennett M.K. 1998. The synaptic SNARE complex is a parallel four-stranded helical bundle.Nat. Struct. Biol. 5:765–769 [DOI] [PubMed] [Google Scholar]
- Reim K., Mansour M., Varoqueaux F., McMahon H.T., Südhof T.C., Brose N., Rosenmund C. 2001. Complexins regulate a late step in Ca2+-dependent neurotransmitter release.Cell. 104:71–81 [DOI] [PubMed] [Google Scholar]
- Rizo J., Rosenmund C. 2008. Synaptic vesicle fusion.Nat. Struct. Mol. Biol. 15:665–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizo J., Südhof T.C. 2002. Snares and munc18 in synaptic vesicle fusion.Nat. Rev. Neurosci. 3:641–653 [DOI] [PubMed] [Google Scholar]
- Rizo J., Chen X., Arac D. 2006. Unraveling the mechanisms of synaptotagmin and SNARE function in neurotransmitter release.Trends Cell Biol. 16:339–350 [DOI] [PubMed] [Google Scholar]
- Rodkey T.L., Liu S., Barry M., McNew J.A. 2008. Munc18a scaffolds SNARE assembly to promote membrane fusion.Mol. Biol. Cell. 19:5422–5434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen J., Tareste D.C., Paumet F., Rothman J.E., Melia T.J. 2007. Selective activation of cognate SNAREpins by Sec1/Munc18 proteins.Cell. 128:183–195 [DOI] [PubMed] [Google Scholar]
- Sollner T., Bennett M.K., Whiteheart S.W., Scheller R.H., Rothman J.E. 1993. A protein assembly-disassembly pathway in vitro that may correspond to sequential steps of synaptic vesicle docking, activation, and fusion.Cell. 75:409–418 [DOI] [PubMed] [Google Scholar]
- Starai V.J., Hickey C.M., Wickner W. 2008. HOPS proofreads the trans-SNARE complex for yeast vacuole fusion.Mol. Biol. Cell. 19:2500–2508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stroupe C., Collins K.M., Fratti R.A., Wickner W. 2006. Purification of active HOPS complex reveals its affinities for phosphoinositides and the SNARE Vam7p.EMBO J. 25:1579–1589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Südhof T.C. 2004. The synaptic vesicle cycle.Annu. Rev. Neurosci. 27:509–547 [DOI] [PubMed] [Google Scholar]
- Sutton R.B., Fasshauer D., Jahn R., Brunger A.T. 1998. Crystal structure of a SNARE complex involved in synaptic exocytosis at 2.4 A resolution.Nature. 395:347–353 [DOI] [PubMed] [Google Scholar]
- Tang J., Maximov A., Shin O.H., Dai H., Rizo J., Südhof T.C. 2006. A complexin/synaptotagmin 1 switch controls fast synaptic vesicle exocytosis.Cell. 126:1175–1187 [DOI] [PubMed] [Google Scholar]
- Togneri J., Cheng Y.S., Munson M., Hughson F.M., Carr C.M. 2006. Specific SNARE complex binding mode of the Sec1/Munc-18 protein, Sec1p.Proc. Natl. Acad. Sci. USA. 103:17730–17735 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toonen R.F., Verhage M. 2003. Vesicle trafficking: pleasure and pain from SM genes.Trends Cell Biol. 13:177–186 [DOI] [PubMed] [Google Scholar]
- Verhage M., Maia A.S., Plomp J.J., Brussaard A.B., Heeroma J.H., Vermeer H., Toonen R.F., Hammer R.E., van den Berg T.K., Missler M., et al. 2000. Synaptic assembly of the brain in the absence of neurotransmitter secretion.Science. 287:864–869 [DOI] [PubMed] [Google Scholar]
- Weninger K., Bowen M.E., Choi U.B., Chu S., Brunger A.T. 2008. Accessory proteins stabilize the acceptor complex for synaptobrevin, the 1:1 syntaxin/SNAP-25 complex.Structure. 16:308–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu M.N., Littleton J.T., Bhat M.A., Prokop A., Bellen H.J. 1998. ROP, the Drosophila Sec1 homolog, interacts with syntaxin and regulates neurotransmitter release in a dosage-dependent manner.EMBO J. 17:127–139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi T., Dulubova I., Min S.W., Chen X., Rizo J., Südhof T.C. 2002. Sly1 binds to Golgi and ER syntaxins via a conserved N-terminal peptide motif.Dev. Cell. 2:295–305 [DOI] [PubMed] [Google Scholar]
- Yang B., Steegmaier M., Gonzalez L.C., Jr., Scheller R.H. 2000. nSec1 binds a closed conformation of syntaxin1A.J. Cell Biol. 148:247–252 [DOI] [PMC free article] [PubMed] [Google Scholar]