Abstract
A putative spindle matrix has been hypothesized to mediate chromosome motion, but its existence and functionality remain controversial. In this report, we show that Megator (Mtor), the Drosophila melanogaster counterpart of the human nuclear pore complex protein translocated promoter region (Tpr), and the spindle assembly checkpoint (SAC) protein Mad2 form a conserved complex that localizes to a nuclear derived spindle matrix in living cells. Fluorescence recovery after photobleaching experiments supports that Mtor is retained around spindle microtubules, where it shows distinct dynamic properties. Mtor/Tpr promotes the recruitment of Mad2 and Mps1 but not Mad1 to unattached kinetochores (KTs), mediating normal mitotic duration and SAC response. At anaphase, Mtor plays a role in spindle elongation, thereby affecting normal chromosome movement. We propose that Mtor/Tpr functions as a spatial regulator of the SAC, which ensures the efficient recruitment of Mad2 to unattached KTs at the onset of mitosis and proper spindle maturation, whereas enrichment of Mad2 in a spindle matrix helps confine the action of a diffusible “wait anaphase” signal to the vicinity of the spindle.
Introduction
The mitotic spindle is composed of dynamic microtubules (MTs) and associated proteins that mediate chromosome segregation during mitosis. The requirement of an additional stationary or elastic structure forming a spindle matrix where molecular motors slide MTs has long been proposed to power chromosome motion and account for incompletely understood features of mitotic spindle dynamics (Pickett-Heaps et al., 1984). However, definitive evidence for its existence in living cells or on its biochemical nature and whether it plays a direct role during mitosis has been missing.
A functional spindle matrix would be expected to (a) form a fusiform structure coalescent with spindle MTs, (b) persist in the absence of MTs, (c) be resilient in response to changes of spindle shape and length, and (d) affect spindle assembly and/or function if one or more of its components are perturbed. In Drosophila melanogaster, a complex of at least four nuclear proteins, Skeletor, Megator (Mtor), Chromator, and EAST (enhanced adult sensory threshold), form a putative spindle matrix that persists in the absence of MTs in fixed preparations (Johansen and Johansen, 2007). From this complex, Mtor is the only protein that shows clear sequence conservation with proteins in other organisms, such as the nuclear pore complex (NPC) protein translocated promoter region (Tpr) in mammals (Cordes et al., 1997; Zimowska et al., 1997), its respective counterparts Mlp1 and Mlp2 in yeast (Strambio-de-Castillia et al., 1999), and nuclear pore anchor in plants (Xu et al., 2007). NPC proteins, including Mtor/Tpr orthologues in yeast, were shown to functionally interact with spindle assembly checkpoint (SAC) components (Iouk et al., 2002; Scott et al., 2005). The SAC ensures correct chromosome segregation by providing time for proper kinetochore (KT) attachments to spindle MTs while inhibiting the activity of the anaphase-promoting complex/cyclosome (Musacchio and Salmon, 2007).
Assuming that any critical function by the spindle matrix is widely conserved, we focus on understanding the mitotic role of Mtor in living Drosophila somatic cells. Our results provide a new conceptual view of a spindle matrix not as a rigid structural scaffold but as a spatial determinant of key mitotic regulators.
Results and discussion
Mtor localizes to a dynamic nuclear derived spindle matrix in living cells
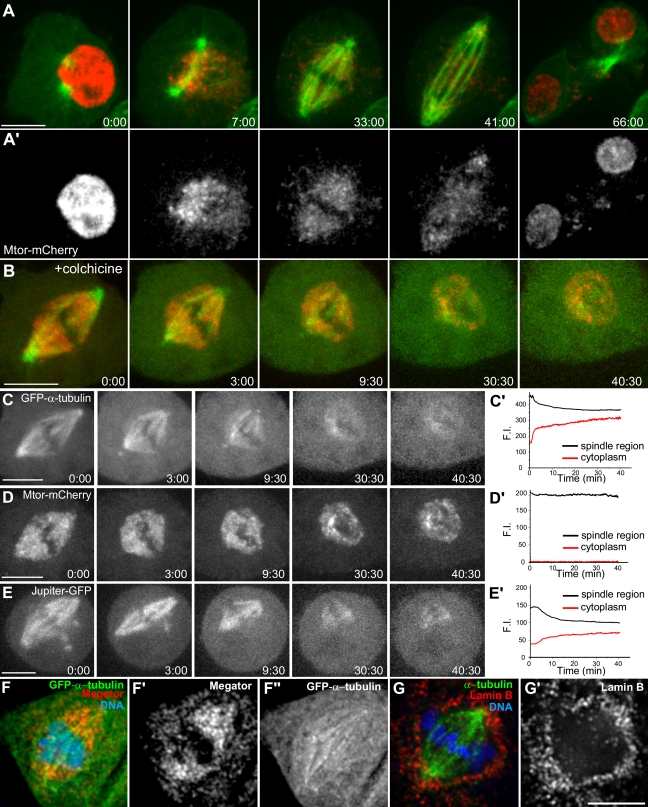
To investigate the localization of Mtor in living cells, we generated a Drosophila S2 cell line stably coexpressing Mtor-mCherry and GFP–α-tubulin. Mtor-mCherry is nuclear in interphase and at nuclear envelope breakdown (NEB) reorganizes into a fusiform structure coalescent with spindle MTs (Fig. 1 A; and Video 1, available at http://www.jcb.org/cgi/content/full/jcb.200811012/DC1). Mtor-mCherry shows a highly adaptable morphology in response to changes in spindle shape and dynamics throughout mitosis, which is inconsistent with a static structure. Similar to endogenous Mtor, Mtor-mCherry retracts and loses the fusiform shape upon MT depolymerization but is retained in a conspicuous milieu around chromosomes (Fig. 1, B–D and F; and Video 2), suggesting that MTs exert a pushing force on the Mtor-defined matrix.
Figure 1.
Mtor is part of a dynamic nuclear derived spindle matrix distinct from MTs. (A) An S2 cell stably expressing Mtor-mCherry (red) and GFP–α-tubulin (green). (A′) The corresponding Mtor-mCherry channel alone. (B) S2 cell stably expressing Mtor-mCherry and GFP–α-tubulin upon colchicine addition (time = 0). (C–E) Live cell analysis of GFP–α-tubulin, Mtor-mCherry, and Jupiter-GFP after colchicine treatment. (C′) Loss of GFP–α-tubulin fluorescence in the spindle is accompanied with equivalent fluorescence gain in the cytoplasm. (D′) Mtor-mCherry fluorescence in the spindle is not affected by MT depolymerization. (E′) Jupiter-GFP fluorescence is lost from the spindle to the cytoplasm after MT depolymerization. (F–F″) Endogenous Mtor after cold-induced MT depolymerization. (G and G′) Lamin B localization around the spindle. F.I., fluorescence intensity. Time is shown in minutes/seconds. Bars, 5 µm.
Previous electron microscopy analysis revealed the existence of a membranous network surrounding the spindle from prophase to metaphase in S2 cells (Maiato et al., 2006). In this study, we used immunofluorescence to show that lamin B is not fully disintegrated at this stage (Fig. 1 G). Similar results have recently been reported in living Drosophila embryos and neuroblasts, where a spindle envelope was proposed to limit the diffusion of nuclear derived Nup107 before anaphase (Katsani et al., 2008). To test whether this membranous network works as a diffusion barrier around the spindle, we compared the dynamic behavior of Mtor-mCherry relative to GFP–α-tubulin and a known MT-associated protein, Jupiter (Karpova et al., 2006), upon colchicine addition. GFP–α-tubulin or Jupiter-GFP fluorescence is gradually lost from the spindle region with an equivalent gain in the cytoplasm (Fig. 1, C and E). In contrast, Mtor-mCherry remains confined to the spindle region with no detectable fluorescence gain in the cytoplasm (Fig. 1 D). These results argue against the existence of a diffusion barrier around the metaphase spindle in Drosophila S2 cells and suggest that Mtor is being selectively retained in this region.
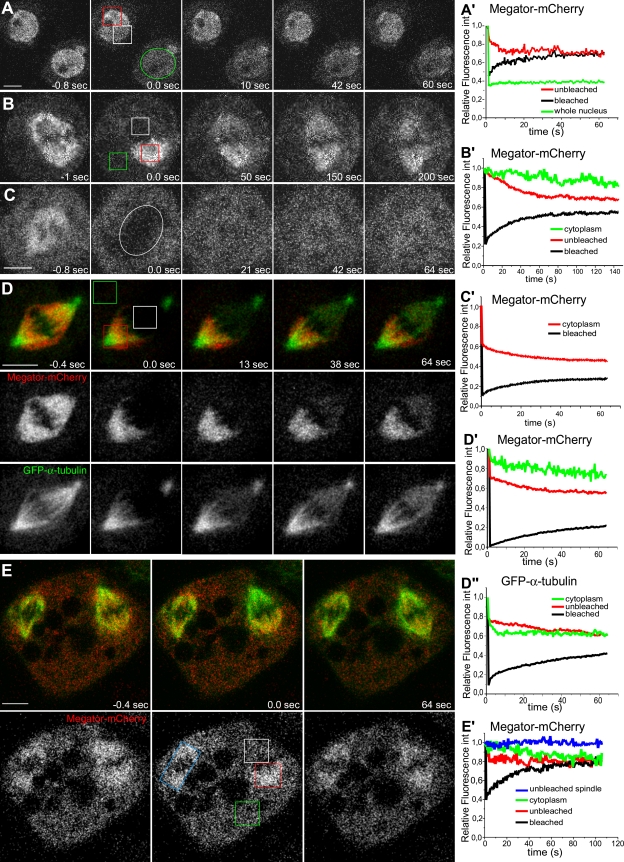
To shed light on the dynamic properties of Mtor, we used FRAP. In interphase nuclei, there is ∼50% recovery of fluorescence in the bleached region with an equivalent loss from a similar unbleached region and undetectable cytoplasmic exchange (Fig. 2, A and A′), suggesting that Mtor in the nucleoplasm is mobile. In mitosis, FRAP of Mtor-mCherry in one half-spindle is mirrored by an equivalent loss of fluorescence from the unbleached half-spindle as if Mtor exchanges between half-spindles (Fig. 2, B and B′). However, this recovery was slower (t1/2 = 18.7 ± 4.3 s, n = 9 cells) than in interphase nuclei (t1/2 = 9.0 ± 5.1 s, n = 3 cells; Fig. 2, A′ and B′) and had a minor contribution from a cytoplasmic pool (Fig. 2, B′–C′). In both interphase and mitosis, the recovery curves of Mtor-mCherry fitted a single exponential, suggesting affinity to a yet unidentified substrate, whereas GFP–α-tubulin in the spindle displayed biphasic recovery kinetics and best fit the sum of two exponentials as result of a rapid diffusion phase (t1/2 = 0.42 ± 0.39 s, n = 3 cells) followed by a slower recovery phase (t1/2 = 28.6 ± 6.7 s) associated with MT turnover (Fig. 2, D–D″). Finally, in S2 cells that sporadically form two spindles in the same cytoplasm, we found fluorescence exchange within the same spindle and from surrounding cytoplasm with no apparent loss from the neighboring unbleached spindle (Fig. 2, E and E′), supporting that Mtor is unable to exchange between two spindles located <10 µm apart. Collectively, these data indicate that Mtor is part of a dynamic, nuclear derived spindle matrix surrounded by a fenestrated membranous system containing lamin B and shows mobility properties that are distinct from MTs and associated proteins.
Figure 2.
Analysis of Mtor-mCherry dynamics by FRAP. (A and A′) FRAP of Mtor-mCherry in interphase nuclei. The white ROI shows fluorescence recovery within a bleached region; the red ROI shows fluorescence loss from an equivalent unbleached region; and the green ROI shows the entire nuclear area bleached in a neighboring cell. (B and B′) FRAP of Mtor-mCherry in one half-spindle (white ROI) and respective fluorescence loss in the other half-spindle (red ROI). The green ROI shows cytoplasmic fluorescence decay. (C and C′) FRAP of Mtor-mCherry in the entire mitotic spindle (white circle). Fluorescence decay in an equivalent area in the cytoplasm is indicated in the graph. (D–D″) Simultaneous FRAP of Mtor-mCherry (red) and GFP–α-tubulin (green) in the mitotic spindle. The corresponding FRAP of Mtor-mCherry and GFP–α-tubulin (white ROI) in the half-spindle was measured and compared with fluorescence loss in the unbleached half-spindle (red ROI) and cytoplasm (green ROI). (E and E′) Cell with two spindles in which Mtor-mCherry was photobleached in one half-spindle (white ROI). FRAP of Mtor-mCherry in this half-spindle was measured and compared with fluorescence loss in the unbleached half-spindle (red ROI), in the entire unbleached spindle (blue ROI), and cytoplasm (green ROI). Time = 0 at first frame after photobleaching. Relative fluorescence int, relative fluorescence intensity. Bars, 5 µm.
Mtor is required for normal mitotic duration and efficient response to spindle damage
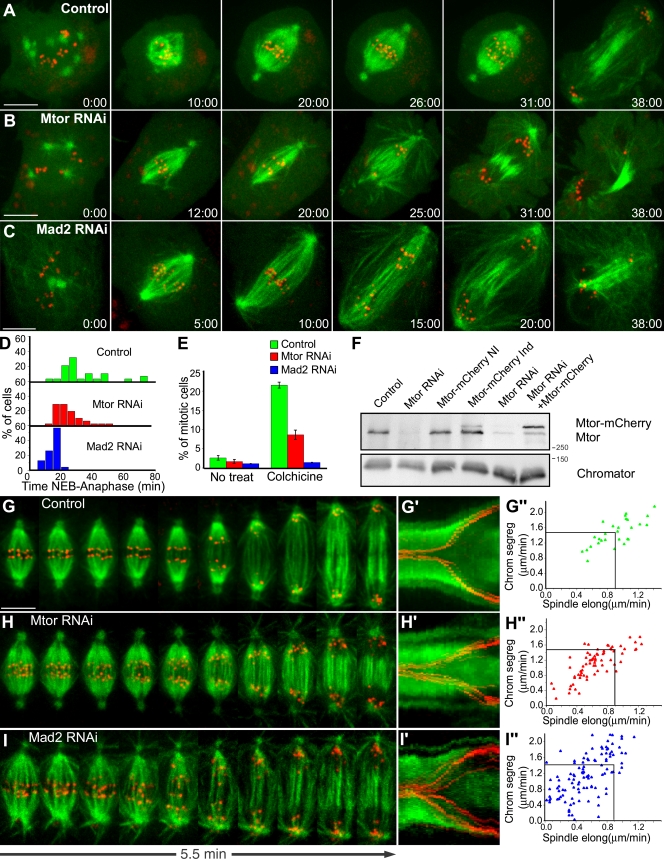
To address the mitotic role of Mtor, we used RNAi in Drosophila S2 cells stably coexpressing GFP–α-tubulin and the KT marker mCherry–centromere identifier (CID). Mtor-depleted cells show no major spindle defects but typically form a poorly defined metaphase plate (Fig. S1, A and A′, available at http://www.jcb.org/cgi/content/full/jcb.200811012/DC1) as the result of progressing ∼15% faster through mitosis (median = 23.5 min, range = 14–50 min, n = 52 cells) when compared with controls (median = 28 min, range = 13.5–70 min, n = 28 cells; Fig. 3, A, B, D, and F). Such problems in completing chromosome congression are corrected if anaphase onset is delayed by treating cells with the proteasome inhibitor MG132 (Fig. S1, B and B′). As in Mtor RNAi, S2 cells depleted of the SAC protein Mad2 undergo a faster mitosis (median = 11.5 min, range = 8–21 min, n = 23 cells; Fig. 3, C and D; Orr et al., 2007). Moreover, Mtor-depleted cells show a lower mitotic index as well as a weakened response to MT depolymerization (Fig. 3 E), suggesting that Mtor is required for proper SAC response.
Figure 3.
Mtor is required for proper mitotic timing and SAC response. (A–C) S2 cells stably expressing GFP–α-tubulin (green) and CID-mCherry (red) were used for live imaging of mitotic progression in control (A), Mtor RNAi (B), and Mad2 RNAi (C). (D) Respective quantification of the time from NEB to anaphase. Mtor and Mad2 RNAi are statistically different from controls (P < 0.05; Dunn's test). Mtor is also statistically different from controls in a pairwise comparison (P = 0.003; Mann-Whitney test). (E) Mitotic index under physiological conditions or after colchicine treatment. Error bars represent SD from the mean obtained from three independent experiments. (F) Western blot analysis of Mtor. (left to right) Control, Mtor RNAi (75% depletion), stable expression of Mtor-mCherry without induction, stable expression of Mtor-mCherry after induction, RNAi using the 3′ UTR region of Mtor as target (86% depletion), and stable expression of Mtor-mCherry after induction and RNAi using the 3′ UTR region of Mtor as target. Chromator was used as loading control. (G–I) Analysis of chromosome and spindle dynamics during anaphase in control (G), Mtor RNAi (H), and Mad2 RNAi cells (I). (G′–I′) The corresponding kymograph analyses are shown. (G″–I″) Half-spindle elongation (spindle elong) and chromosome segregation (chrom segreg) velocities in control, Mtor RNAi, and Mad2 RNAi cells. Black lines indicate reference mean values for control cells. Spindle elongation in Mtor and Mad2 RNAi is statistically different from controls (P < 0.05; Student-Newman-Keuls Method). Time is shown in minutes/seconds. Bars, 5 µm.
Mtor is required for normal anaphase chromosome velocity
Quantitative analysis of anaphase revealed a significant attenuation in the velocity of chromosome separation in Mtor-depleted cells by affecting spindle elongation (mean ± SD; half-spindle elongation rate in controls = 0.9 ± 0.2 µm/min, range = 0.5–1.4 µm/min, n = 28 cells; Mtor RNAi = 0.6 ± 0.3 µm/min, range = 0.1–1.2 µm/min, n = 70 cells; Fig. 3, G–H″). These results could be accounted for if Mtor is part of a structural scaffold where motor proteins assemble to generate force (Pickett-Heaps et al., 1984). However, an alternative hypothesis is that Mtor may function to provide the necessary time for proper maturation of a competent spindle. To test this, we delayed anaphase onset by treating Mtor-depleted cells with MG132 and measured half-spindle elongation velocity after drug washout. We found no difference in half-spindle elongation velocity between Mtor RNAi (0.7 ± 0.1 µm/min) and control cells (0.7 ± 0.2 µm/min) treated with MG132 (mean ± SD; range in Mtor RNAi = 0.5–0.8 µm/min; range in controls = 0.5–1.1 µm/min; n = 7 cells/condition; Fig. S1, B and B′). Additionally, half-spindle elongation in Mad2-depleted cells, which progress faster through mitosis (Fig. 3, C, D, and I), was similar to Mtor-depleted cells, namely 0.5 ± 0.2 µm/min (mean ± SD, range = 0–1.2, n = 19 cells; Fig. 3, I′ and I″), supporting the spindle maturation hypothesis.
Mad2 is part of a spindle matrix and requires Mtor for its efficient recruitment to unattached KTs
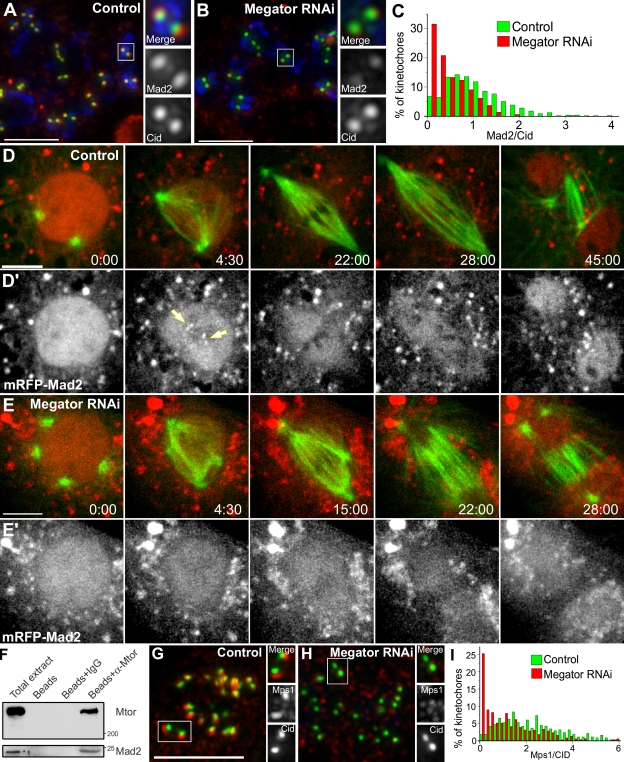
To shed light on the role of Mtor in SAC response, we analyzed the recruitment of Mad2 and BubR1 to unattached KTs after Mtor depletion. We found that although BubR1 was unaltered after Mtor depletion (our unpublished observations), Mad2 KT accumulation was significantly reduced (Fig. 4, A–C). Decreased Mad2 levels at KTs explain why Mtor-depleted cells enter anaphase prematurely, presumably because it requires binding of fewer MTs to remove all Mad2 from KTs and satisfy the SAC, whereas residual Mad2 at KTs may be sufficient to produce a weakened response to colchicine.
Figure 4.
Mad2 associates with and requires Mtor to localize to unattached KTs. (A and B) S2 cells treated with colchicine and processed for immunofluorescence with Mad2 (red) and CID (green) antibodies. DNA (blue) was counterstained with DAPI. (C) Quantification of Mad2/CID pixel intensity at KTs for control (median = 0.946, range = 0–5.52, n = 571 KTs/20 cells) and Mtor RNAi (median = 0.357, range = 0–3.95, n = 515 KTs/20 cells). The two populations are statistically different (P < 0.001; Mann-Whitney test). (D–E′) Mitotic progression in S2 cells stably expressing GFP–α-tubulin (green) and mRFP-Mad2 (red). Arrows indicate KTs. Red cytoplasmic aggregates likely correspond to misfolded mRFP-Mad2. (F and G) Control and Mtor RNAi cells were treated with colchicine and processed for immunofluorescence with Mps1 (red) and CID (green) antibodies. (H) Quantification of Mps1/CID pixel intensity at KTs for control (median = 2.06, range = 0.04–13.1, n = 384 KTs/20 cells) and Mtor RNAi (median = 0.91, range = 0–8.9, n = 391 KTs/20 cells). The two populations are statistically different (P < 0.001; Mann-Whitney test). (I) Co-IP of Mad2 with Mtor in lysates obtained from Drosophila embryos harvested between 0–3 h after egg laying. Time is given in minutes/seconds. Bars, 5 µm.
We next investigated how Mtor regulates the recruitment of Mad2 to KTs in living S2 cells stably coexpressing GFP–α-tubulin and monomeric RFP (mRFP)–Mad2. In interphase, mRFP-Mad2 is nuclear, accumulating at unattached KTs and spindles as cells transit into mitosis (Fig. 4 D; and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200811012/DC1). The spindle accumulation of Mad2 is thought to result from dynein-dependent poleward transport as MTs attach to KTs (Howell et al., 2000; Buffin et al., 2005). Interestingly, however, we found that a distinct pool of mRFP-Mad2 localizes to a nuclear derived spindle matrix even when MTs have just started invading the nuclear space. A similar behavior has been observed in vertebrate cells, where GFP-Mad2 accumulates as an ill-defined nuclear derived matrix during early prometaphase after its initial recruitment to unattached KTs (Howell et al., 2000). Like Mtor, the retention of Mad2 in the spindle matrix is resistant to MT depolymerization (Fig. S1 C and Video 4), suggesting that spindle-associated Mad2 is not freely diffusible. mRFP-Mad2 remains associated with the spindle matrix in the absence of Mtor, but it is unable to accumulate at KTs even after MT depolymerization with colchicine (Fig. 4 E; Fig. S1, D–E; and Videos 5 and 6). Stable expression of Mtor-mCherry (which is RNAi insensitive) rescues normal Mad2 localization at KTs after Mtor RNAi (Fig. 3 F and Fig. S1 F), indicating that the observed phenotype is specific and supporting that Mtor-mCherry is a functional protein. Lastly, Mtor depletion does not affect normal Mad2 expression levels and vice versa (Fig. S2 A), which rules out unspecific effects of Mtor over Mad2 mRNA transport to the cytoplasm.
Mtor/Tpr forms a conserved complex with Mad2
The colocalization of Mtor and Mad2 in the spindle matrix suggests that these proteins may interact. Indeed, Mad2 was found to coimmunoprecipitate with Mtor in lysates obtained from Drosophila embryos harvested between 0–3 h after egg laying (Fig. 4 F). Given that Mtor does not specifically accumulate at KTs, this interaction might represent an important regulatory step for the subsequent recruitment of Mad2 to unattached KTs. Several proteins such as Mad1, Rod, Ndc80, or Mps1 are involved in recruiting Mad2 to unattached KTs (Musacchio and Salmon, 2007). We found that although Mad1, Rod, and Ndc80 are effectively targeted to unattached KTs after Mtor depletion (Fig. S2, B–G′), Mps1 accumulation is significantly reduced (Fig. 4, G–I). Mps1 kinase activity was recently shown to be required to specifically target Mad2 but not Mad1 to unattached KTs in human cells (Tighe et al., 2008). To investigate whether the same regulatory role upon Mad2 is true in Drosophila, we generated an mps1 kinase-dead (mps1KD) allele by homologous recombination in flies (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200811012/DC1). In agreement with the results in human cells, neuroblasts from mps1KD third instar larvae show reduced or undetectable Mad2 accumulation at KTs upon colchicine treatment (Fig. S3). Collectively, these results support that the regulatory role of Mtor upon Mad2 is indirect and may be catalyzed by Mps1.
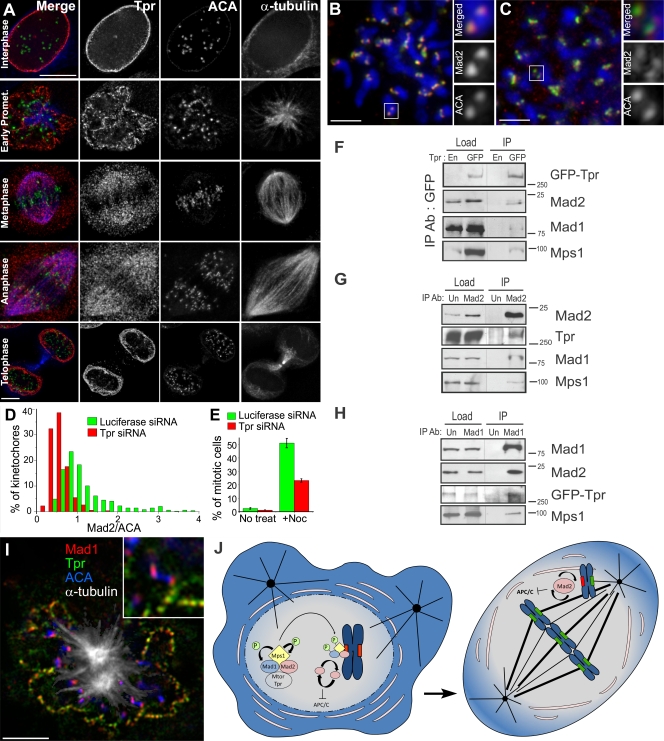
Like Tpr, Mad1, Mad2, and Mps1 localize at the NPC during interphase in human cells. During mitosis, Tpr remains associated with the nuclear envelope until prometaphase (Fig. 5 A and Fig. S2 H). Moreover, a fraction of Tpr is associated with the mitotic spindle from late prometaphase until anaphase and is recruited to the reforming nuclear envelope during telophase. This confirms the previous identification of Tpr in isolated human mitotic spindles (Sauer et al., 2005), but we failed to detect any enriched fraction of Tpr that resists MT depolymerization with nocodazole, including KTs (our unpublished observations). To see whether Tpr has a conserved regulatory role in the recruitment of Mad2 to unattached KTs in human cells, we used RNAi to deplete Tpr in HeLa cells (Fig. S2 I). Like in S2 cells, Tpr RNAi leads to reduced accumulation of Mad2 but not Mad1 to unattached KTs (Fig. 5, B–D; and Fig. S2, J and J′) accompanied by a decrease in the normal mitotic index and a weakened SAC response in the presence of nocodazole (Fig. 5 E). Tpr knockdown does not enrich for cells in G2 and slightly increases the number of cells in G1 (Loiodice et al., 2004), supporting that the lower mitotic index is not caused by the inability of cells to enter mitosis but rather reflects a faster exit. Moreover, Tpr, Mad1, Mad2, and Mps1 coimmunoprecipitate in mitotic enriched HeLa cell extracts prepared in the presence of nocodazole (Fig. 5, F–H), extending the results obtained in Drosophila and reinforcing that this complex forms independently of MTs and an intact nuclear envelope. While this paper was under revision, Tpr was independently found to interact with Mad1 and Mad2 in human cells (Lee et al., 2008). In agreement with our results, the authors propose that Tpr is important for controlling the SAC but reject the possibility that Tpr is playing a role in mitotic timing. However, quantification of the NEB to anaphase duration in Tpr-depleted cells does show a 25% acceleration of mitosis during this period (Lee et al., 2008). Finally, our results are not consistent with a model in which KT-associated Tpr serves as a docking place for Mad1 because we were unable to detect Tpr (or Mtor) at KTs, including those that were positive for Mad1 (Fig. 5 I), and found no impairment in Mad1 KT recruitment in Tpr- or Mtor-depleted cells.
Figure 5.
Human Tpr shares functional conservation with Drosophila Mtor. (A) Immunodetection of endogenous Tpr with a mouse mAb (red), ACA (green), and MTs (blue) in HeLa cells. (B and C) Luciferase (control) and Tpr RNAi cells were treated with nocodazole and processed for immunofluorescence with Mad2 (red) and ACA (green) antibodies. DNA (blue) was counterstained with DAPI. White boxed regions indicate the chromosome that is shown at a higher magnification on the right. (D) Quantification of Mad2/ACA pixel intensity at KTs for luciferase (median = 1.039, range = 0.44–4.45, n = 486 KTs/13 cells) and Tpr RNAi (median = 0.46, range = 0.09–1.69, n = 528 KTs/14 cells). The two populations are statistically different (P < 0.001; Mann-Whitney test). (E) Mitotic index in HeLa cells after luciferase and Tpr RNAi under physiological conditions or 16 h nocodazole treatment. Error bars represent SD from the mean obtained from three independent experiments. (F) IP from mitotic enriched parental HeLa cells (En) or HeLa cells stably expressing EGFP-Tpr (GFP). Load indicates total protein extracts. Purified beads (IP) were subjected to Western blot analysis for detection of interacting proteins. (G and H) IP from mitotic enriched HeLa extracts using an unspecific rabbit IgG (Un), rabbit anti-Mad2, or anti-Mad1 IgGs. (I) Colocalization of Mad1 (red) with Tpr (green) at nuclear pores but not at KTs (blue) during early prometaphase. MTs are shown in white. Inset shows a higher magnification of one KT pair without depicting MTs. (J) Proposed model for the role of Mtor/Tpr in the recruitment of Mad2 and Mps1 to unattached KTs after NEB. Ab, antibody; APC/C, anaphase-promoting complex/cyclosome; P, phosphorylation; Promet, prometaphase. Bars, 5 µm.
The role of a nuclear derived spindle matrix in mitotic control
Overall, our results support a model in which Mtor/Tpr acts as a spatial regulator of SAC, ensuring a timely and effective recruitment of Mad2 and Mps1 to unattached KTs as cells enter mitosis (Fig. 5 J). In budding yeast, Mps1 phosphorylates Mad1 (Hardwick et al., 1996), which is continuously recycled to KTs from Mlps at NPCs, but N-terminal deletion mutants of Mad1 lacking the Mlp-binding domain have a functional SAC (Scott et al., 2005). In humans and Drosophila, Mps1 regulates Mad2 but not Mad1 accumulation at KTs. Because Mad1 localization at KTs does not depend on Mlps/Mtor/Tpr and Mps1 kinase activity, the residual Mad2 at KTs after Mtor/Tpr RNAi possibly corresponds to the Mad1-bound fraction. One possibility is that Mps1 phosphorylation of Mad1 regulates the recruitment of a fast-exchanging pool of Mad2 to KTs (Chung and Chen, 2002; Musacchio and Salmon, 2007). Parallelly, Mtor/Tpr may spatially regulate Mps1 autophosphorylation, which is important for its normal KT accumulation, together with Mad2 (Xu et al., 2008). The presence of Mad2 in the complex may work as a positive feedback mechanism to ensure continuous Mps1 kinase activity upon SAC activation.
SAC proteins evolved from systems with a closed mitosis like budding yeast, where the spindle assembles inside an intact nuclear envelope into more complex systems like animals and plants, where the nuclear envelope is thought to fully or partially disintegrate during spindle formation, justifying the requirement of a nuclear derived spindle matrix for an effective SAC response. What retains matrix components around the spindle in systems where mitosis is thought to be open remains an intriguing question. In this regard, lamin B was proposed to tether several factors that mediate spindle assembly in Xenopus laevis egg extracts and possibly in human cells (Tsai et al., 2006). Additionally, a continuous endoplasmic reticulum surrounding the mitotic spindle is thought to be recycled from the nuclear envelope after its disassembly and has been observed in many systems undergoing an open mitosis, including humans (Ellenberg et al., 1997; McCullough and Lucocq, 2005). Although such fenestrated membranous systems cannot work as diffusion barriers, it is possible that they indirectly help to generate local gradients or concentrate matrix-affine substrates (Fig. 5 J). The enrichment of Mad2 in the spindle matrix provides an explanation for an unsolved SAC paradigm in which the “wait anaphase” signal emanating from unattached KTs must be diffusible to prevent premature anaphase onset of already bioriented chromosomes but at the same time is known to be restricted to the vicinity of the spindle (Rieder et al., 1997).
The proposed role of Mtor/Tpr further supports the necessity of spindle maturation for proper KT–MT attachments and anaphase spindle elongation in which the spindle matrix may help extend the duration of mitosis for the assembly of a competent chromosome segregation machinery. Mtor/Tpr-depleted cells have a weakened SAC response that, as opposed to complete checkpoint loss, may be compatible with cell viability and lead to cancer (Michel et al., 2001). The involvement of Tpr in the activation of several oncogenes (Park et al., 1986; Ishikawa et al., 1987; Greco et al., 1992) may translate into an unfavorable combination that facilitates transformation and tumorigenesis in humans.
Materials and methods
Cell culture, RNAi, drug treatments, and Western blot analysis
Drosophila S2 and Jupiter-GFP cells (provided by A. Debec, Institut Jacques Monod, Paris, France) were cultured in Schneider's (Sigma-Aldrich) or M3 media, respectively, containing 10% FBS (Invitrogen) in a 25°C incubator. The S2 GFP–α-tubulin cell line was provided by R. Vale (University of California, San Francisco, San Francisco, CA). HeLa cells were grown in DME supplemented with 10% FBS in a 37°C incubator with 5% CO2 and humidified atmosphere. RNAi depletion of Drosophila Mtor was performed as previously described (Qi et al., 2004) or using a 256-bp fragment specific for the 3′ untranslated region (UTR) of Mtor cDNA sequence using the following set of primers: 5′-TAATACGACTCACTATAGGGGGCGAGGAGTTCGGCGGACC-3′ and 5′-TAATACGACTCACTATAGGGATCGACAAAATTACACATAT-3′. For the rescue experiment, CuSO4 was added for induction of Mtor-mCherry expression from a metallothionein promoter (pMT) vector (Invitrogen) to a final concentration of 500 µM for 18 h before the analysis. mCherry cDNA was provided by R. Tsien (University of California, San Diego, La Jolla, CA). Mad2 RNAi in S2 cells was performed as previously described (Orr et al., 2007). Depletion of human Tpr was performed using published siRNA oligonucleotides against the targeting sequence 5′-GGTGGAGAGCGAACAACAG-3′ and transfected using HiPerFect (QIAGEN). Phenotypes were analyzed and quantified after 72 h, and protein knockdown was monitored by Western blotting using the following antibodies and dilutions: mouse anti-Mtor 1:1,000, rabbit anti-Tpr 1:1,000 (provided by L. Gerace, The Scripps Research Institute, La Jolla, CA), rabbit anti-CLASP1 1:400 (provided by N. Galjart, Erasmus University Medical Center, Rotterdam, Netherlands), and mouse anti-Chromator 1:2,000. Appropriate secondary antibodies were visualized using the ECL system (GE Healthcare). MT depolymerization in S2 cells was induced by colchicine at 100 µM during live cell recordings or at 30 µM for 18 h for the quantification of mitotic index or immunofluorescence analysis of KT proteins. MG132 was used at 2 µM for 120 min followed by washout with fresh medium while recording. For HeLa cells, nocodazole was used at 1 µM for 16 h for the analysis of Mad1 and Mad2 KT accumulation and mitotic index quantification.
Immunofluorescence microscopy
Immunofluorescence microscopy in Drosophila S2 cells was performed as described previously (Maiato et al., 2006). HeLa cells were grown on poly-l-lysine–coated coverslips and processed for immunofluorescence as described previously (Maiato et al., 2006). The following primary antibodies were used: mouse anti–α-tubulin clone B512 (1:2,000; Sigma-Aldrich), rat anti–α-tubulin YL1/2 1:10 (AbD Serotec), rabbit anti-DmBubR1 and anti-DmMad2 1:2,000 and 1:30, respectively, mouse anti-Mtor 1:10, mouse anti-DmMad1 1:50 (provided by R. Karess, Institut Jacques Monod, Paris, France), mouse anti–lamin B T40 1:200 (provided by P. Symmons, University of Tübingen, Tübingen, Germany), rabbit anti-CID 1:500 (provided by S. Henikoff, Fred Hutchinson Cancer Research Center, Seattle, WA), chicken anti-Ndc80 1:100 (provided by T. Maresca and T. Salmon, University of North Carolina, Chapel Hill, NC) rabbit anti-Rod 1:300 (provided by R. Karess), mouse anti–phosphohistone H3 1:100,000 (Abcam), human anticentromere antibodies (ACA) 1:5,000 (provided by B. Earnshaw, University of Edinburgh, Edinburgh, Scotland, UK), rabbit anti-Tpr 1:500, mouse anti-Tpr 1:500 (provided by V. Cordes, University of Heidelberg, Heidelberg, Germany), rabbit anti-HsMad1 1:500 (provided by P. Meraldi, ETH Zurich Institute of Biochemistry, Zurich, Switzerland), and sheep anti-HsMad2 1:500 (provided by S. Taylor, University of Manchester, Manchester, England, UK). Secondary antibodies used were Alexa Fluor 350, 488, 568, and 647 (1:2,000; Invitrogen) and 1 µg/ml DAPI. For mitotic index analysis in S2 and Hela cells, 1,000 or 500 cells, respectively, were scored in three independent experiments.
Time-lapse microscopy
S2 stable cell lines were grown on concanavalin A–coated coverslips in modified Rose chambers with Schneider's medium containing 10% FBS. The Mtor-mCherry construct was obtained by PCR amplification of the coding region of Mtor cloned into a pMT-mCherry vector in which a blasticidin resistance cassette for stable selection had been previously inserted into SalI site. The mRFP-Mad2 fusion protein under the control of its endogenous promoter (Buffin et al., 2005) was cloned into pAC-HisB vector (Invitrogen) in which a blasticidin resistance cassette had been previously inserted into SalI site. Four dimensional datasets were collected at 25°C with a spinning disc confocal system (Revolution; Andor) equipped with an electron multiplying charge-coupled device camera (iXonEM+; Andor) and a CSU-22 unit (Yokogawa) based on an inverted microscope (IX81; Olympus). Two laser lines (488 and 561 nm) were used for near-simultaneous excitation of GFP and mCherry/mRFP, and the system was driven by iQ software (Andor). Time-lapse imaging of z stacks with 1-µm steps covering the entire volume of the mitotic apparatus were collected every 30 or 5 s, according to the experiments.
Kymography
We used a custom routine written in Matlab to compensate for rotation and translation of the spindle. The direct output is a whole-spindle kymograph resulting from conversion of each time point image matrix into a vector. Chromosome poleward velocity relative to the equator was measured following each CID-mCherry track obtained from kymographs. CID cDNA was provided by G. Karpen (University of California, Berkeley, Berkeley, CA). Velocity was determined measuring the slope of the fastest linear movement after anaphase onset within each half-spindle. Spindle elongation velocity was measured following spindle MT minus ends.
FRAP analysis
FRAP was performed with a spectral confocal (SP2; Leica) with a 63×/1.4 NA objective lens and an additional zoom of 6×. Images were acquired every 422 ms or 1 s. Bleaching was conducted for 1.7 s after two frames of prebleach imaging. GFP intensity of the bleached area was normalized using the intensity of a neighboring cell after background subtraction. Nonlinear (exponential) curve fit was applied to recovery curves (Microcal Origin; OriginLab Corporation) for half-time recovery calculation.
Fluorescence quantification
Mad2 and Mps1 accumulation at KTs was measured for individual KTs by quantification of the pixel gray levels of the focused z plane within a region of interest (ROI). Background was measured outside ROI and was subtracted to the measured fluorescent intensity inside ROI. Results were normalized against a constitutive KT marker (CID for S2 cells or ACA in HeLa cells) using a custom routine written in Matlab (Mathworks). For quantification of mRFP-Mad2, pixel gray levels of the focused z plane within ROIs were defined for the spindle region and cytoplasm of prometaphase cells, and the respective ratio was determined after background subtraction. For quantification of GFP–α-tubulin, Jupiter-GFP, and Mtor-mCherry after MT depolymerization, ROIs were defined for the spindle region and cytoplasm. After background subtraction and bleaching correction, pixel gray levels of the focused z plane within ROIs were quantified over time.
Immunoprecipitations (IPs)
For co-IP experiments in Drosophila, anti-Mtor or control antibodies were bound to protein G beads (Sigma-Aldrich) for 4 h at 4°C on a rotating wheel in IP buffer. Antibody-coupled beads or beads only were incubated overnight at 4°C with 1 ml of 0–3 h embryonic lysate on a rotating wheel. Beads were washed extensively with IP buffer. The resulting bead-bound immunocomplexes were analyzed by SDS-PAGE and Western blotting using dMad2 or dMad1 antibodies. mAb 12F10 was used to detect Mtor to monitor the IP efficiency. Co-IP experiments in human cells were performed with native protein extracts (5 mg of total protein in a total volume of 500 µl of IP buffer obtained from HeLa cells or a derivative clone stably expressing EGFP-Tpr, both enriched for mitotic cells by incubation with nocodazole). Extracts were incubated with the precipitating antibody (rabbit anti-GFP, unspecific rabbit–IgG, or rabbit anti-Mad2; Bethyl Laboratories, Inc.) and 40 µl of protein A–Sepharose slurry overnight at 4°C on a rotating platform. Samples were centrifuged, the supernatant was retained as unbound sample, and the pelleted beads (IP) were washed three times with washing buffer (IP buffer with 250 mM KCl). Precipitated proteins were removed from the beads by boiling 5 min in SDS sample buffer and analyzed by SDS-PAGE and Western blotting with the appropriate antibodies.
Generation of Mps1-GFP and Mps1KD-GFP constructs
The mps1 coding region was PCR amplified from the cDNA LD08595, subcloned into pEGFP-N1 (Clontech Laboratories, Inc.), and the GFP-fusion was transferred to pMTV5-HisB (Invitrogen). Mutagenic PCR to introduce the point mutation for the kinase dead was performed with the following oligonucleotides: 5′-GATCGCTTTTGGCATAGCCAGC-3′ and 5′-GCTGGCTATGCCAAAAGCGATC-3′, corresponding to D478A in Mps1 protein (Tighe et al., 2008). The underlined nucleotides were mutated from the original Mps1 sequence. The resulting fragment was substituted in the pMTV5-Mps1-GFP vector.
Kinase assays
Soluble fractions from S2 cell extracts transiently expressing Mps1-GFP, Mps1KD-GFP, or GFP after 36 h of induction were obtained after centrifugation at 14,000 rpm for 5 min at 4°C. 10 µl of protein A beads (GE Healthcare) was preincubated with anti-GFP antibody (ab290; GE Healthcare) and used to immunoprecipitate 1 mg from the protein extracts obtained before a 2-h incubation at 4°C. Beads were washed with IP buffer supplemented with 300 mM NaCl and subsequently with the kinase buffer. For determination of kinase activity, the extracts were resuspended in kinase buffer supplemented with 3 µCi γ-[ATP32], 30 µM ATP, and 0.5 µg/µl dephosphorylated maltose-binding protein (Sigma-Aldrich). After a 60-min incubation at 25°C, samples were resolved by SDS-PAGE and subsequently exposed to an x-ray film (GE Healthcare).
Generation of mps1KD mutant stock
A point mutation was introduced in the coding region of a genomic clone containing the mps1 gene and promoter region by PCR mutagenesis as described for the generation of Mps1KD-GFP construct. The resulting 4.7-kb fragment was cloned into pTV2 and subsequently microinjected into Drosophila embryos. Gene targeting by homologous recombination was used to generate the mps1KD Drosophila stock as described previously (Rong and Golic, 2000).
Statistical analysis
Statistical analysis was performed using either parametric one-way analysis of variance or nonparametric analysis of variance (Kruskal-Wallis) for multiple group comparisons according to the normality of the distribution. All pairwise multiple comparisons were subsequently analyzed using either posthoc Student-Newman-Keuls (parametric) or Dunn's (nonparametric) tests. Parametric t test or nonparametric Mann-Whitney was used for two group comparisons. All statistical analyses were performed using SigmaStat 3.5 (Systat Software, Inc.).
Online supplemental material
Figs. S1 and S2 present additional characterization of Mtor RNAi phenotype and mRFP-Mad2, Mtor-mCherry, and Tpr localization and function. Fig. S3 provides a detailed characterization of the mps1KD allele. Videos 1–6 show the colocalization of Mtor-mCherry or mRFP-Mad2 with MTs in living cells and their response to MT depolymerization. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200811012/DC1.
Acknowledgments
We thank A.J. Pereira for the development of Matlab routines used in this paper, P. Sampaio for help with FRAP, and R. Vale, G. Karpen, R. Tsien, P. Symmons, L. Gerace, V. Cordes, T. Maresca, T. Salmon, B. Earnshaw, S. Taylor, N. Galjart, R. Karess, P. Meraldi, and A. Debec for the generous gift of reagents.
M. Lince-Faria and S. Maffini hold postdoctoral fellowships (SFRH/BPD/26780/2006 and SFRH/BPD/26780/2006) from Fundação para a Ciência e a Tecnologia of Portugal. Work in the laboratory of C.E. Sunkel is supported by a grant (PTDC/BIA-BCM/64224/2006) from the Fundação para a Ciência e a Tecnologia of Portugal. Work in the laboratory of K.M. Johansen is supported by a National Science Foundation grant (MCB0817107). Work in the laboratory of H. Maiato is supported by grants (PTDC/BIA-BCM/66106/2006 and PTDC/SAU-OBD/66113/2006) from Fundação para a Ciência e a Tecnologia of Portugal, the Gulbenkian Programmes for Research Stimulation and Frontiers in the Life Sciences, and the Luso-American Foundation for Development/National Science Foundation Research Network.
Footnotes
Abbreviations used in this paper: ACA, anticentromere antibodies; CID, centromere identifier; IP, immunoprecipitation; KT, kinetochore; mRFP, monomeric RFP; MT, microtubule; Mtor, Megator; NEB, nuclear envelope breakdown; NPC, nuclear pore complex; ROI, region of interest; SAC, spindle assembly checkpoint; Tpr, translocated promoter region; UTR, untranslated region.
References
- Buffin E., Lefebvre C., Huang J., Gagou M.E., Karess R.E. 2005. Recruitment of Mad2 to the kinetochore requires the Rod/Zw10 complex.Curr. Biol. 15:856–861 [DOI] [PubMed] [Google Scholar]
- Chung E., Chen R.H. 2002. Spindle checkpoint requires Mad1-bound and Mad1-free Mad2.Mol. Biol. Cell. 13:1501–1511 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cordes V.C., Reidenbach S., Rackwitz H.R., Franke W.W. 1997. Identification of protein p270/Tpr as a constitutive component of the nuclear pore complex-attached intranuclear filaments.J. Cell Biol. 136:515–529 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberg J., Siggia E.D., Moreira J.E., Smith C.L., Presley J.F., Worman H.J., Lippincott-Schwartz J. 1997. Nuclear membrane dynamics and reassembly in living cells: targeting of an inner nuclear membrane protein in interphase and mitosis.J. Cell Biol. 138:1193–1206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greco A., Pierotti M.A., Bongarzone I., Pagliardini S., Lanzi C., Della Porta G. 1992. TRK-T1 is a novel oncogene formed by the fusion of TPR and TRK genes in human papillary thyroid carcinomas.Oncogene. 7:237–242 [PubMed] [Google Scholar]
- Hardwick K.G., Weiss E., Luca F.C., Winey M., Murray A.W. 1996. Activation of the budding yeast spindle assembly checkpoint without mitotic spindle disruption.Science. 273:953–956 [DOI] [PubMed] [Google Scholar]
- Howell B.J., Hoffman D.B., Fang G., Murray A.W., Salmon E.D. 2000. Visualization of Mad2 dynamics at kinetochores, along spindle fibers, and at spindle poles in living cells.J. Cell Biol. 150:1233–1250 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iouk T., Kerscher O., Scott R.J., Basrai M.A., Wozniak R.W. 2002. The yeast nuclear pore complex functionally interacts with components of the spindle assembly checkpoint.J. Cell Biol. 159:807–819 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa F., Takaku F., Nagao M., Sugimura T. 1987. Rat c-raf oncogene activation by a rearrangement that produces a fused protein.Mol. Cell. Biol. 7:1226–1232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johansen K.M., Johansen J. 2007. Cell and molecular biology of the spindle matrix.Int. Rev. Cytol. 263:155–206 [DOI] [PubMed] [Google Scholar]
- Karpova N., Bobinnec Y., Fouix S., Huitorel P., Debec A. 2006. Jupiter, a new Drosophila protein associated with microtubules.Cell Motil. Cytoskeleton. 63:301–312 [DOI] [PubMed] [Google Scholar]
- Katsani K.R., Karess R.E., Dostatni N., Doye V. 2008. In vivo dynamics of Drosophila nuclear envelope components.Mol. Biol. Cell. 19:3652–3666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S.H., Sterling H., Burlingame A., McCormick F. 2008. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint.Genes Dev. 22:2926–2931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loiodice I., Alves A., Rabut G., Van Overbeek M., Ellenberg J., Sibarita J.B., Doye V. 2004. The entire Nup107-160 complex, including three new members, is targeted as one entity to kinetochores in mitosis.Mol. Biol. Cell. 15:3333–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiato H., Hergert P.J., Moutinho-Pereira S., Dong Y., Vandenbeldt K.J., Rieder C.L., McEwen B.F. 2006. The ultrastructure of the kinetochore and kinetochore fiber in Drosophila somatic cells.Chromosoma. 115:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullough S., Lucocq J. 2005. Endoplasmic reticulum positioning and partitioning in mitotic HeLa cells.J. Anat. 206:415–425 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Michel L.S., Liberal V., Chatterjee A., Kirchwegger R., Pasche B., Gerald W., Dobles M., Sorger P.K., Murty V.V., Benezra R. 2001. MAD2 haplo-insufficiency causes premature anaphase and chromosome instability in mammalian cells.Nature. 409:355–359 [DOI] [PubMed] [Google Scholar]
- Musacchio A., Salmon E.D. 2007. The spindle-assembly checkpoint in space and time.Nat. Rev. Mol. Cell Biol. 8:379–393 [DOI] [PubMed] [Google Scholar]
- Orr B., Bousbaa H., Sunkel C.E. 2007. Mad2-independent spindle assembly checkpoint activation and controlled metaphase-anaphase transition in Drosophila S2 cells.Mol. Biol. Cell. 18:850–863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M., Dean M., Cooper C.S., Schmidt M., O'Brien S.J., Blair D.G., Vande Woude G.F. 1986. Mechanism of met oncogene activation.Cell. 45:895–904 [DOI] [PubMed] [Google Scholar]
- Pickett-Heaps J., Spurck T., Tippit D. 1984. Chromosome motion and the spindle matrix.J. Cell Biol. 99:137s–143s [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qi H., Rath U., Wang D., Xu Y.Z., Ding Y., Zhang W., Blacketer M.J., Paddy M.R., Girton J., Johansen J., Johansen K.M. 2004. Megator, an essential coiled-coil protein that localizes to the putative spindle matrix during mitosis in Drosophila.Mol. Biol. Cell. 15:4854–4865 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rieder C.L., Khodjakov A., Paliulis L.V., Fortier T.M., Cole R.W., Sluder G. 1997. Mitosis in vertebrate somatic cells with two spindles: implications for the metaphase/anaphase transition checkpoint and cleavage.Proc. Natl. Acad. Sci. USA. 94:5107–5112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rong Y.S., Golic K.G. 2000. Gene targeting by homologous recombination in Drosophila.Science. 288:2013–2018 [DOI] [PubMed] [Google Scholar]
- Sauer G., Korner R., Hanisch A., Ries A., Nigg E.A., Sillje H.H. 2005. Proteome analysis of the human mitotic spindle.Mol. Cell. Proteomics. 4:35–43 [DOI] [PubMed] [Google Scholar]
- Scott R.J., Lusk C.P., Dilworth D.J., Aitchison J.D., Wozniak R.W. 2005. Interactions between Mad1p and the nuclear transport machinery in the yeast Saccharomyces cerevisiae.Mol. Biol. Cell. 16:4362–4374 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strambio-de-Castillia C., Blobel G., Rout M.P. 1999. Proteins connecting the nuclear pore complex with the nuclear interior.J. Cell Biol. 144:839–855 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tighe A., Staples O., Taylor S. 2008. Mps1 kinase activity restrains anaphase during an unperturbed mitosis and targets Mad2 to kinetochores.J. Cell Biol. 181:893–901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai M.Y., Wang S., Heidinger J.M., Shumaker D.K., Adam S.A., Goldman R.D., Zheng Y. 2006. A mitotic lamin B matrix induced by RanGTP required for spindle assembly.Science. 311:1887–1893 [DOI] [PubMed] [Google Scholar]
- Xu Q., Zhu S., Wang W., Zhang X., Old W., Ahn N., Liu X. 2009. Regulation of kinetochore recruitment of two essential mitotic spindle checkpoint proteins by mps1 phosphorylation.Mol. Biol. Cell. 20:10–20 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X.M., Rose A., Muthuswamy S., Jeong S.Y., Venkatakrishnan S., Zhao Q., Meier I. 2007. NUCLEAR PORE ANCHOR, the Arabidopsis homolog of Tpr/Mlp1/Mlp2/megator, is involved in mRNA export and SUMO homeostasis and affects diverse aspects of plant development.Plant Cell. 19:1537–1548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimowska G., Aris J.P., Paddy M.R. 1997. A Drosophila Tpr protein homolog is localized both in the extrachromosomal channel network and to nuclear pore complexes.J. Cell Sci. 110:927–944 [DOI] [PubMed] [Google Scholar]