Abstract
Cyclins are key cell cycle regulators, yet few analyses test their role in timing the events that they regulate. We used RNA interference and real-time visualization in embryos to define the events regulated by each of the three mitotic cyclins of Drosophila melanogaster, CycA, CycB, and CycB3. Each individual and pairwise knockdown results in distinct mitotic phenotypes. For example, mitosis without metaphase occurs upon knockdown of CycA and CycB. To separate the role of cyclin levels from the influences of cyclin type, we knocked down two cyclins and reduced the gene dose of the one remaining cyclin. This reduction did not prolong interphase but instead interrupted mitotic progression. Mitotic prophase chromosomes formed, centrosomes divided, and nuclei exited mitosis without executing later events. This prompt but curtailed mitosis shows that accumulation of cyclin function does not directly time mitotic entry in these early embryonic cycles and that cyclin function can be sufficient for some mitotic events although inadequate for others.
Introduction
Since the discovery of the cell cycle–coupled accumulation and destruction of cyclins (Evans et al., 1983), the increase in these mitotic regulators has been discussed as a possible clock timing cell cycle progression (Murray and Kirschner, 1989, 1991). Although other regulatory inputs (notably relief of inhibitory phosphorylation of the mitotic cyclin partner Cdk1) have been recognized as triggers of mitotic entry (Russell and Nurse, 1986; Edgar and O'Farrell, 1990; O'Farrell, 2001), the realization that the mechanisms controlling progress of the cell cycle change during development opened the question of what controls mitotic entry at other stages (O'Farrell et al., 1989). In particular, early embryonic cell cycles, which typically are exceedingly fast and run independent of new gene expression, occur with little or no Cdk1 inhibitory phosphorylation in frogs and flies (Ferrell et al., 1991; Edgar et al., 1994). These cycles, which also lack gap phases, have been viewed as streamlined cycles that operate without many of the controls that limit cell cycle progress later in development, and it has been asserted that these cycles are driven by a cyclin oscillator (Murray and Kirschner, 1991; Murray, 2004). However, there is very little in vivo experimental support for the influential view that interphase duration of embryonic cycles is determined by the time required to accumulate cyclin to a threshold (Hartley et al., 1996).
Developmental changes in cell cycle regulation have been detailed in Drosophila melanogaster. The cell cycle acquires a G2 and slows at embryonic cycle 14 as a result of increased Cdk1 inhibitory phosphorylation (Edgar and O'Farrell, 1989). Programmed transcription of the activating phosphatase String (Cdc25) times entry into mitosis from this G2 (Edgar and O'Farrell, 1989, 1990). However, string is abundant throughout the earlier mitotic cycles, which occur in a syncytial cytoplasm without cytokinesis and feature rapid oscillation between DNA replication and mitosis. In accord with the consensus view, it has been suggested that cyclin accumulation times these cycles (O'Farrell et al., 1989). Indeed, it has been shown that the dose of maternal genes encoding cyclins influences the speed of the early mitotic cycles (Edgar et al., 1994; Stiffler et al., 1999; Ji et al., 2004; Crest et al., 2007) and that mitotic exit requires cyclin destruction (Su et al., 1998). However, the bulk levels of mitotic cyclin exhibit little oscillation during the earliest embryonic cycles (Edgar et al., 1994), and the influence of cyclin dose on interphase length does not follow the direct proportionality predicted by the cyclin oscillator model. The combination of maternal contribution of cyclins to the egg and germline requirements for cyclin has prevented more complete genetic analyses of cyclin function during the early divisions, but analyses of mutations revealed cyclin roles during later zygotically controlled cycles.
The three mitotic cyclins of Drosophila are degraded in succession, CycA before metaphase, CycB at the metaphase–anaphase transition, and CycB3 during anaphase (Sigrist et al., 1995). Mutants lacking CycB or CycB3 produce viable flies (Lehner and O'Farrell, 1990; Jacobs et al., 1998). CycA is required for mitosis 16 because of an unexpected interphase role rather than a mitotic requirement (Lehner and O'Farrell, 1989; Sigrist et al., 1995; Reber et al., 2006). Despite dispensability of the individual cyclins, the mitotic phenotypes of cyclin double mutants in the postsyncytial divisions (cycles 15 and 16) indicate that individual requirements are partially covered by redundancy (Lehner and O'Farrell, 1990; Knoblich and Lehner, 1993; Jacobs et al., 1998). The mutant phenotypes and the differing consequences of stabilizing each of the cyclins revealed specializations (Sigrist et al., 1995; Jacobs et al., 1998; Parry and O'Farrell, 2001). It was proposed that, rather than acting as generic promoters of mitosis, the specialized actions of the mitotic cyclins are modulated by their schedules of destruction to guide progression through mitosis. However, this model ascribing specific roles to the cyclins is based mostly on fixed analyses, which have left us with only a superficial understanding of each cyclin's function.
In this report, we show the specialized role of each mitotic cyclin based on real-time analysis after cyclin RNAi in the early syncytial embryo. Knockdown of any two cyclins modestly extended interphase but did not prevent mitotic entry. Surprisingly, lowering cyclin function even further by halving the maternal dose of the remaining cyclin did not further extend interphase as would be predicted by the cyclin accumulation model. Instead, nuclei entered a curtailed mitosis, wherein chromosomes temporarily condensed, and centrosomes divided without nuclear envelope breakdown or nuclear division. Apparently, nuclei attempted mitosis while still lacking cyclin function adequate for proper mitotic execution. These data suggest that cyclin accumulation to a specific threshold does not directly time Cdk1 activation.
Results and discussion
Cyclin contributions to mitotic progression
RNAi in Drosophila embryos (Yang et al., 2000; Echard and O'Farrell, 2003) results in particularly rapid, specific, and effective knockdown of mitotic cyclins during the blastoderm divisions (McCleland and O'Farrell, 2008). Embryos (cycles 8–10) were injected at one pole with single and all pairwise combinations of cyclin double-stranded RNA (dsRNA) and imaged in real time through interphase 14. Unlike the triple-cyclin knockdown (McCleland and O'Farrell, 2008), none of these knockdowns prevented mitosis, although it was delayed and its progression was altered (Fig. 1 and Fig. 2 A). Gradations in the severity of phenotypes occurred in accord with proximity to the dsRNA injection site and with time after injection (see Materials and methods).
Figure 1.
Mitotic cyclins provide both redundant and specific function during mitosis. (A–C) Embryos expressing histone-GFP (top) or GFP-Polo (bottom) were imaged through mitosis 13. (A) Control embryos injected with LacI dsRNA. (B) Embryos injected with CycA and CycB dsRNA. Note that chromosomes fail to congress (02:36) or properly recruit GFP-Polo onto kinetochores (02:23 and 04:23). (C) Embryos injected with CycB and CycB3 dsRNA. Note that nuclei exhibit a prolonged metaphase (06:26), extended GFP-Polo concentration on kinetochores in metaphase (08:42), and a truncated anaphase. Arrows highlight kinetochores that fail to reach centrosomes. The times given in parentheses identify the corresponding frames in Videos 1–6 (available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1).
Figure 2.
RNAi of individual and pairwise combinations of mitotic cyclin disrupt mitotic progression. (A) Embryos expressing histone-GFP were injected with the indicated dsRNA and followed in real time from mitosis of cycle 11 to the beginning of cycle 14. Mean times for mitosis 13 are presented. The beginning of prophase was defined by the onset of chromatin condensation, and metaphase was defined by the complete alignment of chromosomes until anaphase onset. The asterisk indicates absence of metaphase. Error bars represent SD. (B) Model for coupling mitotic events to cyclin function and destruction. The three mitotic cyclins are temporally degraded within mitosis: CycA before metaphase, CycB at the metaphase–anaphase transition, and CycB3 during anaphase. Our cyclin RNAi experiments suggest that each cyclin has a specialized function for distinct mitotic steps. Coordinating cyclin function and destruction may guide mitotic progression.
Histone-GFP marks mitotic chromosome behavior, whereas dynamic changes in GFP-Polo mark several events (Fig. 1 and Videos 1 and 2, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1). GFP-Polo is centrosomal in interphase, but at the first sign of mitosis, it enters the nucleus where it decorates kinetochores (Video 2). During chromosome alignment, kinetochore staining declines, and upon anaphase, GFP-Polo accumulates at the spindle midzone (Moutinho-Santos et al., 1999).
After simultaneous knockdown of CycA and CycB, leaving CycB3, mitosis occurred in the absence of metaphase. Chromatin condensed and persisted for ∼3 min in a central spherical mass without forming a characteristic rectangular cluster of metaphase chromosomes, then two chromosomal masses segregated to opposite poles, although the distribution was often unequal, and lagging chromosomes were frequent (Fig. 1 B; Fig. 2 A; and Video 3, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1). During this CycB3-directed mitosis, GFP-Polo entered the nucleus during prophase and localized to the spindle midzone upon mitotic exit (Fig. 1 B and Video 4). Kinetochores accumulated much less GFP-Polo, and they failed to achieve a metaphase alignment. Thus, CycA and CycB normally promote metaphase, and CycB3 lacks this capability under these conditions.
Simultaneous knockdown of CycB and CycB3, leaving CycA, doubled the duration of chromosome alignment and metaphase (Fig. 1; Fig. 2 A; and Video 5, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1). Anaphase chromosome separation was slow, truncated, and accompanied by decondensation. Nuclei typically snapped back together in telophase and subsequently fell to the embryo interior. GFP-Polo displayed robust prometaphase kinetochore localization and faded only slowly during the extended metaphase (Fig. 1 C and Video 6). Often, anaphase kinetochores appeared to remain attached to spindle remnants rather than to centrosomes (Fig. 1 C, arrows). The prolongation of early mitotic events, slowed chromosome separation, and truncated mitotic exit suggest that CycB and/or CycB3 normally promote the transition to and execution of anaphase.
Embryos injected with CycA and CycB3 dsRNA, leaving CycB, exhibited a more mild disruption of mitotic coordination. Chromosome alignment was slightly delayed and metaphase duration doubled, but nonetheless, nuclei successfully completed mitosis (Fig. 2 A). Thus, CycB can mediate the key events of mitosis. Knockdown of just CycB interfered with chromosome congression and resulted in anaphase failures (Fig. S1 and Videos 7 and 8, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1). The resulting nuclei ultimately fell to the embryo interior. Thus, CycB is essential for the syncytial mitotic cycles.
Advances in confocal microscopy, RNAi methodology, and GFP biomarkers have allowed us to capture temporal records of the altered mitoses resulting from cyclin reduction. In this way, our data go beyond the previous analysis of cyclin mutants using fixed embryos. Furthermore, we have characterized cyclin function during the early rapid mitotic cycles, whereas previous work focused on cyclin roles during later cycles (Knoblich and Lehner, 1993; Jacobs et al., 1998) in which mitotic entry is governed by String expression. Perhaps because of differences in methodology or stage, two of our observations differ from previous findings. First, we show that CycB is essential for the success of syncytial mitosis but that it is zygotically dispensable. Second, we observed the unusual metaphaseless mitosis upon knockdown of CycA and CycB, whereas the double mutant of CycA and CycB arrested in interphase (Knoblich and Lehner, 1993; Jacobs et al., 1998).
It is also interesting to compare our findings to the gain of function phenotypes observed upon expression of individual stabilized cyclins (Sigrist et al., 1995; Parry and O'Farrell, 2001). Each stabilized cyclin arrested progress of mitosis at different points, stable CycA in metaphase, and stabilized CycB or CycB3 in distinct stages of anaphase (Parry and O'Farrell, 2001; Parry et al., 2003). Progress to anaphase in the presence of a stabilized cyclin has generally been interpreted to mean that anaphase onset is independent of cyclin degradation and requires only separase activation (Holloway et al., 1993); however, the particular capability of the tested cyclin to support different mitotic stages will influence the result. We find that CycB3 cannot support metaphase. Furthermore, reduction of CycB3 alone or in combination with other cyclins extended metaphase, suggesting that CycB3 promotes the transition to anaphase (Fig. 2 A). These findings support an interpretation of the stable cyclin results that emphasizes the distinctions in their activities and suggests that the normal successive destruction of the different cyclins modulates cyclin/Cdk1 activity so as to promote the normal sequence of mitotic events (Fig. 2 B; Jacobs et al., 1998).
In conclusion, our data suggest specializations in the roles of cyclins, especially CycA and CycB3, which appear to be particularly involved in early and late mitotic events, respectively. Though strikingly evident in the data, such specialization is incomplete because redundancies imply overlap in many activities of the cyclins (Knoblich and Lehner, 1993). Clearly, the origins of cyclin specialization are complex and will not only reflect specific differences in cyclin activity (Parry et al., 2003) but also each cyclin's spatial and temporal function (Royou et al., 2008).
Embryos operating on a single dose of cyclin enter mitosis punctually but abort execution
The accumulation of mitotic cyclins to a threshold has been suggested to directly time Cdk activation and mitotic entry (Murray and Kirschner, 1991). Cyclins do influence mitotic timing in early syncytial Drosophila embryos, as shown previously by alterations in interphase length after changes in cyclin maternal gene dose (Edgar et al., 1994; Stiffler et al., 1999; Ji et al., 2004; Crest et al., 2007) and by our finding that RNAi knockdown of any two cyclins prolonged interphase, in the most dramatic case, to 180% of normal in cycle 13 (after knockdown of CycB and CycB3; unpublished data). However, the previous dose experiments did not change interphase in proportion to the change of gene dose as might be expected by a direct regulation by accumulation of the cognate cyclin. This lack of parallel might be explained by the ability of all three cyclins to induce mitosis. Accordingly, previous dose changes would be expected to give complex results because they would shift the balance among different cyclins, each with a seemingly different ability to induce mitosis. To simplify the situation and thereby test whether the quantity of cyclin times mitotic entry, we knocked down two of the cyclins and reduced the dose of the third.
Embryos derived from a mother heterozygous for a CycB mutation (1× CycB) carry a correspondingly reduced level of CycB protein (Stiffler et al., 1999). Wild-type (2× CycB) and 1× CycB embryos were injected in one pole at approximately cycle 9 with CycA and CycB3 dsRNA and followed in real time. The regions in which dsRNA was injected will be referred to as 2× CycB only and 1× CycB only. In the 2× CycB–only case (Fig. 2 A), the nuclei go through mitosis, whereas in our previous work, we showed that knockdown of all three mitotic cyclins arrested nuclei in interphase (McCleland and O'Farrell, 2008). The intermediate 1× CycB–only region displayed a new phenotype not previously observed.
We quantified timing through cycle 12. Interphase of 1× CycB–only regions was essentially the same length as that of 2× CycB–only embryos (Fig. 3 A), after which the nuclei began to condense into mitotic chromosomes, a hallmark of mitotic entry (Fig. 3 and Video 9, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1). However, rather than progressing through mitosis, after formation of prophase chromosomes (Fig. 3 C, 08:01), 1× CycB–only nuclei appeared to decondense and revert back to interphase (Fig. 3 C, 12:31). Prophase chromatin condensation was only evident for ∼2 min. The failure to see a significant interphase extension upon reduction of CycB dose and the abortive attempt at mitosis challenge two ideas: (1) that cyclin accumulation directly times mitotic entry and (2) that all mitotic events occur at a single threshold of cyclin/Cdk.
Figure 3.
Embryos relying on one maternal dose of CycB exhibit timely but abortive mitosis. (A) Embryos expressing histone-GFP and containing either one or two maternal doses of CycB were injected with CycA and CycB3 dsRNA (represented as 1× CycB or 2× CycB, respectively). Mean cell cycle times during cycle 12 are presented. Embryos injected with control LacI dsRNA are presented for reference. (B) Behavior of mitosis 12 in different regions of a 1× CycB embryo after CycA and CycB3 dsRNA injection at one pole (left). (C) Embryos as in B were imaged at a higher resolution during cycle 12 near the injection site. (D) Single z sections from C during the atypical interphase (12:31).
Nuclei exhibited an unusual morphology after the aborted mitosis. The majority of chromatin was localized at the nuclear periphery (Fig. 3 D). Although appearing somewhat condensed, this chromatin did not stain positive for phosphohistone H3 (Ser10), suggesting that nuclei were not in an extended prophase (Fig. S2 B, top, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1).
Nuclei remained in this atypical state for approximately the same time as the previous interphase (∼12 min) at which time mitotic chromosomes reformed, phosphohistone H3 staining was evident, and nuclei progressed through mitosis (Fig. 3; and Fig. S2 B, bottom). When the 1× CycB–only region finally progressed through mitosis, timing defects in mitosis were very similar to those observed in 2× CycB–only regions (Fig. 3 A) except that chromosomes from neighboring nuclei frequently collided during anaphase (Fig. 3 C and Video 9). Our interpretation of this successful second try at mitosis is that after two cell cycle intervals, a single dose of cyclin is able to provide a level of cyclin function achieved by two doses in one cell cycle interval. Accordingly, the second mitosis is much like that seen in 2× CycB–only regions except for the colliding anaphases, which we attribute to the centrosome division that accompanies the abortive mitosis (see Centrosome division accompanies…).
Using a similar strategy, we examined 1× CycA and 1× CycB3 embryos (Fig. S3, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1). 1× CycB3 embryos, when injected with CycA and CycB RNAi, exhibited a curtailed nuclear division similar to that described in 1× CycB–only regions (Fig. S3 B). 1× CycA embryos injected with CycB and CycB3 RNAi exhibited a slight interphase delay compared with 2× CycA–only embryos but nonetheless completed nuclear division (Fig. S3). Because the timing of mitotic entry as judged by chromosome condensation is relatively unaffected by the change in gene dose, whereas the success of mitosis is affected, we conclude that a rise in cyclin function to a threshold is not the direct timer of mitotic entry in Drosophila embryos.
Centrosome division accompanies the abortive mitosis supported by a single maternal dose of CycB
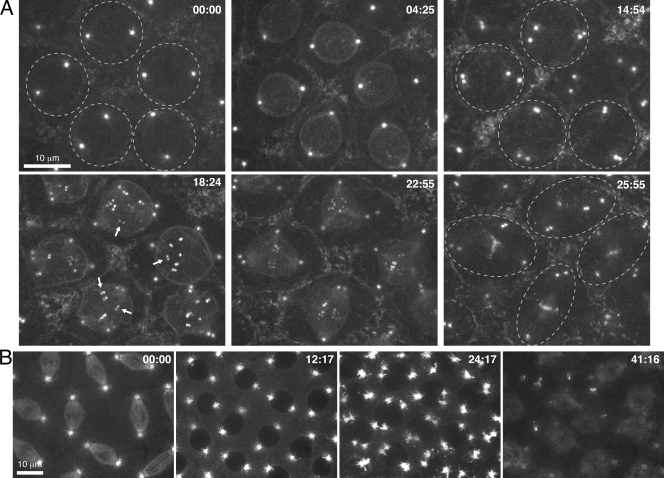
To examine centrosome behavior in conjunction with the abortive mitosis, we constructed flies with a single dose of maternal CycB that expressed either GFP-Polo or transforming acidic coiled-coil (TACC)–GFP to mark centrosomes. In 1× CycB–only embryos, GFP-Polo moved into the nucleus and faintly decorated kinetochores during the aborted mitosis in cycle 12 just as GFP-Polo does at the start of normal mitosis (Fig. 4 A, 04:25; and Video 10, left). GFP-Polo subsequently retreated from the nucleus without evident kinetochore separation (Fig. 4 A, 14:54). Centrosomes divided following retreat of GFP-Polo from the nucleus ∼8 min after the initial GFP-Polo nuclear influx (Fig. 4 A, 14:54; and Video 10, left). Some sister centrosomes separated only 2 µm, whereas others separated and moved around the nucleus. Embryos expressing TACC-GFP corroborated these results and additionally showed the persistent exclusion of TACC-GFP in the nucleus, suggesting that nuclear envelope breakdown did not occur (Fig. 4 B; and Video 10, right, available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1). Thus, 1× CycB–only regions exhibit prophase chromatin condensation, GFP-Polo movements, and centrosome division without reaching a cyclin threshold for nuclear division.
Figure 4.
Centrosome division occurs in concert with the abortive attempt at mitosis in 1× CycB–only embryos. (A) 1× CycB embryos expressing GFP-Polo were injected with CycA and CycB3 dsRNA. Dashed circles highlight individual nuclei and associated centrosomes. Arrows point to paired kinetochores during the second successful mitoses. (B) 1× CycB embryos expressing TACC-GFP were injected with CycA and CycB3 dsRNA.
When nuclei reentered mitosis after the initial aborted attempt, they did so with an extra pair of centrosomes and often had multipolar spindles (Fig. 4 A, 18:24 and 22:55). Each of the four centrosomes split again in late anaphase, indicating that centrioles had replicated during the atypical interphase (Video 10). In contrast, based on the number of kinetochores, it did not appear that chromosomes had replicated again. Moreover, GFP-Polo–labeled kinetochores were paired, suggesting that sister chromatid cohesion was not removed during the aborted mitosis 12 (Fig. 4 A).
In this report, we have examined the cell cycle consequences of progressive reductions in mitotic cyclin during the Drosophila blastoderm divisions. When we nominally reduced cyclin function to the contribution of a single maternal gene, embryos attempted mitosis without triggering or completing all mitotic events. Therefore, there is not a common threshold of cyclin function for all mitotic events. Moreover, because mitosis is attempted at about the normal time despite cyclin knockdown, the accumulation of cyclin does not appear to be the direct timer of mitosis. It remains unclear what times interphase in the Drosophila blastoderm cycles, but inputs from S phase and/or centrosome duplication might well have important roles (Sibon et al., 1997; Ji et al., 2004; Crest et al., 2007; McCleland and O'Farrell, 2008).
Materials and methods
Fly stocks
Drosophila strains were grown as described previously (McCleland and O'Farrell, 2008). Flies expressing histone H2AvD-GFP, GFP-Polo (Flytrap no. CC01326), and TACC-GFP were used for live embryo analysis (Clarkson and Saint, 1999; Barros et al., 2005; Buszczak et al., 2007). Mitotic cyclin gene mutants CycAC8LR1, CycB2, and CycB32 were used in experiments in which maternal gene dose was reduced to a single copy (Knoblich and Lehner, 1993; Jacobs et al., 1998). Each mutant was crossed to flies containing H2AvD-GFP to generate the following stocks: (a) w; ; CycAC8LR1, H2AvD-GFP/TM6,Tb, (b) w; CycB2, H2AvD-GFP/Cyo, and (c) w; ; CycB32/TM3, Ser, H2AvD-GFP. To facilitate these crosses, the original p-element containing H2AvD-GFP (Clarkson and Saint, 1999) was mobilized from the third chromosome onto other chromosomes by crossing these flies to flies expressing the Δ2-3 transposase. CycB2 mutants were crossed to flies expressing GFP-Polo and TACC-GFP to generate the following stocks: (a) w; CycB2/Cyo; GFP-Polo/TM6, Ser, Tb and (b) w, TACC-GFP; CycB2/Cyo.
Embryo manipulation and dsRNA production
Embryos were collected and processed for injection as described previously (McCleland and O'Farrell, 2008). dsRNA was generated as described previously (McCleland and O'Farrell, 2008).
Live analysis of cell cycle progression in embryos
In most experiments shown, embryos were injected at one pole. The severity of phenotypes increased with time after injection. For example, after RNAi of CycA and CycB (Video 3), the first mitosis showed a shortened metaphase, whereas in the next mitosis (cycle 13), there was no obvious metaphase. Additionally, phenotypes were most severe near the point of injection. In some cases, marked differences occurred in the behavior of nuclei at the interface between affected and unaffected territories. For example, the interface nuclei between 1× CycB–only regions and the regions unaffected by the RNAi did not reenter mitosis (Fig. 3 B and Fig. S2 A). We imagine that the neighboring nuclei in the uninjected region influence cell cycle behavior at the edges of the 1× CycB–only region, perhaps by locally promoting cyclin destruction in conjunction with their mitotic program. In other experiments, we controlled for such neighbor effects by depositing dsRNA along the length of the embryos. These experiments gave a uniform phenotype identical to that observed near the point of injection after local injection. This control indicates that the neighbor effects do not extend long distances and do not contribute to the phenotypes described.
Embryos were imaged on an inverted microscope (DM 1RB; Leica) equipped with a spinning disk confocal unit (CSU10; Yokagowa), 100× Plan Fluotar 1.3 NA and 40× Plan Fluotar 0.7 NA objectives (Leica), a camera (Orca AG; Hamamatsu Photonics), and Volocity 4 acquisition software (PerkinElmer). All images and videos were processed in Volocity 4 software and unless otherwise noted are presented as extended focused images. Image capture rates are indicated on each video. Time is displayed in hours:minutes:seconds. Most image stacks were collected at 0.5–1.5 µm over a 5–10-µm range using a controlled stage (MS-2000; Applied Scientific Instrumentation).
At least three independent injections were performed for each experiment shown. In each injection/experiment, an x-y stage facilitated the filming of multiple embryos (usually greater than five embryos). We used a minimum of four embryos in experiments in which cell cycle times were quantified (Figs. 2, 3, and S3). Usually, times were also collected from the uninjected end of the embryo and used for an internal reference.
Fixed analysis and immunofluorescence
Embryos injected with dsRNA were incubated for the appropriate time, fixed in a mixture of 37% formaldehyde and heptane for 10 min, and transferred to methanol before embryo hand devitellinization. Embryos were rehydrated in phosphate-buffered saline with 0.1% Triton X-100 and processed for immunofluorescence. Embryos were labeled with phosphohistone H3 (Ser10) antibody (Millipore) and Alexa Fluor 546–conjugated goat anti–rabbit antibody (Invitrogen). DNA was labeled with Hoescht 33258 (Invitrogen).
Online supplemental material
Fig. S1 illustrates mitotic defects upon knockdown of CycB. Figs. S2 and S3 provide further characterization of embryos operating on one dose of mitotic cyclin. Videos 1–8 show real-time videos of embryos in Figs. 1 and S1 progressing through mitosis after cyclin knockdown. Videos 9 and 10 are real-time videos of embryos in Figs. 3 and 4 in which nuclei attempt mitosis with a single maternal dose of CycB. Online supplemental material is available at http://www.jcb.org/cgi/content/full/jcb.200810012/DC1.
Acknowledgments
We thank the O'Farrell laboratory (especially Tony Shermoen and Soo-Jung Lee) for comments.
This work was supported by a National Institutes of Health training grant (GM078710) to M.L. McCleland and a National Institutes of Health grant (GM037193) to P.H. O'Farrell.
Footnotes
Abbreviations used in this paper: dsRNA, double-stranded RNA; TACC, transforming acidic coiled coil.
References
- Barros T.P., Kinoshita K., Hyman A.A., Raff J.W. 2005. Aurora A activates D-TACC–Msps complexes exclusively at centrosomes to stabilize centrosomal microtubules.J. Cell Biol. 170:1039–1046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buszczak M., Paterno S., Lighthouse D., Bachman J., Planck J., Owen S., Skora A.D., Nystul T.G., Ohlstein B., Allen A., et al. 2007. The carnegie protein trap library: a versatile tool for Drosophila developmental studies.Genetics. 175:1505–1531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clarkson M., Saint R. 1999. A His2AvDGFP fusion gene complements a lethal His2AvD mutant allele and provides an in vivo marker for Drosophila chromosome behavior.DNA Cell Biol. 18:457–462 [DOI] [PubMed] [Google Scholar]
- Crest J., Oxnard N., Ji J.Y., Schubiger G. 2007. Onset of the DNA replication checkpoint in the early Drosophila embryo.Genetics. 175:567–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echard A., O'Farrell P.H. 2003. The degradation of two mitotic cyclins contributes to the timing of cytokinesis.Curr. Biol. 13:373–383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., O'Farrell P.H. 1989. Genetic control of cell division patterns in the Drosophila embryo.Cell. 57:177–187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., O'Farrell P.H. 1990. The three postblastoderm cell cycles of Drosophila embryogenesis are regulated in G2 by string.Cell. 62:469–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edgar B.A., Sprenger F., Duronio R.J., Leopold P., O'Farrell P.H. 1994. Distinct molecular mechanism regulate cell cycle timing at successive stages of Drosophila embryogenesis.Genes Dev. 8:440–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evans T., Rosenthal E.T., Youngblom J., Distel D., Hunt T. 1983. Cyclin: a protein specified by maternal mRNA in sea urchin eggs that is destroyed at each cleavage division.Cell. 33:389–396 [DOI] [PubMed] [Google Scholar]
- Ferrell J.E., Jr., Wu M., Gerhart J.C., Martin G.S. 1991. Cell cycle tyrosine phosphorylation of p34cdc2 and a microtubule-associated protein kinase homolog in Xenopus oocytes and eggs.Mol. Cell. Biol. 11:1965–1971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartley R.S., Rempel R.E., Maller J.L. 1996. In vivo regulation of the early embryonic cell cycle in Xenopus.Dev. Biol. 173:408–419 [DOI] [PubMed] [Google Scholar]
- Holloway S.L., Glotzer M., King R.W., Murray A.W. 1993. Anaphase is initiated by proteolysis rather than by the inactivation of maturation-promoting factor.Cell. 73:1393–1402 [DOI] [PubMed] [Google Scholar]
- Jacobs H.W., Knoblich J.A., Lehner C.F. 1998. Drosophila cyclin B3 is required for female fertility and is dispensable for mitosis like cyclin B.Genes Dev. 12:3741–3751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji J.Y., Squirrell J.M., Schubiger G. 2004. Both cyclin B levels and DNA-replication checkpoint control the early embryonic mitoses in Drosophila.Development. 131:401–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoblich J.A., Lehner C.F. 1993. Synergistic action of Drosophila cyclins A and B during the G2-M transition.EMBO J. 12:65–74 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner C.F., O'Farrell P.H. 1989. Expression and function of Drosophila cyclin A during embryonic cell cycle progression.Cell. 56:957–968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lehner C.F., O'Farrell P.H. 1990. The roles of Drosophila cyclins A and B in mitotic control.Cell. 61:535–547 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCleland M.L., O'Farrell P.H. 2008. RNAi of mitotic cyclins in Drosophila uncouples the nuclear and centrosome cycle.Curr. Biol. 18:245–254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moutinho-Santos T., Sampaio P., Amorim I., Costa M., Sunkel C.E. 1999. In vivo localisation of the mitotic POLO kinase shows a highly dynamic association with the mitotic apparatus during early embryogenesis in Drosophila.Biol. Cell. 91:585–596 [PubMed] [Google Scholar]
- Murray A.W. 2004. Recycling the cell cycle: cyclins revisited.Cell. 116:221–234 [DOI] [PubMed] [Google Scholar]
- Murray A.W., Kirschner M.W. 1989. Cyclin synthesis drives the early embryonic cell cycle.Nature. 339:275–280 [DOI] [PubMed] [Google Scholar]
- Murray A.W., Kirschner M.W. 1991. What controls the cell cycle? Sci. Am. 264:56–63 [DOI] [PubMed] [Google Scholar]
- O'Farrell P.H. 2001. Triggering the all-or-nothing switch into mitosis.Trends Cell Biol. 11:512–519 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Farrell P.H., Edgar B.A., Lakich D., Lehner C.F. 1989. Directing cell division during development.Science. 246:635–640 [DOI] [PubMed] [Google Scholar]
- Parry D.H., O'Farrell P.H. 2001. The schedule of destruction of three mitotic cyclins can dictate the timing of events during exit from mitosis.Curr. Biol. 11:671–683 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parry D.H., Hickson G.R., O'Farrell P.H. 2003. Cyclin B destruction triggers changes in kinetochore behavior essential for successful anaphase.Curr. Biol. 13:647–653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reber A., Lehner C.F., Jacobs H.W. 2006. Terminal mitoses require negative regulation of Fzr/Cdh1 by cyclin A, preventing premature degradation of mitotic cyclins and string/Cdc25.Development. 133:3201–3211 [DOI] [PubMed] [Google Scholar]
- Royou A., McCusker D., Kellogg D.R., Sullivan W. 2008. Grapes(Chk1) prevents nuclear CDK1 activation by delaying cyclin B nuclear accumulation.J. Cell Biol. 183:63–75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell P., Nurse P. 1986. cdc25+ functions as an inducer in the mitotic control of fission yeast.Cell. 45:145–153 [DOI] [PubMed] [Google Scholar]
- Sibon O.C., Stevenson V.A., Theurkauf W.E. 1997. DNA-replication checkpoint control at the Drosophila midblastula transition.Nature. 388:93–97 [DOI] [PubMed] [Google Scholar]
- Sigrist S., Jacobs H., Stratmann R., Lehner C.F. 1995. Exit from mitosis is regulated by Drosophila fizzy and the sequential destruction of cyclins A, B and B3.EMBO J. 14:4827–4838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stiffler L.A., Ji J.Y., Trautmann S., Trusty C., Schubiger G. 1999. Cyclin A and B functions in the early Drosophila embryo.Development. 126:5505–5513 [DOI] [PubMed] [Google Scholar]
- Su T.T., Sprenger F., DiGregorio P.J., Campbell S.D., O'Farrell P.H. 1998. Exit from mitosis in Drosophila syncytial embryos requires proteolysis and cyclin degradation, and is associated with localized dephosphorylation.Genes Dev. 12:1495–1503 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang D., Lu H., Erickson J.W. 2000. Evidence that processed small dsRNAs may mediate sequence-specific mRNA degradation during RNAi in Drosophila embryos.Curr. Biol. 10:1191–1200 [DOI] [PubMed] [Google Scholar]