Abstract
Haematopoiesis is the process whereby blood stem cells give rise to at least fourteen functionally distinct mature cell types, and represents the best characterized mammalian adult stem cell system. Here we introduce the BloodExpress database, the first public resource integrating mouse blood cell expression profiles. BloodExpress enables the searching of data from individual studies in a single database accessible through a user-friendly web interface. Microarray datasets have been processed uniformly to allow their comparison on the BloodExpress platform. BloodExpress covers the majority of murine blood cell types, including both progenitors and terminally differentiated cells. This allows for the identification of dynamic changes in gene expression as cells differentiate down the well-defined haematopoietic hierarchy. A gene-centric interface returns haematopoietic expression patterns together with functional annotation and a list of other genes with similar expression patterns. A cell type-centric interface allows the identification of genes expressed at specific points of blood development, with the additional and useful capability of filtering by specific gene functional categories. BloodExpress thus constitutes a platform for the discovery of novel gene functions across the haematopoietic tree. BloodExpress is freely accessible at http://hscl.cimr.cam.ac.uk/bloodexpress/.
INTRODUCTION
Haematopoiesis is the process whereby haematopoietic stem cells (HSCs) differentiate into at least 14 types of mature blood cells, all of which have distinct microscopic appearance and perform different essential functions. Like other stem cells, HSCs possess the capacity for multi-lineage differentiation and are unique among blood cells, in that they have the ability to produce identical copies of themselves for the entire lifetime of the organism (‘self-renewal’). HSCs are extremely rare (1–10 HSCs per 100 000 cells in adult murine bone marrow) and during ontogeny derive from mesoderm, with sequential sites of haematopoiesis including the yolk sac, the aorta-gonad mesonephros region (AGM, an area surrounding the dorsal aorta), the placenta, fetal liver, thymus and finally the bone marrow in the adult animal.
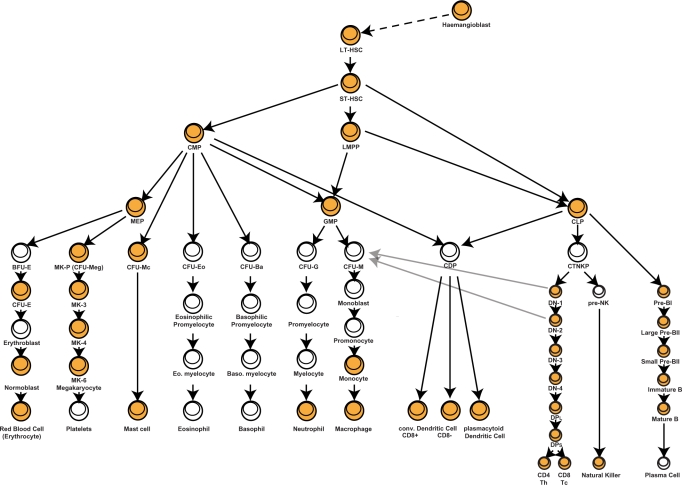
A major strength of the mouse blood system is the ability to isolate and intravenously transplant distinct subpopulations and assess the functional readout in vivo. As a consequence, many paradigms of the wider field of stem cell biology were first established using the haematopoietic stem cell system (1). Progress in the development of monoclonal antibodies directed at cell surface antigens, followed by fluorescence-activated cell sorting (FACS), has permitted the isolation of many haematopoietic subpopulations with differing lineage capabilities. In adult haematopoiesis, long-term HSCs (LT-HSCs) give rise to short-term HSCs (ST-HSCs), which in turn produce common myeloid progenitors (CMPs), common lymphoid progenitors (CLPs) and lymphoid primed multipotent progenitors (LMPPs). CLPs eventually give rise to B- and T cells, and also to natural killer (NK) and dendritic cells. CMPs give rise to megakaryocyte/erythroid (MEPs), granulocyte/macrophage (GMPs), and eosinophil (CFU-Eo) and basophil (CFU-Ba) progenitors. GMPs give rise to the committed precursors of neutrophils and macrophages (Figure 1).
Figure 1.
Overview of adult haematopoiesis in the mouse. The classic scheme of haematopoiesis features a tree that splits into the myeloid and lymphoid lineages. The progressive restriction of developmental fates is carried out by lineage-specific transcription factors that control lineage-specific expression profiles. Of note, the previously recognized distinction between the myeloid and lymphoid lineages has recently been disputed by the finding that T-cells precursors retain the ability to ultimately give rise to macrophages (31,32). The haemangioblast is a multipotent cell and a common precursor to both haematopoietic and endothelial cells (33). The haemangioblast has been isolated from gastrulating mouse (34) and zebrafish (35) embryos as mesodermal subpopulations, although their in vivo functionality awaits demonstration. Abbreviations—Progenitors: LT-HSC (long-term haematopoietic stem cell), ST-HSC (short-term haematopoietic stem cell), LMPP (lymphoid primed multipotent progenitor); Lymphoid: CLP (common lymphoid progenitor), CTNKP (common T and natural killer progenitor), DN-x (double negative ‘x’), DPL (double positive large), DPS (double positive small); Myeloid: CMP (common myeloid progenitor), MEP (megakaryocyte and erythrocyte progenitor), BFU-E (blast-forming unit erythrocyte), CFU-E (colony-forming unit erythrocyte), MK-P (megakaryocyte progenitor), CFU-Mc (colony-forming unit mast cell), CFU-Eo (colony-forming unit eosinophil), CFU-Ba (colony-forming unit basophil), GMP (granulocyte and macrophage progenitor), CFU-G (colony-forming unit granulocyte), CFU-M (colony-forming unit macrophage); Dendritic: CDP (common dendritic progenitor). The coloured cell types are those included in BloodExpress.
Haematopoiesis involves a progressive restriction of differentiation potential, mirrored at the molecular level by the establishment of lineage-specific expression profiles which are controlled by combinatorial interactions of transcription factors (TFs). Conventional gene knock-out experiments have been used to illustrate the stages at which haematopoiesis is blocked in the absence of specific TFs. Interestingly, mis-expression of many of these ‘master regulators’ is associated with oncogenesis, often involving chromosomal translocations (1–3). This has the effect of deregulating the expression of the locus (e.g. Scl/Tal1 and Lmo2 in T-cell acute leukaemia) or generating chimeric fusion proteins, as in myeloid and lymphoid leukaemias (Mll, Runx1 and Tel/Etv6).
Public databases that address the need to integrate disparate information on blood development have typically focused on a specific subset of blood cells, or on a specific aspect of gene behaviour. For example, HemoPDB (4) focuses on promoters and TFs that are active during haematopoiesis in human, mouse and rat. EpoDB (5) is built upon a controlled vocabulary of keywords important for erythropoiesis (the development of a red blood cell from a BFU-E cell) to mine public databases. Those genes eventually considered to be relevant to vertebrate erythropoiesis are integrated into the database together with their structural, functional, regulatory and gene expression information. Hembase (6) harbours ESTs and genes from mRNA gene libraries from developmentally staged, primary human erythroblasts. Finally, LymphTF-DB (7) holds information on murine TFs and their specific targets at various points in B- and T-cell development.
At present, no publicly available database has integrated expression profiles across all major branches of the haematopoietic hierarchy. Reconstruction of the regulatory networks that govern the differentiation of haematopoietic stem cells towards the multiple mature lineages will depend on ready access to comprehensive expression datasets. Here we present BloodExpress, an integrated platform and database of mouse blood expression with flexible search capability that covers the most immature stem cells, intermediate multipotent progenitors and mature blood cell types.
MATERIALS AND METHODS
Contents of BloodExpress
BloodExpress integrates expression data from 271 individual array experiments derived from 15 distinct studies: Akashi et al. (8), Chambers et al. (9), Chen et al. (10), Dudziak et al. (11), Ficara et al. (12), Hoffmann et al. (13,14), Jankovic et al. (15), Lugus et al. (16), Mansson et al. (17), McNagny (18), Nykter et al. (19), Robbins et al. (20), Simonis et al. (21) and Terszowski et al. (22). These datasets represent 37 distinct blood cell types, including all major precursors and mature cell types (Figure 1). Tables S1 and S2 summarize the technical details of the expression studies and the purification strategies for the above cell types, respectively.
The expression data of the above studies were obtained via the Gene Expression Omnibus (23) and StemBase (24), or were kindly supplied by the authors. Microarray probes were mapped to the mouse ENSEMBL gene database (NCBI m37 assembly) (25) and discretized to present (P), absent (A), or unknown (U) values (if the gene was not represented on the array). To do this, all 271 distinct arrays were processed with the mas5calls method of the affy library (26), which performs a Wilcoxon signed rank test on every probe set. The P ⩽ 0.04 as recommended by Affymetrix is indicative of specific signal, a P ⩾ 0.06 indicates lack of specific signal, and a P-value of between 0.04–0.06 is typically flagged as ‘marginal’. However, in order to diminish the number of false positives, all probe sets with P > 0.04 were annotated as ‘A’. For those studies that included biological replicates, the following strategy was adopted: if the P-values of a given probe were ⩽0.04 in >50% of biological replicates, then the probe in question was flagged as ‘P’ in that particular cell type. Most genes on the Affymetrix arrays present in the database are represented by a single probe. As a second step for those genes represented by multiple probes, only if >50% probes were labelled as ‘P’ from the previous step, then the gene was recorded as ‘P’.
We compared the results of our P-value-based discretization strategy with the published dataset by Chambers et al. (9), where a normalization score ⩾5 for a given Affymetrix probe is reported by the authors as indicative of specific signal. The Chambers et al. (9) dataset covers eight cell types and contains two biological replicates per cell type.
In order to compare the specific signal as provided by the Wilcoxon signed rank test (P-value) and the normalized scores as provided in Chambers et al. (9), the two datasets were discretized as described above: only if >50% of probes across biological replicates had a P ⩽ 0.04 or normalized score ⩾5.0, would the probe be annotated as ‘P’. Also, for those genes with multiple probes, only if >50% of probes from the previous probe-level annotation step were flagged as ‘P’, would the gene ultimately be annotated as ‘P’. When this discretization strategy was applied uniformly to the Chambers et al. dataset (9), both with the P-value and normalized score cutoffs, the overlap between the two matrices was found to be 91%. No significant improvement in overlap was observed by varying the recommended P-value cutoff of 0.04 or the cutoff for specific signal from the normalized scores. It is likely that a larger number of biological replicates in the Chambers et al.'s study (9) would have resulted in a greater overlap.
Moreover, the expression patterns of some genes known to be involved in haematopoietic processes were inspected. These included the B-cell-specific factor Pax5, the transcriptional regulators Bcl11a/Bcl11b, which control B- and T-cell development, respectively, and the Gfi1/Gfi1b transcriptional repressors involved in lymphoid versus erythroid development. Presence/absence calls for these genes across the haematopoietic hierarchy were found to be in good agreement with published experimental data.
Annotation of transcription factors was carried out by mapping all mouse ENSEMBL genes to the DNA-binding domain (DBD) database (27). The DBD is a database of predicted sequence-specific DNA-binding domains based on a hidden Markov model library of the SUPERFAMILY and Pfam databases. Many potential users of BloodExpress may want to be able to focus their analyses on transcription factors because transcriptional regulation is a key factor controlling haematopoiesis, a fact underlined by the large number of transcription factor genes that play key roles in normal haematopoiesis and/or the development of leukaemia (1–3).
Information stored for each mouse ENSEMBL gene includes its annotated name in the ENSEMBL database, its description, alternative gene names according to the Mouse Genome Database (28), and its chromosomal location. Moreover, the PANTHER database (29) was used to retrieve the ontology annotation for the ENSEMBL genes.
The BloodExpress web interface
The BloodExpress database can be accessed through a user-friendly web interface (http://hscl.cimr.cam.ac.uk/bloodexpress/) with two principal modes of searching the database: gene-centric and cell type-centric.
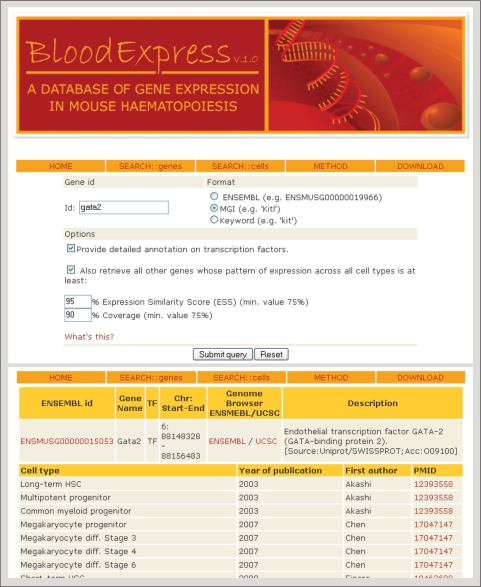
For gene-centric searches, the SEARCH::genes interface (Figure 2) allows the user to input a gene ID in ENSEMBL or MGI formats, and thus retrieve the blood cell types where the gene in question is expressed. In those cases where an MGI gene name is shared by more than one gene (e.g. ‘Sly’), the user is presented with a number of annotated candidates to choose from. Keyword searches are also allowed, in which case the input is matched against the ENSEMBL gene name and description fields. As before, a number of candidates are returned for the user to choose from. The information ultimately returned on every gene includes the ENSEMBL gene ID (hyperlinked to the ENSEMBL database for further exploratory analysis), the ENSEMBL gene name, its chromosomal coordinates, the ENSEMBL gene description and whether it is a predicted TF according to the DBD database. If the gene in question is a predicted TF, the SEARCH::genes interface also allows the user to optionally retrieve detailed SUPERFAMILY/Pfam protein family annotation. This is followed by a detailed list of cell types where the gene has been found to be expressed, with reference to the microarray paper by means of a hyperlink to PubMed (http://www.pubmed.org/).
Figure 2.
Snapshot of the gene-centric SEARCH::genes web interface. The user can search a gene's expression pattern by entering its MGI or ENSEMBL id, or by specifying a keyword that is searched against a list of gene names and gene descriptions before a list of candidates is returned to the user. The user can also specify a similarity threshold to retrieve genes with similar expression patterns.
While it is useful to know the cell type(s) where a particular gene of interest is expressed, it is also valuable to ascertain what other genes follow a similar pattern of expression across the haematopoietic hierarchy. Since a gene's expression pattern in our database is laid out as a vector where the columns are the distinct cell types, vector comparisons can be employed to identify identical and similar patterns of expression. The metric we have used to compare vectors relies on the Hamming distance (HD; 30). In information theory, the HD between two vectors of equal length is the number of positions for which the corresponding symbols are different. The percentage similarity between the genes in our database is calculated as follows in what we call the ‘expression similarity score’ (ESS): ESS = [(l − HD)/l] × 100, where l is the number of cell types being compared and HD is the number of cell types where the expression calls for the two genes being compared are different. The SEARCH::genes interface allows the user to specify a level of similarity between the query gene and other genes in the database. The query gene is compared on-the-fly against the rest of the genes in the database, and those genes with an ESS above the cutoff specified by the user are returned. In those cases where the gene of interest has been covered in a limited number of cell types in the original studies, the ESS will only be based on a limited number of comparisons. An additional score, the coverage score, is provided and which divides the ESS described above by the fraction of cell types upon which the comparison has been performed out of all the distinct cell types in the database. The coverage score thus provides an estimate of how general the expression pattern in question is across the haematopoietic hierarchy.
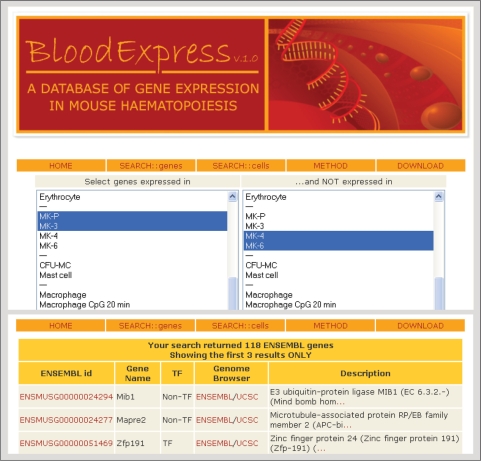
For cell-type-centric searches, the user can select a set of cell types in the SEARCH::cells interface (Figure 3). A list of all the ENSEMBL genes expressed in the specified cell types is returned with their detailed annotation and hyperlinked to the mouse ENSEMBL database. Moreover, the user can further refine queries by excluding those genes that are expressed in a second list of cell types. The resulting gene list can be further filtered by considering only predicted transcription factors as annotated in the DBD database (27) and/or those genes with specific ontology terms based on the annotation provided by the PANTHER database (29). Each of the three main levels of annotation of the PANTHER database (‘biological process’, ‘pathway’ and ‘molecular function’) contains a number of second-level categories (31, 152 and 29 categories, respectively). The user is thus able to filter those genes with a specific ontology annotation such as ‘Defense/immunity protein (MF00173)’, ‘T-cell activation (P00053)’ or ‘Oncogenesis (BP00281)’.
Figure 3.
Snapshot of the cell type-centric SEARCH::cells web interface. The user can select a cell type to return a list of genes expressed therein. Queries can be refined further by excluding those genes that are expressed in the second list of cell types. The resulting gene lists can be filtered further by considering only predicted transcription factors, or genes with specific ontology terms.
Implementation
BloodExpress is stored as a MySQL relational database (http://www.mysql.com/). The server is implemented as a set of Perl CGI scripts running under Apache (http://www.apache.org/).
SUMMARY AND FUTURE DEVELOPMENTS
BloodExpress provides the first comprehensive integration of published expression datasets, covering the majority of mouse blood cell types and accessible through a user-friendly web interface (http://hscl.cimr.cam.ac.uk/bloodexpress/). The gene-centric and cell-type-centric interfaces allow the distinctive and important advantage of identifying gene expression patterns across the well-defined haematopoietic hierarchy. The flexibility of filtering by gene functional category enhances the capability of the BloodExpress platform for discovering previously uncharacterized genes with expression patterns similar to those of genes already known to be important in blood development. Transcriptional regulation is key in controlling haematopoiesis as many TFs, or their regulation, are mutated in leukaemias. The reconstruction of regulatory networks that govern the differentiation of haematopoietic cells towards the multiple mature cell lineages, in both normal and pathological states, will be facilitated by the large-scale exploration of gene expression patterns as provided in BloodExpress. The datasets included in BloodExpress can aid in the reconstruction and/or analysis of regulatory networks by providing gene lists for gene set enrichment analysis as well as the analysis of consistency of inferred networks across multiple lineages. Therefore, BloodExpress fills a previously unmet need for the large haematopoiesis research community. Moreover, the underlying database structure and web server implementation is such that it could be readily adapted for studying regulatory networks in other biological contexts and stem cell systems.
Future developments of BloodExpress include the integration of additional mouse blood cell expression datasets as they become available and, most importantly, those of mouse models of various types of leukaemia and other haematopoietic diseases.
SUPPLEMENTARY DATA
Supplementary data are available at NAR Online.
FUNDING
The Leukaemia Research Fund, Medical Research Council and the Leukemia and Lymphoma Society. Funding for open access charge: Leukemia and Lymphoma Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We wish to thank Drs Emmanuel Levy, David M. A. Martin and Derek Wilson for advice with the web server.
REFERENCES
- 1.Orkin SH, Zon LI. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 2008;132:631–644. doi: 10.1016/j.cell.2008.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rosenbauer F, Tenen DG. Transcription factors in myeloid development: balancing differentiation with transformation. Nat. Rev. Immunol. 2007;7:105–117. doi: 10.1038/nri2024. [DOI] [PubMed] [Google Scholar]
- 3.Orkin SH, Zon LI. SnapShot: hematopoiesis. Cell. 2008;132:712. doi: 10.1016/j.cell.2008.02.013. [DOI] [PubMed] [Google Scholar]
- 4.Pohar TT, Sun H, Davuluri RV. HemoPDB: Hematopoiesis Promoter Database, an information resource of transcriptional regulation in blood cell development. Nucleic Acids Res. 2004;32:D86–D90. doi: 10.1093/nar/gkh056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Stoeckert CJ, Jr., Salas F, Brunk B, Overton GC. EpoDB: a prototype database for the analysis of genes expressed during vertebrate erythropoiesis. Nucleic Acids Res. 1999;27:200–203. doi: 10.1093/nar/27.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Goh SH, Lee YT, Bouffard GG, Miller JL. Hembase: browser and genome portal for hematology and erythroid biology. Nucleic Acids Res. 2004;32:D572–D574. doi: 10.1093/nar/gkh129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Childress PJ, Fletcher RL, Perumal NB. LymphTF-DB: a database of transcription factors involved in lymphocyte development. Genes Immun. 2007;8:360–365. doi: 10.1038/sj.gene.6364386. [DOI] [PubMed] [Google Scholar]
- 8.Akashi K, He X, Chen J, Iwasaki H, Niu C, Steenhard B, Zhang J, Haug J, Li L. Transcriptional accessibility for genes of multiple tissues and hematopoietic lineages is hierarchically controlled during early hematopoiesis. Blood. 2003;101:383–389. doi: 10.1182/blood-2002-06-1780. [DOI] [PubMed] [Google Scholar]
- 9.Chambers SM, Boles NC, Lin KY, Tierney MP, Bowman TV, Bradfute SB, Chen AJ, Merchant AA, Sirin O, Weksberg DC, et al. Hematopoietic Fingerprints: an Expression Database of Stem Cells and Their Progeny. Cell Stem Cell. 2007;1:578–591. doi: 10.1016/j.stem.2007.10.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen Z, Hu M, Shivdasani RA. Expression analysis of primary mouse megakaryocyte differentiation and its application in identifying stage-specific molecular markers and a novel transcriptional target of NF-E2. Blood. 2007;109:1451–1459. doi: 10.1182/blood-2006-08-038901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dudziak D, Kamphorst AO, Heidkamp GF, Buchholz VR, Trumpfheller C, Yamazaki S, Cheong C, Liu K, Lee HW, Park CG, et al. Differential antigen processing by dendritic cell subsets in vivo. Science. 2007;315:107–111. doi: 10.1126/science.1136080. [DOI] [PubMed] [Google Scholar]
- 12.Ficara F, Murphy MJ, Lin M, Cleary ML. Pbx1 regulates self-renewal of long-term hematopoietic stem cells by maintaining their quiescence. Cell Stem Cell. 2008;2:484–496. doi: 10.1016/j.stem.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hoffmann R, Seidl T, Neeb M, Rolink A, Melchers F. Changes in gene expression profiles in developing B cells of murine bone marrow. Genome Res. 2002;12:98–111. doi: 10.1101/gr.201501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoffmann R, Bruno L, Seidl T, Rolink A, Melchers F. Rules for gene usage inferred from a comparison of large-scale gene expression profiles of T and B lymphocyte development. J. Immunol. 2003;170:1339–1353. doi: 10.4049/jimmunol.170.3.1339. [DOI] [PubMed] [Google Scholar]
- 15.Jankovic V, Ciarrocchi A, Boccuni P, DeBlasio T, Benezra R, Nimer SD. Id1 restrains myeloid commitment, maintaining the self-renewal capacity of hematopoietic stem cells. Proc. Natl Acad. Sci. USA. 2007;104:1260–1265. doi: 10.1073/pnas.0607894104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lugus JJ, Chung YS, Mills JC, Kim SI, Grass J, Kyba M, Doherty JM, Bresnick EH, Choi K. GATA2 functions at multiple steps in hemangioblast development and differentiation. Development. 2007;134:393–405. doi: 10.1242/dev.02731. [DOI] [PubMed] [Google Scholar]
- 17.Mansson R, Hultquist A, Luc S, Yang L, Anderson K, Kharazi S, Al-Hashmi S, Liuba K, Thoren L, Adolfsson J, et al. Molecular evidence for hierarchical transcriptional lineage priming in fetal and adult stem cells and multipotent progenitors. Immunity. 2007;26:407–419. doi: 10.1016/j.immuni.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 18.McNagny. Comparison of Hematopoietic Stem Cell, Mast Cell Precursor and Mature Mast Cell Gene Expression. Ontario Genomics Innovation Centre (OGIC) StemBase, experiment Id: E194. http://www.stembase.ca.
- 19.Nykter M, Price ND, Aldana M, Ramsey SA, Kauffman SA, Hood LE, Yli-Harja O, Shmulevich I. Gene expression dynamics in the macrophage exhibit criticality. Proc. Natl Acad. Sci. USA. 2008;105:1897–1900. doi: 10.1073/pnas.0711525105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Robbins SH, Walzer T, Dembele D, Thibault C, Defays A, Bessou G, Xu H, Vivier E, Sellars M, Pierre P, et al. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 2008;9:R17. doi: 10.1186/gb-2008-9-1-r17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Simonis M, Klous P, Splinter E, Moshkin Y, Willemsen R, de Wit E, van Steensel B, de Laat W. Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C) Nat. Genet. 2006;38:1348–1354. doi: 10.1038/ng1896. [DOI] [PubMed] [Google Scholar]
- 22.Terszowski G, Waskow C, Conradt P, Lenze D, Koenigsmann J, Carstanjen D, Horak I, Rodewald HR. Prospective isolation and global gene expression analysis of the erythrocyte colony-forming unit (CFU-E) Blood. 2005;105:1937–1945. doi: 10.1182/blood-2004-09-3459. [DOI] [PubMed] [Google Scholar]
- 23.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Porter CJ, Palidwor GA, Sandie R, Krzyzanowski PM, Muro EM, Perez-Iratxeta C, Andrade-Navarro MA. StemBase: a resource for the analysis of stem cell gene expression data. Methods Mol. Biol. 2007;407:137–148. doi: 10.1007/978-1-59745-536-7_11. [DOI] [PubMed] [Google Scholar]
- 25.Flicek P, Aken BL, Beal K, Ballester B, Caccamo M, Chen Y, Clarke L, Coates G, Cunningham F, Cutts T, et al. Ensembl 2008. Nucleic Acids Res. 2008;36:D707–D714. doi: 10.1093/nar/gkm988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gautier L, Cope L, Bolstad BM, Irizarry RA. affy–analysis of Affymetrix GeneChip data at the probe level. Bioinformatics. 2004;20:307–315. doi: 10.1093/bioinformatics/btg405. [DOI] [PubMed] [Google Scholar]
- 27.Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD–taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36:D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mi H, Guo N, Kejariwal A, Thomas PD. PANTHER version 6: protein sequence and function evolution data with expanded representation of biological pathways. Nucleic Acids Res. 2007;35:D247–D252. doi: 10.1093/nar/gkl869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hamming RW. Error detecting and error correcting codes. Bell System Technical Journal. 1950;26:147–160. [Google Scholar]
- 31.Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. doi: 10.1038/nature06840. [DOI] [PubMed] [Google Scholar]
- 32.Wada H, Masuda K, Satoh R, Kakugawa K, Ikawa T, Katsura Y, Kawamoto H. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. doi: 10.1038/nature06839. [DOI] [PubMed] [Google Scholar]
- 33.Choi K, Kennedy M, Kazarov A, Papadimitriou JC, Keller G. A common precursor for hematopoietic and endothelial cells. Development. 1998;125:725–732. doi: 10.1242/dev.125.4.725. [DOI] [PubMed] [Google Scholar]
- 34.Huber TL, Kouskoff V, Fehling HJ, Palis J, Keller G. Haemangioblast commitment is initiated in the primitive streak of the mouse embryo. Nature. 2004;432:625–630. doi: 10.1038/nature03122. [DOI] [PubMed] [Google Scholar]
- 35.Vogeli KM, Jin SW, Martin GR, Stainier DY. A common progenitor for haematopoietic and endothelial lineages in the zebrafish gastrula. Nature. 2006;443:337–339. doi: 10.1038/nature05045. [DOI] [PubMed] [Google Scholar]