Abstract
Four hundred and eighty-one ultraconserved sequences (UCRs) longer than 200 bases were discovered in the genomes of human, mouse and rat. These are DNA sequences showing 100% identity among the three species. UCRs are frequently located at genomic regions involved in cancer, differentially expressed in human leukemias and carcinomas and in some instances regulated by microRNAs (miRNAs). Here we present UCbase & miRfunc, the first database which provides ultraconserved sequences data and shows miRNA function. Also, it links UCRs and miRNAs with the related human disorders and genomic properties. The current release contains over 2000 sequences from three species (human, mouse and rat). As a web application, UCbase & miRfunc is platform independent and it is accessible at http://microrna.osu.edu/.UCbase4.
INTRODUCTION
UCbase & miRfunc is a database of (i) human, mouse and rat ultraconserved elements and (ii) microRNAs (miRNAs).
The database has three main functions:
It provides ultraconserved sequence data, annotation, secondary structure and information about the disorder related to the band. Furthermore, it shows references and links to other resources for all UCRs published by Bejerano et al. (1).
It provides miRNA sequence data, annotation, secondary structure, information about disorder related to the band, references and links to other resources for all published miRNAs in the genomes of human, mouse and rat. Primary features of the nomenclature scheme, sequences and genomics coordinates are retrieved from miRBase (2).
It provides information about miRNAs and ultraconserved elements function and expression profiles.
This is made possible by a few automated Perl scripts linked to miRBase (http://microrna.sanger.ac.uk) (2), NCBI (http://www.ncbi.nlm.nih.gov) (3) and UCSC server (http://genome.ucsc.edu) (4).
ULTRACONSERVED SEQUENCES
Ultraconserved elements (UCRs) were discovered in 2004 by Bejerano and colleagues by bioinformatics comparisons of the genomes of mouse, rat and human (1). Four hundred and eighty-one UCR sequences are 200–779 bp in length showing 100% identity among the three species. Some of them contain protein coding sequences, but over half are not predicted to codify any protein (1).
Previous studies have suggested an important role for these noncoding sequences both in promoting the expression of several genes and in regulation of alternative splicing (5). Probably many of the UCRs date from a very early period in vertebrates evolution, as they have no orthologous counterparts in sea squirts, flies or worms even if at least one group of these UCRs evolved from a novel retrotransposon family that was active in lobe-finned fishes, and is still active today in the ‘living fossil’ Coelacanth (Latimeria chalumnae), the ancient link between marine and land vertebrates (5).
Recently, Calin et al. (6) identified a functional role for miRNAs in the transcriptional regulation of cancer-associated UCRs. They proved in tumors that differentially expressed UCRs could alter the functional characteristics of malignant cells. By combining this data with the much more elaborate model involving miRNAs in human tumorigenesis, they propose that alteration in both coding and noncoding RNAs cooperate in the initiation and/or progression of malignancy.
miRNAs
miRNAs were first described in 1993 by Lee and colleagues (7), yet the term ‘microrna’ was only introduced years later in 2001 in an article published in Science (8). Findings over the past 5 years supported a role for miRNAs in the regulation of crucial processes such as cell proliferation (9), apoptosis (10), development (11), differentiation (12) and metabolism (13). Each miRNA is supposed to target several hundreds of transcripts (14), making miRNAs one of the main genome regulators.
miRNAs regulate their targets by direct cleavage of the mRNA and by inhibition of protein synthesis, depending on the degree of complementarities with their target 3′-UTR regions (15). miRNAs are processed from primary transcripts known as pri-miRNAs (16) but not translated into protein. A portion of this primary transcript is recognized and cleaved by the enzyme Drosha into a miRNA precursor (pre-miRNA) (17) and finally processed to functional miRNA.
These pre-miRNAs are then processed to mature miRNAs in the cytoplasm by interaction with the endonuclease Dicer, which also initiates the formation of the RNA-induced silencing complex (RISC) (18). This complex is responsible for the gene silencing observed due to miRNA expression and RNAs interference. The pathway in plants varies slightly due to their lack of Drosha homologs (19).
At the time of this writing, the miRBase server version 11 contains 678 human pre-miRNAs (48–150 nt long) and 847 mature miRNAs (17–28 nt long).
Pre-miRNAs do not have a perfect double-stranded RNA structure and they are topped by a terminal loop (hairpin shape). Most of them are conserved between classes, the free energy is often less than −25 Kcal/mol, the GC-Ratio is usually 30–70% and the entropy is between 0 and 2 (20).
Recently, miRNA expression has been linked to cancer. The first evidence came from the finding that miR-15a and miR-16–1 are downregulated or deleted in most patients with chronic lymphocytic leukemia (CLL) (21). Several other groups have then studied the miRNA expression in cancer patients and found that miRNAs are differentially expressed in normal and tumor tissues (22–26) and, in some cases, are associated with prognosis (27).
QUERIES
The UCbase & miRfunc database contains three principal types of search:
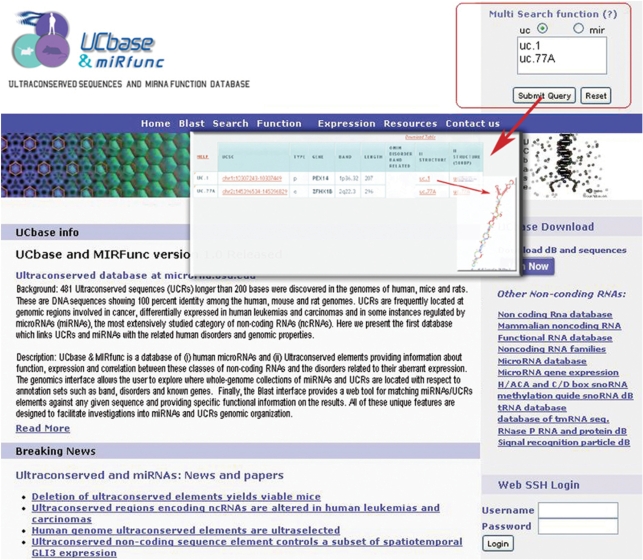
Multiple queries can be performed typing the names of selected miRNAs or UCRs and obtaining as output the band, the gene name, the coordinates, the human diseases (28) related to the band and the secondary structure performed using UNAfold (29) (Figure 1). These tables can be also downloaded in Excel format. A complete list of UCRs and miRNAs gene names is available clicking on the link above the multiple queries text box.
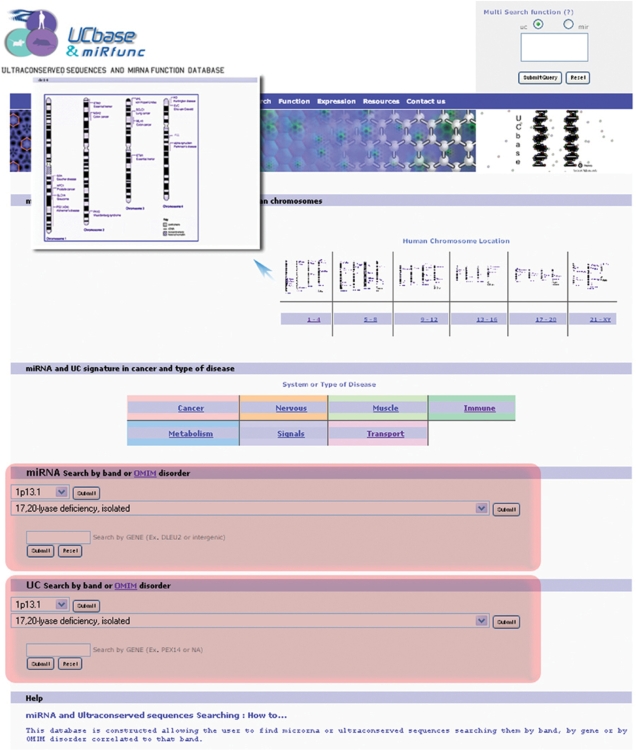
miRNA and UCRs information can also be retrieved searching for band, human Mendelian inheritance disorders (28) related to that band or just typing the name of a gene (Figure 2).
The use of an interactive map of biological systems and diseases allows to link miRNAs and ultraconserved sequences to the related disorders (Figure 2).
Figure 1.
Multi-queries text box (red square). Multiple queries can be performed typing the names of selected miRNAs or UCRs and obtaining as output the band, the gene name, the coordinates, the human diseases related to the band and the secondary structure up to 500 bp up/downstream.
Figure 2.
Search Page. miRNA and UCRs information can be retrieved searching for band, human Mendelian inheritance disorders related to that band or just typing the name of a gene (red highlighted boxes). The use of an interactive map of biological systems and diseases allows to link miRNAs and ultraconserved sequences to the related disorders (blue arrow).
FUNCTION
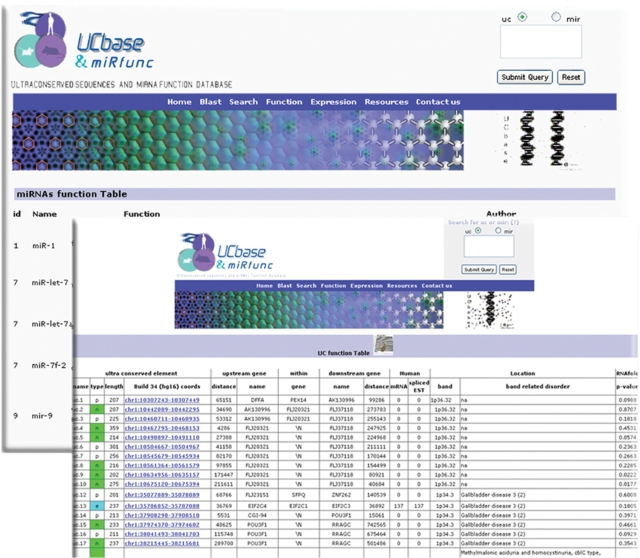
One of the most useful peculiarities of UCbase & miRfunc is that it provides tables showing ultraconserved elements properties (enhancer activity, alternative splicing, splicing NMD regulation and transcription evidence) and miRNA ‘experimentally proved’ function, addressing a number of common questions pertaining to these noncoding RNA classes (Figure 3). We decided to show the experiments that provide molecular biology evidence of a particular miRNA function. Paper references supplying only miRNA data were not included in the table.
Figure 3.
Function page. It provides tables showing ultraconserved elements properties (enhancer activity, alternative splicing, splicing NMD regulation and transcription evidence) and miRNA ‘experimentally proved’ function.
All of this information is periodically retrieved (every 2 months) using a Perl scripts implemented with WWW::Search::Pubmed (http://search.cpan.org/~gwilliams/WWW-Search-PubMed-1.004/) and WWW::Mechanize (http://search.cpan.org/dist/WWWMechanize/) modules which provide an API search engine linked to the Pubmed database (30).
SEQUENCE COMPARISON
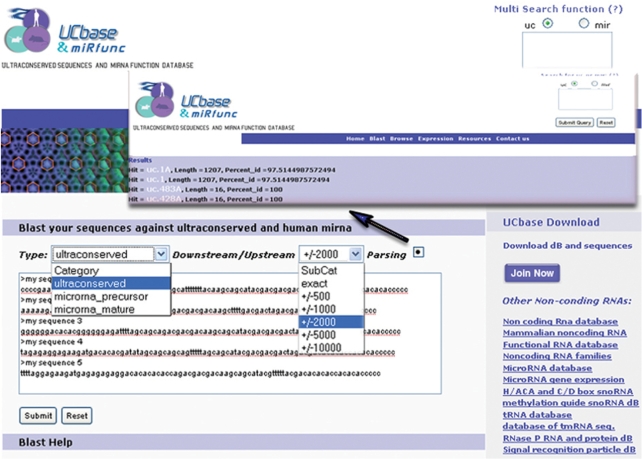
The sequence comparison page allows the researchers to match selected sequences against miRNAs and ultraconserved elements (exact/500/1000/2000/5000/10 000 bp up/downstream) (Figure 4) using The Basic Local Alignment Search Tool (BLAST) which finds regions of local similarity between sequences (31).
Figure 4.
Sequence comparison page. This page allows the researchers to match selected sequences against miRNAs and ultraconserved elements (exact/500/1000/2000/5000/10 000 bp up/downstream). Checking the parsing option it is possible to turn the output into a single-line summary (blue arrow).
The researchers can also align miRNAs and ultraconserved elements versus TRANSFAC (32) and CpG Island Searcher databases (33). Checking the parsing option it is possible to turn the output into a single-line summary. This function is invoked internally by a Perl script which uses the Bio::SearchIO module (http://search.cpan.org/~birney/bioperl-1.2.3/Bio/SearchIO.pm).
Moreover it is possible to download a RepeatMasker (34) miRNAs/UCRs table showing the repetitive miRNAs and ultraconserved sequences in human genome.
EXPRESSION
UCbase & miRfunc database is linked to microrna.org (35) to retrieve miRNA gene expression data sets; whereas, it collects ultraconserved sequences microarray expression data from ArrayExpress (36), GEO (37) and microrna.osu.edu server.
DATABASE INTERFACE
Cascading Style Sheets (CSS) were used for UCbase & miRfunc web development to style web pages written in HTML. The CSS specifications are maintained by the World Wide Web Consortium (W3C). Internet media type (MIME type) text/css is registered for use with CSS by RFC 2318 (March 1998).
UCbase & miRfunc was created using MySQL database under Debian Etch Linux OS installed on a Quad Core Processor machine with 32 GB RAM.
UCbase & miRfunc database can be accessed by Perl programs running on the web server. The MySQL Perl API is mainly provided with the use of CGI (Common Gateway Interface) which is a standard for external gateway programs to interface with information servers such as HTTP servers (Apache2 in this particular case). A CGI object (cgi.pm) was used to handle POST and GET methods correctly, and distinguish between scripts called from ISINDEX documents and form-based documents.
DATA SOURCES AND IMPLEMENTATION
Almost all the code used to develop the database and obtain miRNAs and ultraconserved elements information was written in Perl.
Perl (www.perl.org) is a programming language originally created for text manipulation and for a wide range of tasks including system administration, web development, network programming and GUI development. It is one of the most common language programming in bioinformatics, where it is valued for rapid application development and deployment, and the ability to handle large data sets. Perl is also used for the web automation.
Web automation can automate all the web processes from simple filling of forms to more complicated tasks for data transfer, web data extraction, image recognition and performing tasks based on it, scheduling tasks, batch process and file management. A Perl module called WWW::Mechanize was used to extract the data from NCBI (search_pubmed_nuovo.pl) and miRBase (www_mechanize_sanger_total.pl) pages; whereas, a script written in Pascal (http://www.newbie.com) was used to extract miRNAs and ultraconserved sequences (500/1000/2000/5000/10 000 bp up/downstream) from UCSC server (ucsc.nbl).
In particular, all the utilities that come with WWW::Mechanize print the names and elements of every form and provide all the needed information when searching for form using regular expressions (http://www.opengroup.org/onlinepubs/007908799/xbd/re.html).
However, using that data in the code means to cut and paste multiple entire blocks of code. For this reason, it is useful to install HTTP::Recorder to set up an HTTP proxy. HTTP::Recorder saves each action as WWW::Mechanize code. Before running the recording script (recorder.pl), the browser proxy has to be configured properly. Running recorder.pl starts an HTTP proxy daemon that the browser uses to make requests. The proxy uses HTTP::Recorder agent to log these requests. It saves a logfile as a.t file, which is specified when creating the HTTP::Recorder object.
Moreover, a window will appear displaying the content of.t file which should be a series of statements involving a hypothetical WWW::Mechanize object. These scripts were added to the final Perl code file called www_mechanize_sanger_total.pl.
WWW::Search::PubMed version 1.004 provides instead a backend to the WWW::Search module allowing searches of the National Library of Medicine's PubMed biomedical citation database (30) (pubmed.pl).
All the miRNAs genomic information are obtained using an automatic script linked to miRBase ftp server (ftp://ftp.sanger.ac.uk/pub/mirbase/sequences/CURRENT/database_files) which runs periodically using CRON, a time-based scheduling service in Unix-like computer operating systems (http://packages.debian.org/etch/cron).
CONCLUSIONS
UCbase & miRfunc is the first database containing the long 481 ultraconserved sequences discovered in the genomes of human, mouse and rat by Bejerano et al. (1) and identified as disregulated in cancer by Calin et al. (6). In addition, UCbase & miRfunc is the first database that provides miRNA function, which is a particularly important feature due to the increasing output of miRNA data. The goal of this study is to offer the researchers an advanced set of tools that supply information about correlation between miRNAs, UCRs and the disorders related to their aberrant expression.
The first version of UCbase & miRfunc includes new methods of queries such as search for band, disorders and genes. It has the capability of using multiple queries and combining the results from several miRNA/UCRs input. In addition, the system has a new tool for the RNA secondary structure prediction (up to 500 bp up/downstream), which allows innovative visualization of miRNAs and ultraconserved elements. This feature improves presentation of data output that is especially useful when several structures are obtained using multiple queries.
The sequence comparison tool, which is also available, allows researchers to match selected sequences that are 500/1000/2000/5000/10 000 bp upstream/downstream of all defined miRNAs and UCRs in the database.
Additionally, UCbase & miRfunc has adopted several strategies to integrate search tools. It provides updated data by using automate web scripts.
The system will be regularly upgraded with new human, mouse and rat miRNAs/UCRs by providing the corresponding reference sequences and annotations, thus allowing the data to be refined continuously with every new miRBase and UCSC database version. It also provides complete human, mouse and rat miRBase table as a single file (available from the resource web page), a feature that is particularly useful if one wants to use regular expressions to identify the segment of the input file associated with sub-field of interest.
Although other alternatives are available for retrieval of miRNA nomenclature, sequence data and annotation, UCbase & miRfunc is unique combination of features that provide biologists many options for data analysis and discovery of relationships between miRNAs and ultraconserved sequences.
FUTURE DIRECTIONS
The improvements to a new set of OMIM (28) and HGMD (38) databases needed for an overall plan for collection (39), hopefully should enable UCbase & miRfunc users, in the next year, to fully exploit the relation between miRNAs/UCRs and the mutation or chromosome aberrations related to their genomic location.
SUPPLEMENTARY DATA
Supplementary data and programming codes are available at http://web.unife.it/utenti/cristian.taccioli/software/. The user has to rename the codes files into.pl format because of security reasons we have uploaded them as.txt.
Free web tools written for miRNA and UCRs microarray analysis can also be downloaded at http://web.unife.it/utenti/cristian.taccioli/mix/examples.html.
FUNDING
Funding for open access charge: OSU Human Cancer Genetics Program.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We gratefully acknowledge the support of all members of the Department of Molecular Virology, Immunology and Medical Genetics of The Ohio State University. We also appreciate the technical support of Melissa Dickman, Daniela Taccioli, Ivan Tassani, Stefan Costinean, Nicola Zanesi, Pierluigi Gasparini and Gianpiero Di Leva.
REFERENCES
- 1.Bejerano G, Pheasant M, Makunin I, Stephen S, Kent WJ, Mattick JS, Haussler D. Ultraconserved elements in the human genome. Science. 2004;304:1321–1325. doi: 10.1126/science.1098119. [DOI] [PubMed] [Google Scholar]
- 2.Griffiths-Jones S. miRBase: the microRNA sequence database. Methods Mol. Biol. 2006;342:129–138. doi: 10.1385/1-59745-123-1:129. [DOI] [PubMed] [Google Scholar]
- 3.Pruitt KD, Tatusova T, Maglott DR. NCBI Reference Sequence (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2005;33:D501–D504. doi: 10.1093/nar/gki025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kent WJ, Sugnet CW, Furey TS, Roskin KM, Pringle TH, Zahler AM, Haussler D. The human genome browser at UCSC. Genome Res. 2002;12:996–1006. doi: 10.1101/gr.229102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bejerano G, Lowe CB, Ahituv N, King B, Siepel A, Salama SR, Rubin EM, Kent WJ, Haussler D. A distal enhancer and an ultraconserved exon are derived from a novel retroposon. Nature. 2006;441:87–90. doi: 10.1038/nature04696. [DOI] [PubMed] [Google Scholar]
- 6.Calin GA, Liu CG, Ferracin M, Hyslop T, Spizzo R, Sevignani C, Fabbri M, Cimmino A, Lee EJ, Wojcik SE, et al. Ultraconserved regions encoding ncRNAs are altered in human leukemias and carcinomas. Cancer Cell. 2007;12:215–229. doi: 10.1016/j.ccr.2007.07.027. [DOI] [PubMed] [Google Scholar]
- 7.Lee RC, Feinbaum RL, Ambros V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell. 1993;75:843–854. doi: 10.1016/0092-8674(93)90529-y. [DOI] [PubMed] [Google Scholar]
- 8.Ruvkun G. Molecular biology. Glimpses of a tiny RNA world. Science. 2001;294:797–799. doi: 10.1126/science.1066315. [DOI] [PubMed] [Google Scholar]
- 9.Cheng AM, Byrom MW, Shelton J, Ford LP. Antisense inhibition of human miRNAs and indications for an involvement of miRNA in cell growth and apoptosis. Nucleic Acids Res. 2005;33:1290–1297. doi: 10.1093/nar/gki200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu P, Guo M, Hay BA. MicroRNAs and the regulation of cell death. Trends Genet. 2004;20:617–624. doi: 10.1016/j.tig.2004.09.010. [DOI] [PubMed] [Google Scholar]
- 11.Karp X, Ambros V. Developmental biology. Encountering microRNAs in cell fate signaling. Science. 2005;310:1288–1289. doi: 10.1126/science.1121566. [DOI] [PubMed] [Google Scholar]
- 12.Chen CZ, Li L, Lodish HF, Bartel DP. MicroRNAs modulate hematopoietic lineage differentiation. Science. 2004;303:83–86. doi: 10.1126/science.1091903. [DOI] [PubMed] [Google Scholar]
- 13.Poy MN, Eliasson L, Krutzfeldt J, Kuwajima S, Ma X, Macdonald PE, Pfeffer S, Tuschl T, Rajewsky N, Rorsman P, et al. A pancreatic islet-specific microRNA regulates insulin secretion. Nature. 2004;432:226–230. doi: 10.1038/nature03076. [DOI] [PubMed] [Google Scholar]
- 14.Rajewsky N, Socci ND. Computational identification of microRNA targets. Dev. Biol. 2004;267:529–535. doi: 10.1016/j.ydbio.2003.12.003. [DOI] [PubMed] [Google Scholar]
- 15.Lai EC. Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nat. Genet. 2002;30:363–364. doi: 10.1038/ng865. [DOI] [PubMed] [Google Scholar]
- 16.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 18.Kurihara Y, Watanabe Y. Arabidopsis micro-RNA biogenesis through Dicer-like 1 protein functions. Proc. Natl Acad. Sci. USA. 2004;101:12753–12758. doi: 10.1073/pnas.0403115101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bernstein E, Caudy AA, Hammond SM, Hannon GJ. Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature. 2001;409:363–366. doi: 10.1038/35053110. [DOI] [PubMed] [Google Scholar]
- 20.Nam JW, Kim J, Kim SK, Zhang BT. ProMiR II: a web server for the probabilistic prediction of clustered, nonclustered, conserved and nonconserved microRNAs. Nucleic Acids Res. 2006;34:W455–W458. doi: 10.1093/nar/gkl321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K, et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl Acad. Sci. USA. 2002;99:15524–15529. doi: 10.1073/pnas.242606799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Volinia S, Calin GA, Liu CG, Ambs S, Cimmino A, Petrocca F, Visone R, Iorio M, Roldo C, Ferracin M, et al. A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl Acad. Sci. USA. 2006;103:2257–2261. doi: 10.1073/pnas.0510565103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Calin GA, Cimmino A, Fabbri M, Ferracin M, Wojcik SE, Shimizu M, Taccioli C, Zanesi N, Garzon R, Aqeilan RI, et al. MiR-15a and miR-16-1 cluster functions in human leukemia. Proc. Natl Acad. Sci. USA. 2008;105:5166–5171. doi: 10.1073/pnas.0800121105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Iorio MV, Ferracin M, Liu CG, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M, et al. MicroRNA gene expression deregulation in human breast cancer. Cancer Res. 2005;65:7065–7070. doi: 10.1158/0008-5472.CAN-05-1783. [DOI] [PubMed] [Google Scholar]
- 25.Iorio MV, Visone R, Di Leva G, Donati V, Petrocca F, Casalini P, Taccioli C, Volinia S, Liu CG, Alder H, et al. MicroRNA signatures in human ovarian cancer. Cancer Res. 2007;67:8699–8707. doi: 10.1158/0008-5472.CAN-07-1936. [DOI] [PubMed] [Google Scholar]
- 26.Visone R, Pallante P, Vecchione A, Cirombella R, Ferracin M, Ferraro A, Volinia S, Coluzzi S, Leone V, Borbone E, et al. Specific microRNAs are downregulated in human thyroid anaplastic carcinomas. Oncogene. 2007;26:7590–7595. doi: 10.1038/sj.onc.1210564. [DOI] [PubMed] [Google Scholar]
- 27.Calin GA, Croce CM. MicroRNA-cancer connection: the beginning of a new tale. Cancer Res. 2006;66:7390–7394. doi: 10.1158/0008-5472.CAN-06-0800. [DOI] [PubMed] [Google Scholar]
- 28.Hamosh A, Scott AF, Amberger J, Valle D, McKusick VA. Online Mendelian Inheritance in Man (OMIM) Hum. Mutat. 2000;15:57–61. doi: 10.1002/(SICI)1098-1004(200001)15:1<57::AID-HUMU12>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- 29.Markham NR, Zuker M. UNAFold: software for nucleic acid folding and hybridization. Methods Mol. Biol. 2008;453:3–31. doi: 10.1007/978-1-60327-429-6_1. [DOI] [PubMed] [Google Scholar]
- 30.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 32.Wingender E, Dietze P, Karas H, Knuppel R. TRANSFAC: a database on transcription factors and their DNA binding sites. Nucleic Acids Res. 1996;24:238–241. doi: 10.1093/nar/24.1.238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takai D, Jones PA. The CpG island searcher: a new WWW resource. In Silico Biol. 2003;3:235–240. [PubMed] [Google Scholar]
- 34.Zhi D, Raphael BJ, Price AL, Tang H, Pevzner PA. Identifying repeat domains in large genomes. Genome Biol. 2006;7:R7. doi: 10.1186/gb-2006-7-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Betel D, Wilson M, Gabow A, Marks DS, Sander C. The microRNA.org resource: targets and expression. Nucleic Acids Res. 2008;36:D149–D153. doi: 10.1093/nar/gkm995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Brazma A, Parkinson H. ArrayExpress service for reviewers/editors of DNA microarray papers. Nat. Biotechnol. 2006;24:1321–1322. doi: 10.1038/nbt1106-1321. [DOI] [PubMed] [Google Scholar]
- 37.Barrett T, Suzek TO, Troup DB, Wilhite SE, Ngau WC, Ledoux P, Rudnev D, Lash AE, Fujibuchi W, Edgar R. NCBI GEO: mining millions of expression profiles–database and tools. Nucleic Acids Res. 2005;33:D562–D566. doi: 10.1093/nar/gki022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Stenson PD, Ball EV, Mort M, Phillips AD, Shiel JA, Thomas NS, Abeysinghe S, Krawczak M, Cooper DN. Human Gene Mutation Database (HGMD): 2003 update. Hum. Mutat. 2003;21:577–581. doi: 10.1002/humu.10212. [DOI] [PubMed] [Google Scholar]
- 39.George RA, Smith TD, Callaghan S, Hardman L, Pierides C, Horaitis O, Wouters MA, Cotton RG. General mutation databases: analysis and review. J. Med. Genet. 2008;45:65–70. doi: 10.1136/jmg.2007.052639. [DOI] [PubMed] [Google Scholar]